Abstract
We have cloned T-cell factor 4N (TCF-4N), an alternative isoform of TCF-4, from developing pituitary and 3T3-L1 preadipocytes. This protein contains the N-terminal interaction domain for β-catenin but lacks the DNA binding domain. While TCF-4N inhibited coactivation by β-catenin of a TCF/lymphoid-enhancing factor (LEF)-dependent promoter, TCF-4N potentiated coactivation by β-catenin of several non-TCF/LEF-dependent promoters. For example, TCF-4N synergized with β-catenin to activate the α-inhibin promoter through functional and physical interactions with the orphan nuclear receptor steroidogenic factor 1 (SF-1). In addition, TCF-4N and β-catenin synergized with the adipogenic transcription factor CCAAT/enhancer binding protein α (C/EBPα) to induce leptin promoter activity. The mechanism by which β-catenin and TCF-4N coactivated C/EBPα appeared to involve p300, based upon synergy between these important transcriptional regulators. Consistent with TCF-4N′s redirecting the actions of β-catenin in cells, ectopic expression of TCF-4N in 3T3-L1 preadipocytes partially relieved the block of adipogenesis caused by β-catenin. Thus, we propose that TCF-4N inhibits coactivation by β-catenin of TCF/LEF transcription factors and potentiates the coactivation by β-catenin of other transcription factors, such as SF-1 and C/EBPα.
Wnts are a family of secreted proteins that play important roles in essentially all aspects of cell fate. Wnts act through Frizzled receptors and low-density lipoprotein receptor-related protein coreceptors to activate several signaling pathways (19, 41). In the canonical pathway, Wnt signaling inhibits glycogen synthase kinase 3, resulting in hypophosphorylation and subsequent stabilization of β-catenin. After translocation to the nucleus, β-catenin binds to and coactivates members of the T-cell factor/lymphoid-enhancing factor (TCF/LEF) family of transcription factors, thus mediating the effects of Wnt on gene expression. Wnts regulate the development of many cell types (2, 14, 29, 45). In addition to regulating differentiation of lymphocytes, intestinal crypt cells, and keratinocytes, we recently established that Wnts are key regulators of preadipocyte differentiation (1, 37). Wnt10b is the best candidate for the endogenous inhibitory Wnt because this protein blocks adipocyte conversion and expression of Wnt10b is high in preadipocytes and declines upon induction of differentiation. In addition to regulation of adipogenesis, Wnts play a larger role in preadipocytes, as Wnt1 inhibits preadipocyte apoptosis through induction of insulin-like growth factors (20).
The genetic program of adipogenesis has been studied extensively in vitro, with embryonic fibroblasts and preadipocyte lines, and in vivo, with transgenic and knockout mouse models (21, 22, 26, 32, 34), and a paradigm for the cascade of genetic events has emerged. After induction of differentiation, there is a rapid and transient induction of CCAAT/enhancer-binding protein beta (C/EBPβ) and C/EBPδ. These transcription factors then activate expression of both C/EBPα and peroxisome proliferator-activated receptor gamma (PPARγ), which then reinforce each other's expression through a positive feedback loop. Activation of the canonical Wnt signaling pathway by Wnt10b, dominant stable β-catenin, or inhibition of glycogen synthase kinase 3 inhibits adipogenesis by blocking expression of C/EBPα and PPARγ (1, 37). While expression of Wnt10b declines during adipogenesis, levels of nuclear β-catenin remain elevated until after C/EBPα and PPARγ are induced, suggesting that β-catenin has additional roles other than inhibition of adipogenesis through TCF/LEF transcription factors. In support for this idea, β-catenin coactivates non-TCF/LEF transcription factors such as androgen receptor, retinoic acid receptor, and steroidogenic factor 1 (SF-1) (4, 25, 39, 48).
Endogenous Wnt signaling inhibits adipogenesis through activation of the canonical pathway and activation of TCF/LEF transcription factors (37). The four members of the mammalian TCF/LEF family of transcription factors all have an N-terminal β-catenin-interacting domain and a highly conserved HMG box DNA binding domain (46). Heterogeneity in TCF/LEFs arises through multiple mechanisms. For example, forms of TCF-1 and LEF-1 that lack the β-catenin-interacting domain result from alternative promoter usage (13, 33). Similar forms of TCF-4 and LEF-1 that have been engineered to lack the β-catenin-interacting domain are commonly used as dominant negative inhibitors of TCF/LEF action. In addition, members of this family are also subject to extensive alternative splicing C-terminal to the HMG box. We recently reported several alternatively spliced isoforms of mouse TCF-4 that are truncated N-terminal to the HMG box domain (3). One of these isoforms, TCF-4N, was identified in developing pituitary and subsequently in 3T3-L1 preadipocytes and adipocytes. While TCF-4N does not contain a DNA binding domain, it retains the region involved in binding β-catenin. A similar alternatively spliced LEF-1 was identified in humans, but a function was not reported (18).
Here we report that TCF-4N has multiple roles in regulation of transcription. While TCF-4N interacts with β-catenin and inhibits a TCF/LEF-responsive promoter, TCF-4N also interacts with β-catenin to activate promoters that are not responsive to TCF/LEFs. For example, TCF-4N synergizes with β-catenin to coactivate the orphan nuclear receptor SF-1. In addition, TCF-4N interacts functionally with β-catenin and p300 to coactivate the adipogenic transcription factor C/EBPα. Ectopic expression of TCF-4N in 3T3-L1 preadipocytes partially relieves the block of adipogenesis caused by dominant stable β-catenin, consistent with a model in which TCF-4N regulates Wnt signaling by redirecting β-catenin from TCF/LEF transcription factors to other transcription factors, including SF-1 and C/EBPα.
MATERIALS AND METHODS
Cell culture.
Mouse 3T3-L1 preadipocytes and human embryonic kidney 293T cells were maintained in Dulbecco's modified Eagle's medium (Life Technologies, Inc.) containing 10% calf serum (Sigma) as previously described (42). Y1 adrenocortical carcinoma cells were cultured in Dulbecco's modified Eagle's medium containing 7.5% horse serum (Invitrogen) and 2.5% fetal bovine serum (Invitrogen). 3T3-L1 preadipocytes were induced to differentiate into adipocytes 2 days after confluence as described previously (11). To visualize cytoplasmic lipid accumulation, 3T3-L1 cells were stained with Oil Red-O (31). To quantify retention of Oil Red-O, stained adipocytes were extracted with 1 ml of 4% Igepal CA-630 (Sigma) in isopropanol for 15 min, and absorbance was measured by spectrophotometry at 520 nm (1). Nuclear extracts were prepared essentially as described previously (5).
TCF-4N expression and retroviral plasmids.
TCF-4N (see Fig. 1A) was amplified from embryonic day 12.5 developing mouse pituitary gland by reverse transcription-PCR and subcloned into pcDNA3.1+ (Invitrogen) as described previously (3). An N-terminal deletion of the first 31 amino acids (ΔNTCF-4N) was generated by PCR with TCF-4N as a template (see Fig. 1B). The forward primer (5′-GGATCCACCATGTCGGAAAACTCCTC-3′) included a consensus start site of translation downstream of a BamHI restriction site, while the reverse primer (5′-TTATACCCGCACATGTCCAC-3′) recognized the unique 3′ untranslated region of TCF-4N. The resulting PCR fragment was subcloned into pCR2.1 (Invitrogen). The BamHI-XhoI fragment was then subcloned into the BamHI and XhoI sites of pcDNA3-AU1/Flag. To create a retroviral expression vector for TCF-4N, the HindIII-XhoI fragment from TCF-4N in pcDNA3.1+ was subcloned into the HindIII and XhoI sites of pTS13 containing a HindIII linker.
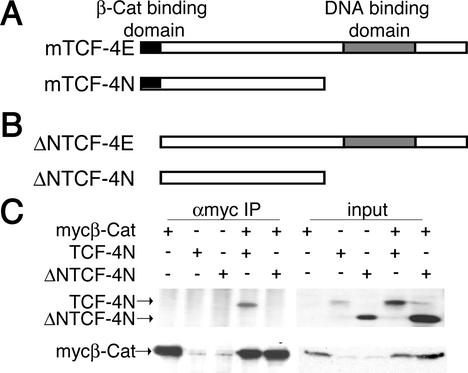
FIG. 1.
TCF-4N, an alternatively spliced isoform of TCF-4, lacks DNA binding domain but retains β-catenin-interacting domain. (A) The amino acid sequence of TCF-4N (GenBank accession number AF363725), a novel TCF-4 isoform independently cloned from developing mouse pituitary and 3T3-L1 preadipocytes, is schematically diagramed relative to full-length TCF-4E. The β-catenin interaction domain is depicted as a black box, and the DNA binding domain is depicted as a gray box. (B) Engineered forms of TCF-4E and TCF-4N designed to lack the N-terminal β-catenin interaction domain (ΔNTCF-4E and ΔNTCF-4N) are diagramed. (C) 293T cells in 10-cm plates were transfected with 10 μg each of the indicated expression vectors by calcium phosphate coprecipitation. Cells were lysed after 48 h, and Myc-tagged β-catenin was immunoprecipitated (IP) with an anti-Myc (αmyc) antibody. Immunoprecipitated β-catenin complexes were separated by SDS-PAGE, followed by immunoblot analysis for TCF-4 and β-catenin. These results are representative of at least three independent experiments.
Luciferase reporter gene assays.
The following reporter genes were used: pTOPFLASH and pFOPFLASH (Upstate Biotechnology), cyclin D1-luciferase, mutant TCF(1) and mutant TCF(0-4) (human cyclin D1 [−961]; Frank McCormick, University of California-San Francisco), (Gal4)5-SV40-luciferase (Mitchell A. Lazar, University of Pennsylvania), α-inhibin-luciferase (Kelly Mayo, Northwestern University), LexA-luciferase (Holly Ingraham, University of Californi-San Francisco), and leptin-luciferase and mutant leptin-luciferase (M. Daniel Lane, Johns Hopkins University). Expression constructs for mutant human β-catenin containing an in-frame N-terminal deletion of amino acids 29 to 48 and ΔNTCF-4E were provided by Frank McCormick (University of California-San Francisco). The expression construct for TCF-4N was described previously (3), and the expression vector for TCF-4E was from Gregory Dressler (University of Michigan).
The mouse C/EBPα expression vector was described previously (36). An expression construct for a fusion protein of the Gal4 DNA binding domain and β-catenin, Gal4-βCat, was provided by Kun-Liang Guan (University of Michigan). Expression vectors for mouse SF-1 and a fusion between the LexA DNA binding domain and SF-1 were from Holly Ingraham (University of California-San Francisco). The expression vector encoding a fusion protein of the Gal4 DNA binding domain and the C/EBPα transactivation domain, Gal4-C/EBPα, was described previously (5). Expression vectors for a deletion mutant of C/EBPα lacking all conserved regions but CR2 (CR2-C/EBPα) and a deletion mutant lacking CR2 (CR1/3/4-C/EBPα) were described previously (5). The human p300 expression vector pVR1012-p300 was obtained from Gary Nabel (National Institutes of Health).
Human embryonic kidney 293T cells (40-mm plates) were transiently transfected by calcium phosphate coprecipitation with 4 μg of total DNA, including the luciferase reporter gene (as indicated), and 250 ng of cytomegalovirus β-galactosidase. Additional plasmid DNAs were varied, based upon experimental conditions, and are documented in the figure legends. A constant amount of total cytomegalovirus promoter (pcDNA3.1; Invitrogen) was maintained to control for potential squelching of the transcriptional machinery. After transfection, cells were incubated for 48 h and lysed. Luciferase activity was measured, and variations in transfection efficiency were accounted for by normalization against β-galactosidase activity (5).
Retroviral infection of 3T3-L1 preadipocytes.
293T cells (10-cm plates) were transfected by calcium phosphate coprecipitation with control (pTS13) or TCF-4N retroviral vectors and the viral packaging vectors SV-E-MLV-env and SVψ-E-MLV (7.5 μg of each). Virus-containing medium was collected 16 h after transfection and passed through a 0.45-μm syringe filter. Polybrene (hexadimethrine bromide; Sigma) was added to a final concentration of 8 μg/ml. This medium was then applied to subconfluent (≈30%) 3T3-L1 preadipocytes (10-cm plates). The infection protocol was repeated every 8 to 16 h until cells were ≈80% confluent. These cells were trypsin treated and replated in Dulbecco's modified Eagle's medium supplemented with 10% calf serum and 150 μg of hygromycin (Invitrogen) per ml for 5 days. Cells were then infected with control (PGS-CMV-CITE-neo) or S33Y β-catenin-expressing retroviruses (Eric Fearon, University of Michigan) and selected with 400 μg of geneticin (Life Technologies, Inc.) per ml, and the ability to undergo adipogenesis was evaluated.
Immunoprecipitation and immunoblot analyses.
For immunoprecipitation of TCF-4N with β-catenin, 293T cells (10-cm plates) were transiently transfected with 10 μg of expression vectors for Myc-tagged mutant β-catenin, TCF-4N, or ΔNTCF-4N as indicated. Cells were lysed in 50 mM HEPES (pH 7.5)-125 mM NaCl-1 mM EDTA (pH 8.0)-1 mM dithiothreitol-1% Igepal CA-630 and protease inhibitors. Lysates were cleared with protein G plus agarose (Santa Cruz) and incubated with 4 μl of anti-Myc tag clone 9E10 (Upstate Biotechnology). Protein G plus agarose was added to samples, and after incubation for 1 h, pellets were washed and resuspended in sodium dodecyl sulfate (SDS) loading buffer. Immunoblot analysis was performed with mouse monoclonal immunoglobulin G against TCF-3 and TCF-4 (Upstate Biotechnology) and β-catenin (Transduction Laboratories).
For immunoprecipitation of β-catenin with SF-1, nuclear lysates were generated from Y1 adrenocortical carcinoma cells. Briefly, cell membranes were disrupted in a buffer containing 25 mM HEPES (pH 7.6), 5 mM KCl, 0.5 mM MgCl2, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, and 0.5% Igepal CA-630. Nuclei were lysed for 1 h in a buffer containing 25 mM HEPES, 10% sucrose, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 0.01% Igepal CA-630, and 350 mM NaCl, and insoluble proteins were removed by centrifugation. Nuclear lysates were cleared with protein A/G-agarose for 30 min and incubated with antihemagglutinin (anti-HA) antibody (Santa Cruz Biotechnology) or anti-SF-1 antibody (Upstate Biotechnology). Protein A/G-agarose was added to the samples, and after incubation for 1 h, pellets were washed and resuspended in SDS loading buffer. Immunoblot analysis was performed with mouse monoclonal antibody against β-catenin (Transduction Laboratories).
Immunoprecipitations of C/EBPα with β-catenin were done essentially as described previously (27). After transient transfection of 293T cells (10-cm plates) with 5 μg of expression vectors for S33Y β-catenin-Flag (Eric Fearon, University of Michigan), p42C/EBPα, and CR1/3/4-C/EBPα, cells were lysed in 0.5 ml of buffer A (20 mM HEPES [pH 7.9], 350 mM NaCl, 30 mM MgCl2, 1 mM EDTA [pH 8.0], 0.1 mM EGTA [pH 8.0], 20% glycerol, 0.5% Igepal CA-630, 1 mM dithiothreitol, and protease inhibitors). After centrifugation, lysates were diluted with 0.75 ml of buffer B (20 mM HEPES [pH 7.9], 30 mM MgCl2, 1 mM EDTA [pH 8.0], 0.1 mM EGTA [pH 8.0], 20% glycerol, 0.2% Igepal CA-630, 1 mM dithiothreitol, and protease inhibitors). Lysates were cleared with protein A-Sepharose (Pharmacia), split in half, and incubated overnight with either 1 μl of anti-Flag M2 (Sigma) or 1 μl of negative control immunoglobulin G (PPARγ mouse monoclonal; Santa Cruz) as indicated. Protein A-Sepharose was added to the samples, and after incubation for 1 h, pellets were washed with wash buffer A containing 150 mM NaCl. Proteins were eluted by incubation with competitive Flag peptide (Sigma) overnight, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and transferred to a polyvinylidene difluoride membrane. After blocking, the membrane was incubated with mouse monoclonal antibody against β-catenin (Transduction Laboratories) or rabbit polyclonal antibody against C/EBPα (Daniel Lane, Johns Hopkins University).
RESULTS
TCF-4N, an alternatively spliced isoform of TCF-4, lacks DNA binding domain but retains β-catenin-interacting domain.
Novel isoforms of mouse TCF-4 lacking the HMG box DNA binding domain were first identified in embryonic mouse pituitary gland (3). We also cloned a similar isoform of TCF-4 from 3T3-L1 preadipocytes. The mRNA for this novel isoform contained a stop codon that terminated translation of the protein prior to the DNA binding domain. We designated this protein TCF-4N because it contained only the N terminus (Fig. 1A). Transfection of a fusion protein containing TCF-4N and green fluorescent protein indicated expression in both nuclear and cytoplasmic compartments (data not shown). In addition, we identified a bovine expressed sequence tag (EST) (GenBank accession number BE749318) with homology to TCF-3 that contains a stop codon at the same site as in TCF-4N. These data, together with the identification of a similar isoform of human LEF-1 (18), suggest a conserved role for isoforms such as TCF-4N in regulating transcription.
TCF-4N retains the β-catenin-interacting domain at the N terminus.
To determine whether TCF-4N interacts physically with β-catenin, we performed a coimmunoprecipitation analysis (Fig. 1C). Expression constructs for TCF-4N and ΔNTCF-4N, an N-terminal deletion mutant (Fig. 1B), were cotransfected into 293T cells with an expression construct for dominant stable Myc-tagged β-catenin. As expected, TCF-4N was immunoprecipitated with β-catenin (Fig. 1C), similar to full-length TCF-4E (data not shown). However, ΔNTCF-4N was not immunoprecipitated by β-catenin (Fig. 1C), suggesting that TCF-4N interacts with β-catenin via an interaction at the N terminus.
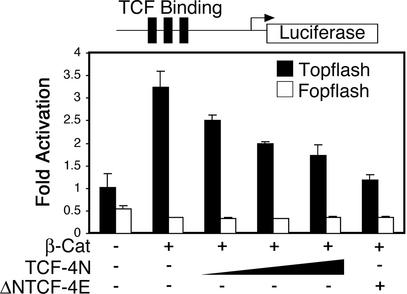
TCF-4N inhibits activation by β-catenin of a TCF-responsive reporter gene.
β-Catenin is a coactivator of TCF/LEF transcription factors. Unlike previously described TCF/LEF isoforms, TCF-4N lacks a DNA binding domain; however, TCF-4N retains its ability to interact with β-catenin (Fig. 1C). We hypothesized that TCF-4N acts as an endogenous inhibitor of β-catenin by competing with DNA-bound TCF/LEF transcription factors for binding to β-catenin. To determine if TCF-4N inhibits transcriptional coactivation by β-catenin, we performed luciferase reporter assays with pTOPFLASH, a TCF-responsive reporter gene that contains multimerized TCF binding sites upstream of a minimal promoter and the coding sequence for luciferase.
Expression constructs for dominant stable β-catenin, TCF-4N, and ΔNTCF-4E (Fig. 1B) were transiently transfected into 293T cells along with pTOPFLASH or the negative control pFOPFLASH. As expected, ectopic expression of β-catenin increased reporter gene activity (Fig. 2). Cotransfection of increasing amounts of TCF-4N progressively decreased the reporter activity in response to β-catenin, similar to the inhibitory effect of the well-established dominant negative form of TCF, ΔNTCF-4E (Fig. 2) (44). TCF-4N had no effect on reporter activity in the absence of ectopic β-catenin (data not shown). In addition, neither β-catenin nor TCF-4N influenced gene expression from pFOPFLASH, in which the TCF binding sites are mutated (Fig. 2). These results indicate that TCF-4N acts through a dominant negative mechanism to inhibit activation by β-catenin of a TCF-responsive promoter.
FIG. 2.
TCF-4N inhibits activation by β-catenin of a TCF-responsive reporter gene. 293T cells were transfected with pTOPFLASH (25 ng), which is a TCF-responsive reporter construct (solid bars), along with expression vectors for β-catenin (10 ng), TCF-4N (25, 50, or 100 ng) and ΔNTCF-4E (100 ng) as indicated. pFOPFLASH (25 ng), a reporter containing mutated TCF consensus binding sites, was transfected as a control (open bars). Samples were normalized to β-galactosidase activity to correct for variations in transfection efficiency. Luciferase activity is reported as activation (mean ± standard deviation) relative to pTOPFLASH alone. These results are representative of at least three independent experiments.
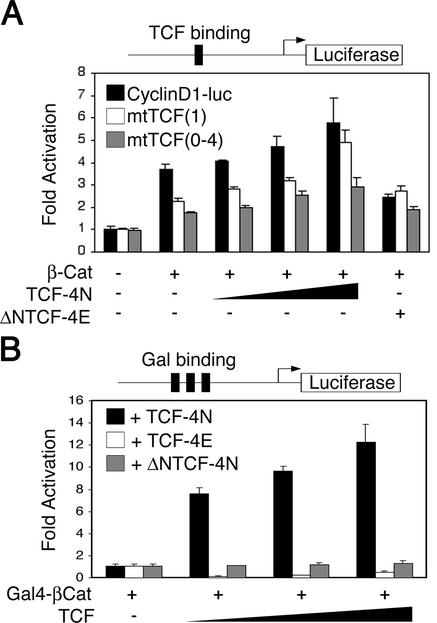
TCF-4N and β-catenin synergize to activate transcription from non-TCF-responsive promoters.
TCF-4N inhibits activation by β-catenin of the TCF-responsive promoter gene pTOPFLASH (Fig. 2). To determine the effect of TCF-4N and β-catenin on what has been reported to be a TCF-dependent promoter, we performed luciferase reporter assays with the human cyclin D1 promoter (cyclin D1-luc), which contains one TCF/LEF consensus site and four cryptic binding sites (40, 44). While β-catenin activated the cyclin D1 promoter (Fig. 3A), TCF-4N alone had no effect (data not shown). In contrast to our hypothesis that TCF-4N would inhibit cyclin D1 promoter activity, cotransfection of TCF-4N with β-catenin caused an increase in reporter gene expression even in the absence of TCF binding sites, i.e., mtTCF(1) and mtTCF(0-4) (Fig. 3A). Cotransfection of ΔNTCF-4N had no effect on β-catenin activation of the cyclin D1 promoter (data not shown), indicating that the N terminus of TCF-4N is necessary for its effect. β-Catenin activated the mutant cyclin D1 reporter genes independent of DNA binding by TCF/LEF, because cotransfection of ΔNTCF-4E did not reduce promoter activity for either mutant reporter gene (Fig. 3A). Furthermore, the increase in promoter activity by cotransfection of TCF-4N, despite mutations in the putative TCF binding sites, suggests that β-catenin activates transcription via interactions with transcription factors other than TCF/LEFs.
FIG. 3.
TCF-4N and β-catenin synergize to activate transcription from non-TCF-responsive promoters. (A) 293T cells were transfected with 5 ng of a cyclin D1 promoter-luciferase construct (cyclinD1-luc; black bars), a cyclin D1-luciferase construct with mutations in the consensus TCF binding site, mtTCF(1) (open bars), or with mutations in the one consensus site and four cryptic TCF binding sites, mtTCF(0-4) (grey bars). Reporter genes were cotransfected with expression vectors for β-catenin (250 ng), TCF-4N (125, 250, and 500 ng), and ΔNTCF-4E (500 ng) as indicated. Samples were normalized to β-galactosidase activity to correct for variations in transfection efficiency. Luciferase activity is reported as activation (mean ± standard deviation) relative to the reporter gene alone. (B) 293T cells were transfected with a Gal4-responsive reporter gene (250 ng), which contains multimerized Gal4 binding sites upstream of a minimal promoter and the luciferase gene. Cells were cotransfected with expression constructs for the Gal4 DNA binding domain fused to β-catenin (Gal4-βCat; 250 ng) and either 125, 250, or 500 ng of TCF-4N (solid bars), TCF-4E (open bars), or ΔNTCF-4N (gray bars) as indicated. Luciferase activity is reported as activation (mean ± standard deviation) relative to the reporter gene with Gal4-βCat. These results are representative of at least three independent experiments.
To verify that TCF-4N enhances transcriptional coactivation by β-catenin independently of TCF/LEF transcription factors, we performed reporter gene assays with a Gal4 DNA binding domain-β-catenin fusion protein (Gal4-βCat) and a Gal4-responsive reporter gene (Fig. 3B). While transfection of Gal4-βCat activated the Gal4-responsive promoter by approximately100-fold, TCF-4N had no effect on basal promoter activity (data not shown). When TCF-4N was cotransfected with Gal4-βCat, TCF-4N synergized with β-catenin, resulting in a dramatic 8- to 12-fold increase in reporter gene activity over that observed with Gal4-βCat alone (Fig. 3B). Cotransfection of full-length TCF-4E decreased reporter gene activity, presumably by sequestering Gal4-βCat. ΔNTCF-4N had no effect on promoter activity, indicating that the N terminus of TCF-4N is required for its functional interaction with β-catenin. Taken together, these data indicate that for certain non-TCF/LEF-dependent promoters, TCF-4N and β-catenin synergize to activate gene expression.
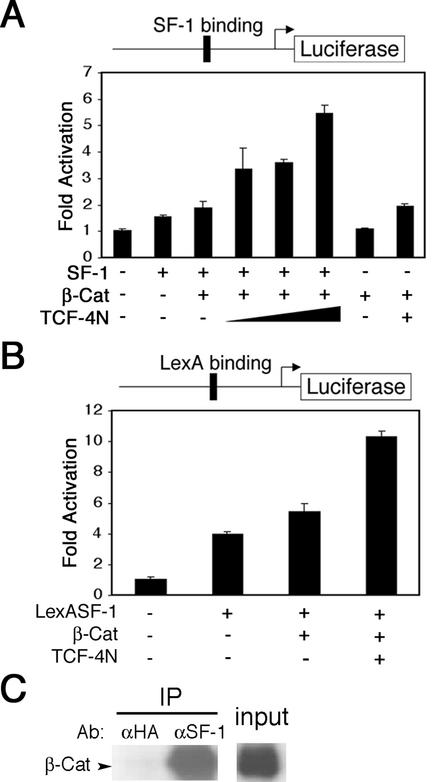
β-Catenin and TCF-4N synergize to coactivate SF-1.
Our results with the mutant cyclin D1 promoter suggest that β-catenin interacts functionally with transcription factors other than TCF/LEFs (Fig. 3A). Recent evidence with the Dax-1 promoter supports a role for β-catenin as a coactivator of SF-1 (25). Expression of this orphan nuclear receptor is restricted to a subset of endocrine tissues, including gonads, adrenal cortex, ventromedial hypothalamus, and pituitary gonadotropes (8). Interestingly, TCF-4N and similar isoforms were first identified in developing and adult pituitary (3).
To explore the possibility that TCF-4N potentiates coactivation by β-catenin of SF-1, we performed reporter assays with the SF-1-dependent inhibin promoter (inhibin-luc) (16, 23). Expression of SF-1 induced inhibin promoter activity, and cotransfection of β-catenin resulted in an increase above that with SF-1 alone (Fig. 4A). Cotransfection of increasing amounts of TCF-4N synergized with β-catenin and SF-1 to stimulate a greater than 2.5-fold increase in inhibin promoter activity. The induction of inhibin promoter activity by TCF-4N, β-catenin, and SF-1 was not influenced by coexpression with the well-established dominant negative TCF ΔNTCF-4E (data not shown), indicating that the effects are not mediated through TCF/LEF DNA binding sites. The effects of β-catenin and TCF-4N were largely dependent upon the presence of SF-1 (Fig. 4A). Our data suggest that β-catenin and TCF-4N synergize to coactivate the orphan nuclear receptor SF-1.
FIG. 4.
TCF-4N and β-catenin synergize to coactivate SF-1. (A) 293T cells were transfected with an inhibin-luciferase reporter gene (300 ng). Expression constructs for SF-1 (75 ng), β-catenin (300 ng), and TCF-4N (125, 250, and 500 ng) were cotransfected as indicated. Samples were normalized to β-galactosidase activity to correct for variations in transfection efficiency. Luciferase activity is reported as activation (mean ± standard deviation) relative to the reporter gene alone. (B) 293T cells were cotransfected with a LexA-responsive reporter gene (100 ng) and a LexASF-1 expression construct (10 ng). Expression constructs for β-catenin (500 ng) and TCF-4N (500 ng) were cotransfected as indicated. Luciferase activity is reported as activation (mean ± standard deviation) relative to the reporter gene alone. (C) SF-1 was immunoprecipitated from nuclear lysates of Y1 adrenocortical carcinoma cells. Immunoprecipitated SF-1 complexes were separated by SDS-PAGE, followed by immunoblot analysis for β-catenin. These results are representative of at least three independent experiments.
To verify the functional interaction between SF-1 and a complex containing β-catenin and TCF-4N, we used a LexA-dependent reporter gene and a fusion construct in which the SF-1 DNA binding domain was replaced with the LexA DNA binding domain (LexASF-1). Expression of LexASF-1 stimulated an increase in reporter gene activity, and this was further increased by cotransfection of LexASF-1 with β-catenin (Fig. 4B). Coactivation of LexASF-1 by β-catenin was almost doubled by coexpression of TCF-4N, consistent with our prior observations with the inhibin promoter (Fig. 4A). In contrast, cotransfection of β-catenin and TCF-4N without SF-1 resulted in no increase in promoter activity (data not shown). These data demonstrate that TCF-4N potentiates the coactivation of SF-1 by β-catenin.
The ability of β-catenin and TCF-4N to coactivate LexASF-1 confirms that the synergy between β-catenin, TCF-4N, and SF-1 is independent of DNA binding by TCF/LEF transcription factors, raising the possibility that there is a direct physical interaction between SF-1 and β-catenin. To determine whether β-catenin and SF-1 interact in vivo, we used immunoprecipitation assays with nuclear lysates prepared from Y1 adrenocortical carcinoma cells, which express both SF-1 and β-catenin. Consistent with the hypothesis that β-catenin and SF-1 interact physically, endogenous β-catenin was coimmunoprecipitated with SF-1 (Fig. 4C). These data indicate that β-catenin and TCF-4N coactivate SF-1 through direct physical interactions.
β-Catenin and TCF-4N synergize to coactivate C/EBPα.
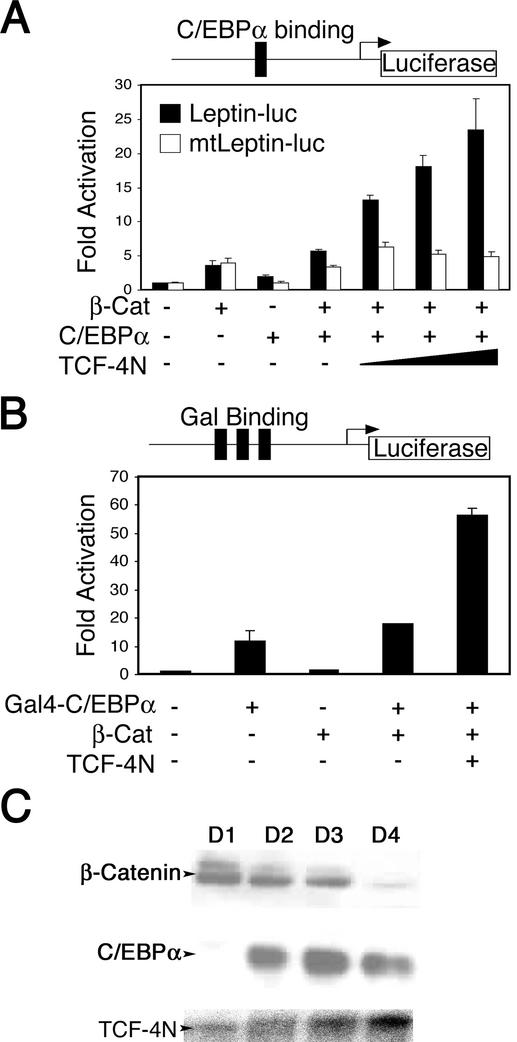
Given the emerging concept that β-catenin coactivates transcription factors other than TCF/LEFs, we explored the possibility that β-catenin and TCF-4N coactivate C/EBPα, a transcription factor important for adipogenesis. To determine if β-catenin and TCF-4N functionally interact with C/EBPα, we performed reporter gene assays with the leptin promoter (Leptin-luc and mtLeptin-luc). The leptin promoter contains a C/EBP binding site and has been shown to be C/EBPα responsive (12, 15). Expression of C/EBPα increased leptin reporter gene activity, and coexpression of C/EBPα and β-catenin resulted in an additive increase (Fig. 5A). Cotransfection of TCF-4N with C/EBPα and β-catenin resulted in a synergistic activation of the leptin promoter that was dependent upon the C/EBPα binding site (Fig. 5A). The well-established dominant negative TCF ΔNTCF-4E did not influence the increase of reporter activity by TCF-4N, C/EBPα, and β-catenin (data not shown). In addition, TCF-4N had no effect on C/EBPα activation of the leptin promoter in the absence of exogenous β-catenin (data not shown). The lack of inhibition by ΔNTCF-4E suggests that β-catenin and TCF-4N enhance C/EBPα activity through a mechanism that is independent of TCF/LEF DNA binding. Because 293T cells express little or no endogenous C/EBPα and β-catenin increased leptin promoter activity even in the absence of C/EBPα binding sites (Fig. 5A), it appears that β-catenin also interacts with other transcriptional regulators of the leptin promoter. Our data suggest that a complex containing β-catenin and TCF-4N induces leptin promoter activity specifically through coactivation of C/EBPα.
FIG. 5.
TCF-4N and β-catenin synergize to coactivate C/EBPα. (A) 293T cells were transfected with a leptin promoter reporter gene (250 ng; solid bars) or mtLeptin-luc (open bars), containing a mutated C/EBPα binding site. Expression constructs for β-catenin (250 ng), C/EBPα (50 ng), and TCF-4N (125, 250, and 500 ng) were cotransfected as indicated. Samples were normalized to β-galactosidase activity to correct for variations in transfection efficiency. Luciferase activity is reported as activation (mean ± standard deviation) relative to the reporter gene alone. (B) 293T cells were cotransfected with a Gal4-responsive reporter gene (250 ng) and an expression construct for the Gal4 DNA binding domain fused to the transactivation and basic region of C/EBPα (5 ng). Expression constructs for β-catenin (250 ng) and TCF-4N (500 ng) were cotransfected as indicated. Luciferase activity is reported as activation (mean ± standard deviation) relative to the reporter gene alone. These results are representative of at least three independent experiments. (C) Nuclear extracts (20 μg) prepared 1 to 4 days after induction of 3T3-L1 cell differentiation were analyzed by immunoblot for expression of β-catenin and C/EBPα. RNA prepared 1 to 4 days after induction of 3T3-L1 cell differentiation was analyzed for expression of TCF-4N by RNase protection assay with a riboprobe specific for the unique 3′ untranslated region.
To verify the functional interactions between β-catenin, TCF-4N, and C/EBPα, we performed luciferase assays with a Gal4-C/EBPα expression construct and a Gal4-responsive reporter gene (Fig. 5B). The Gal4-C/EBPα chimeric protein consists of the Gal4 DNA binding domain and the C/EBPα transactivation domain truncated prior to the leucine zipper. Transfection of Gal4-C/EBPα increased reporter gene activity by almost 10-fold (Fig. 5B). While coexpression of β-catenin with Gal4-C/EBPα did not greatly alter promoter activity, TCF-4N synergized with these factors to dramatically increase promoter activity. The effects of TCF-4N required expression of both Gal4-C/EBPα and β-catenin (data not shown). These data provide strong evidence that β-catenin and TCF-4N are transcriptional coactivators for C/EBPα.
Regulation of C/EBPα activity by β-catenin in transfected cells raises the issue of whether the endogenous proteins interact within differentiating preadipocytes. To ascertain whether β-catenin and C/EBPα are present at the same time and in the same cellular compartment, we examined expression of these proteins in nuclear extracts prepared from 3T3-L1 cells at various times after induction of differentiation. Nuclear β-catenin was elevated for the first 3 days of differentiation but declined dramatically by day 4 (Fig. 5C). C/EBPα was expressed by day 2 and was maximal by day 3. Thus, β-catenin is present in the nucleus during the time that C/EBPα is inducing expression of PPARγ and other adipocyte proteins. In addition, RNase protection analysis demonstrated that TCF-4N was expressed throughout differentiation (Fig. 5C).
β-Catenin and TCF-4N interact with C/EBPα and p300 to activate the leptin promoter.
Our laboratory and others have shown that the coactivators p300 and CREB binding protein (CBP) functionally interact with C/EBPα (5, 9, 38). Given that β-catenin also interacts with p300/CBP (10, 24, 43), we hypothesized that p300 is a component of the β-catenin/TCF-4N complex that coactivates C/EBPα. Previous analysis of the N-terminal transactivation domain of C/EBPα indicated that there were four main conserved regions, designated CR1 to CR4 (Fig. 6A) (6). Of these four regions, CR2 (amino acids 55 to 108) is sufficient to activate transcription and to interact functionally with p300 (5).
FIG. 6.
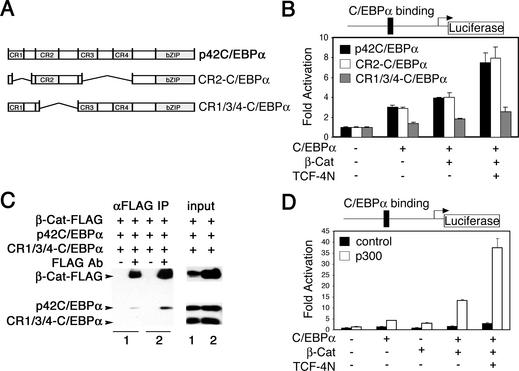
β-Catenin and TCF-4N interact with C/EBPα and p300 to activate the leptin promoter. (A) The amino acid sequence of mouse C/EBPα is schematically diagrammed, with the four conserved regions within the transactivation domain indicated. The CR2-C/EBPα and CR1/3/4-C/EBPα deletion constructs are shown schematically relative to full-length p42C/EBPα. (B) 293T cells were transfected with Leptin-luc (250 ng) and 50 ng of expression vectors for full-length C/EBPα (solid bars), CR2-C/EBPα (open bars), or CR1/3/4-C/EBPα (gray bars). Expression constructs for β-catenin (250 ng) and TCF-4N (500 ng) were cotransfected as indicated. Samples were normalized to β-galactosidase activity to correct for variations in transfection efficiency. Luciferase activity is reported as fold activation (mean ± standard deviation) relative to the reporter gene alone. (C) 293T cells in 10-cm plates were transfected with 5 μg each of the indicated expression vectors by calcium phosphate coprecipitation. Cells were lysed after 48 h, and β-catenin was immunoprecipitated with anti-Flag antibody. Immunoprecipitated β-catenin complexes were separated by SDS-PAGE, followed by immunoblot analysis for C/EBPα and β-catenin. (D) 293T cells were transfected with Leptin-luc (250 ng) and expression constructs for C/EBPα (50 ng), β-catenin (250 ng), and TCF-4N (500 ng) as indicated. Cells were cotransfected with 250 ng of either empty vector (solid bars) or human p300 (open bars). Luciferase activity is reported as activation (mean ± standard deviation) relative to the reporter gene alone. Results are representative of at least three independent experiments.
To determine the domains of C/EBPα that mediate functional interactions with the β-catenin/TCF-4N complex, we performed luciferase assays with the leptin reporter gene (Leptin-luc) and mutants of C/EBPα (Fig. 6B). Cotransfection of a C/EBPα mutant consisting of CR2 and the bZIP domain (CR2-C/EBPα) with β-catenin/TCF-4N increased leptin reporter activity similar to that in full-length C/EBPα, suggesting that CR2 is sufficient for functional interactions between C/EBPα and the β-catenin/TCF-4N complex. Deletion of CR2 (CR1/3/4-C/EBPα) largely reduced the functional interaction of C/EBPα with β-catenin/TCF-4N (Fig. 6B). Thus, it appears that CR2 is sufficient to mediate coactivation not only by p300, but also by β-catenin/TCF-4N.
Our data suggest that the CR2 region of C/EBPα is required for functional interactions with β-catenin and TCF-4N. To determine whether the CR2 region of C/EBPα is necessary for physical interaction between C/EBPα and β-catenin, we performed immunoprecipitation assays with lysates from 293T cells. We cotransfected 293T cells with β-catenin, p42 C/EBPα and CR1/3/4-C/EBPα, which lacks the CR2 domain. Full-length C/EBPα (p42) was immunoprecipitated with β-catenin, but CR1/3/4-C/EBPα was not (Fig. 6C). Using similar methods, we also found that β-catenin was immunoprecipitated with p42C/EBPα (data not shown). Taken together, these data suggest that functional interactions between C/EBPα and β-catenin are mediated through direct physical interactions.
Our prior findings indicate that CR2 is the strongest transactivation domain within C/EBPα and acts, in part, through the transcriptional coactivator p300 (5). Given that CR2 interacts with β-catenin/TCF-4N (Fig. 6B and C) and that β-catenin interacts with p300 (10, 24, 43), it is possible that the transcriptional complex for C/EBPα contains β-catenin, TCF-4N, and p300. To test whether p300 interacts with β-catenin/TCF-4N to coactivate C/EBPα, we performed luciferase assays on cells transfected with the leptin reporter gene and expression constructs for C/EBPα, β-catenin, TCF-4N, and p300, as indicated (Fig. 6D). Cotransfection of p300 resulted in a synergistic increase in promoter activity over that observed with C/EBPα and β-catenin/TCF-4N. A similar but less robust increase was observed with p300, C/EBPα, and β-catenin in the absence of TCF-4N, suggesting a role for TCF-4N in stabilizing interactions between p300 and the transcriptional complex. Taken together, these data support a model in which β-catenin coactivates C/EBPα through a complex that contains TCF-4N and p300.
TCF-4N partially rescues the block of differentiation by ectopic expression of β-catenin.
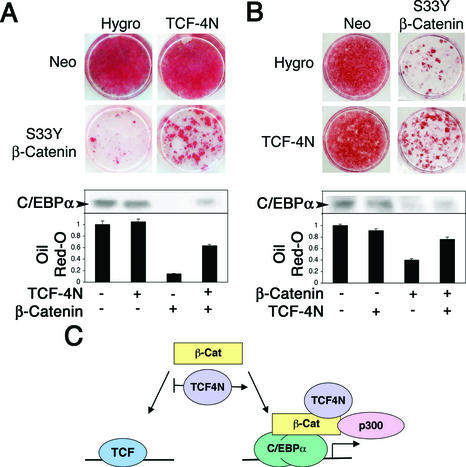
Activation of the canonical Wnt signaling pathway by inhibition of glycogen synthase kinase 3 or enforced expression of β-catenin blocks adipogenesis through activation of TCF/LEF target genes (1, 35, 37). Our results indicate that TCF-4N decreases coactivation of TCF/LEF family members by β-catenin yet enhances coactivation of other transcription factors, including C/EBPα. To determine if TCF-4N alters the ability of β-catenin to block differentiation, we stably expressed β-catenin alone or β-catenin with TCF-4N by sequential retroviral infections (Fig. 7A and B). Control 3T3-L1 preadipocytes and those expressing TCF-4N differentiated fully in response to inducers of adipogenesis (Fig. 7A). While expression of β-catenin blocked the differentiation of 3T3-L1 preadipocytes, coexpression with TCF-4N resulted in more cells undergoing adipogenesis than observed with β-catenin alone (Fig. 7A). Expression of exogenous β-catenin in both cell lines was similar by immunoblot analysis (data not shown).
FIG. 7.
TCF-4N partially rescues the block of differentiation caused by ectopic expression of β-catenin. (A) 3T3-L1 preadipocytes were infected with a control retrovirus (Hygro) or a retrovirus containing the coding region for TCF-4N. Control and TCF-4N-expressing cells were reinfected with a control retrovirus (Neo) or a retrovirus containing the coding region for S33Y β-catenin. Two days after confluence, cells were treated with inducers of adipogenesis. Two weeks later, cells were stained with Oil Red-O to visualize the degree of lipid accumulation. The amount of Oil Red-O was quantified after extraction and is displayed as fold absorbance relative to the empty vector control. Cells were lysed, and expression of C/EBPα was analyzed by immunoblot (inset). (B) Control (Neo) and S33Y β-catenin-expressing cells were reinfected with a retrovirus alone (Hygro) or a retrovirus containing the coding region for TCF-4N and analyzed as in A. (C) Model for the regulation of β-catenin by TCF-4N. Expression of TCF-4N inhibits interactions between β-catenin and TCF/LEF transcription factors. Complexes containing β-catenin, TCF-4N, and p300 coactivate multiple transcription factors, including the adipogenic transcription factor C/EBPα.
We also performed a reciprocal experiment in which control cells and β-catenin-expressing cells were infected with control or TCF-4N retroviruses (Fig. 7B). This experiment produced similar results, with increased differentiation in cells that expressed β-catenin and TCF-4N compared to preadipocytes that expressed β-catenin alone. After extraction and quantification of Oil Red-O, we found that the amount of stain was at least twofold higher in cells that coexpressed β-catenin and TCF-4N compared to cells that expressed β-catenin alone (Fig. 7A and B). In addition to increasing amounts of triacylglycerol in these cells, TCF-4N rescued expression of C/EBPα (Fig. 7A and B, insets), which is an indicator of adipogenesis. These results are consistent with our reporter gene assays, which indicate that TCF-4N inhibits β-catenin activation of TCF/LEF target genes and that TCF-4N enhances coactivation by β-catenin of other transcription factors, such as the adipogenic factor C/EBPα.
DISCUSSION
In this study we characterized the function of an alternatively spliced isoform of TCF-4, which we named TCF-4N because it contains the N terminus of the protein. TCF-4N has a translational stop site prior to the DNA binding domain, resulting in a stable protein that retains the ability to bind β-catenin (Fig. 1C). TCF-4N inhibits the activation by β-catenin of TOPFLASH, a TCF-dependent promoter, similar to the well-established dominant negative TCF ΔNTCF-4E (Fig. 2). However, TCF-4N enhances the activation by β-catenin of TCF-independent promoters, such as the mutant cyclin D1 promoters (Fig. 3A), the SF-1-dependent inhibin promoter (Fig. 4B), the LexA-responsive promoter as activated by LexASF-1 (Fig. 4B), the Gal4-responsive promoter as activated by Gal4-βCat or Gal4-C/EBPα (Fig. 3A and 5B), and the C/EBPα-dependent leptin promoter (Fig. 5A). The mechanism by which the β-catenin/TCF-4N complex coactivates C/EBPα appears to involve p300, based upon synergy between these important transcriptional regulators (Fig. 5 and 6). Finally, ectopic expression of TCF-4N in 3T3-L1 preadipocytes partially relieves the block of differentiation caused by β-catenin (Fig. 7A and B). Thus, we propose that TCF-4N inhibits coactivation by β-catenin of TCF/LEF transcription factors and enhances the coactivation by β-catenin of other transcription factors, such as SF-1 (Fig. 4) and the adipogenic transcription factor C/EBPα (Fig. 7C).
The ability of TCF-4N to partially relieve the inhibition of 3T3-L1 adipogenesis caused by β-catenin supports the evidence from reporter gene assays that TCF-4N potentiates coactivation by β-catenin of non-TCF/LEF transcription factors, including the positive regulator of adipogenesis C/EBPα (Fig. 5). Rescue of the inhibition of β-catenin by TCF-4N could potentially be due to inhibition of TCF/LEF-dependent transcription and/or activation of C/EBPα and other adipogenic transcription factors. Coactivation of C/EBPα by β-catenin is paradoxical, given the inhibitory effect of Wnt signaling and dominant-stable β-catenin on adipogenesis. Interestingly, nuclear β-catenin levels remained elevated for 3 days after induction of adipocyte differentiation (Fig. 5C), despite a decline in the Wnt10b and Frizzled receptors (1). Levels of the adipogenic transcription factors C/EBPα and PPARγ increased during this time and peaked 3 days after induction of differentiation. Since β-catenin declined after induction of many adipocyte markers and the accumulation of lipid, it appears that β-catenin has a more complex role during adipogenesis than the strictly antagonistic function predicted by its role in the canonical Wnt signaling pathway.
Studies that examined the structure of TCF/LEF transcription factors bound to β-catenin suggest that they bind as a heterodimer, with a 1:1 stoichiometry (7, 30). As TCF-4N lacks the DNA binding domain and a putative nuclear localization signal, we postulated that TCF-4N could inhibit transcriptional coactivation by β-catenin by blocking interactions of β-catenin with DNA-bound TCF/LEF transcription factors. However, the mechanism by which TCF-4N enhances transcriptional coactivation by β-catenin of target genes is unknown. Protein levels of total exogenous β-catenin are unaltered by TCF-4N, suggesting that TCF-4N does not increase the stability of β-catenin (data not shown). β-Catenin has been reported to be tyrosine phosphorylated, which decreases its affinity for E-cadherin and increases its affinity for TATA binding protein (28); however, we saw no change in tyrosine phosphorylation of β-catenin when it was coexpressed with TCF-4N (data not shown). However, other important posttranslational modifications of β-catenin, such as sumoylation or acetylation, may be regulated by TCF-4N (17, 47). Another possibility is that TCF-4N alters the cellular localization of β-catenin through regulation of the cytosolic/nuclear pool or the membrane-bound pool by an unknown mechanism. However, expression of TCF-4N does not appear to alter the localization of fluorescently tagged β-catenin (data not shown). Furthermore, the positive effect of TCF-4N on the activity of the Gal4-βCat fusion protein (Fig. 3B) suggests that the effects of TCF-4N may not involve differences in subcellular localization, as Gal4-βCat is nuclear due to the Gal4 nuclear localization signal.
While the mechanism whereby TCF-4N potentiates transcriptional coactivation by β-catenin does not appear to involve regulation of β-catenin stability, tyrosine phosphorylation, or subcellular localization, the positive effect of TCF-4N on β-catenin may be through the recruitment of transcriptional coactivators. It is well established that β-catenin physically and functionally interacts with p300 and CBP (10, 24, 43). TCF-4N may stabilize these interactions, because TCF-4N greatly increased the coactivation of C/EBPα by p300 and β-catenin (Fig. 6D). Furthermore, TCF-4N may recruit additional coactivators to the complex that are not recruited by β-catenin or p300 alone.
While we identified and characterized TCF-4N as encoding the N terminus of TCF-4 in the mouse, we also identified by database searches a similar isoform of TCF-3 in cows. Strikingly, the alternative splice site and stop codon are completely conserved between mouse TCF-4N and bovine TCF-3. In addition, another group recently cloned a similarly truncated LEF-1 isoform in humans (18). In contrast to TCF-4N and the putative TCF-3N, which are alternatively spliced and truncated by insertion of a stop codon immediately after the exon junction (3), the novel LEF-1 isoform contains an alternative exon coding for a novel string of amino acids prior to a stop codon. Similar isoforms of LEF-1 in humans, TCF-4 in mice, and TCF-3 in cows suggest a conserved and important role for N-terminal isoforms in regulation of transcription. In this paper we have shown that TCF-4N inhibits TCF/LEF-dependent transcription and stimulates the coactivation by β-catenin and p300 of other transcription factors, including C/EBPα. The effects of TCF-4N on adipogenesis demonstrate its ability to influence cell growth and differentiation.
Acknowledgments
This work was supported by National Institutes of Health research grant DK51563 (O.A.M.), Cellular and Molecular Approaches to Systems and Integrative Biology Training Grant T32-GM08322 (J.A.K. and G.M.W.), and Cellular and Molecular Biology Training Grant GM07315-26 (J.A.K. and E.E.O.).
REFERENCES
- 1.Bennett, C. N., S. E. Ross, K. A. Longo, L. Bajnok, N. Hemati, K. W. Johnson, S. D. Harrison, and O. A. MacDougald. 2002. Regulation of Wnt signaling during adipogenesis. J. Biol. Chem. 277:30998-31004. [DOI] [PubMed] [Google Scholar]
- 2.Cadigan, K. M., and R. Nusse. 1997. Wnt signaling: a common theme in animal development. Genes Dev. 11:3286-3305. [DOI] [PubMed] [Google Scholar]
- 3.Douglas, K. R., M. L. Brinkmeier, J. A. Kennell, P. Eswara, T. A. Harrison, A. I. Patrianakos, B. S. Sprecher, M. A. Potok, R. H. Lyons, Jr., O. A. MacDougald, and S. A. Camper. 2001. Identification of members of the Wnt signaling pathway in the embryonic pituitary gland. Mamm. Genome 12:843-851. [DOI] [PubMed] [Google Scholar]
- 4.Easwaran, V., M. Pishvaian, Salimuddin, and S. Byers. 1999. Cross-regulation of beta-catenin-LEF/TCF and retinoid signaling pathways. Curr. Biol. 9:1415-1418. [DOI] [PubMed] [Google Scholar]
- 5.Erickson, R. L., N. Hemati, S. E. Ross, and O. A. MacDougald. 2001. p300 coactivates the adipogenic transcription factor C/EBPα. J. Biol. Chem. 276:16348-16355. [DOI] [PubMed] [Google Scholar]
- 6.Erickson, R. L., K. A. Longo, S. E. Ross, N. Hemati, and O. A. MacDougald. 2000. Structure and function of C/EBPα, p. 79-90. In J. M. Ntambi (ed.), Adipocyte biology and hormone signaling. IOS Press, Amsterdam, The Netherlands.
- 7.Graham, T. A., C. Weaver, F. Mao, D. Kimelman, and W. Xu. 2000. Crystal structure of a β-catenin/Tcf complex. Cell 103:885-896. [DOI] [PubMed] [Google Scholar]
- 8.Hammer, G. D., and H. A. Ingraham. 1999. Steroidogenic factor-1: its role in endocrine organ development and differentiation. Front. Neuroendocrinol. 20:199-223. [DOI] [PubMed] [Google Scholar]
- 9.Hartman, H. B., X. Hu, K. X. Tyler, C. K. Dalal, and M. A. Lazar. 2002. Mechanisms regulating adipocyte expression of resistin. J. Biol. Chem. 277:19754-19761. [DOI] [PubMed] [Google Scholar]
- 10.Hecht, A., K. Vleminckx, M. P. Stemmler, F. van Roy, and R. Kemler. 2000. The p300/CBP acetyltransferases function as transcriptional coactivators of beta-catenin in vertebrates. EMBO J. 19:1839-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hemati, N., S. E. Ross, R. L. Erickson, G. E. Groblewski, and O. A. MacDougald. 1997. Signaling pathways through which insulin regulates CCAAT/enhancer binding protein alpha (C/EBPα) phosphorylation and gene expression in 3T3-L1 adipocytes. Correlation with GLUT4 gene expression. J. Biol. Chem. 272:25913-25919. [DOI] [PubMed] [Google Scholar]
- 12.Hollenberg, A. N., V. S. Susulic, J. P. Madura, B. Zhang, D. E. Moller, P. Tontonoz, P. Sarraf, B. M. Spiegelman, and B. B. Lowell. 1997. Functional antagonism between CCAAT/enhancer binding protein-α and peroxisome proliferator-activated receptor-gamma on the leptin promoter. J. Biol. Chem. 272:5283-5290. [DOI] [PubMed] [Google Scholar]
- 13.Hovanes, K., T. W. Li, J. E. Munguia, T. Truong, T. Milovanovic, J. Lawrence Marsh, R. F. Holcombe, and M. L. Waterman. 2001. Beta-catenin-sensitive isoforms of lymphoid enhancer factor-1 are selectively expressed in colon cancer. Nat. Genet. 28:53-57. [DOI] [PubMed] [Google Scholar]
- 14.Huelsken, J., and W. Birchmeier. 2001. New aspects of Wnt signaling pathways in higher vertebrates. Curr. Opin. Genet. Dev. 11:547-553. [DOI] [PubMed] [Google Scholar]
- 15.Hwang, C. S., S. Mandrup, O. A. MacDougald, D. E. Geiman, and M. D. Lane. 1996. Transcriptional activation of the mouse obese (ob) gene by CCAAT/enhancer binding protein alpha. Proc. Natl. Acad. Sci. USA 93:873-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ito, M., Y. Park, J. Weck, K. E. Mayo, and J. L. Jameson. 2000. Synergistic activation of the inhibin alpha-promoter by steroidogenic factor-1 and cyclic adenosine 3′,5′-monophosphate. Mol. Endocrinol. 14:66-81. [DOI] [PubMed] [Google Scholar]
- 17.Kadoya, T., Y. H. Suzuki, T., Yukita, A. Fukui, A. Michiue, T. Asahara, T. Tanaka, K. Asashima, and M. Kikuchi. 2002. Desumoylation activity of axam, a novel axin-binding protein, is involved in downregulation of β-catenin. Mol. Cell. Biol. 22:3803-3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobielak, A., K. Kobielak, and W. H. Trzeciak. 2001. A novel isoform of human lymphoid enhancer-binding factor-1 (LEF-1) gene transcript encodes a protein devoid of HMG domain and nuclear localization signal. Acta Biochim. Pol. 48:221-226. [PubMed] [Google Scholar]
- 19.Kuhl, M., L. C. Sheldahl, M. Park, J. R. Miller, and R. T. Moon. 2000. The Wnt/Ca2+ pathway: a new vertebrate Wnt signaling pathway takes shape. Trends Genet. 16:279-283. [DOI] [PubMed] [Google Scholar]
- 20.Longo, K. A., J. A. Kennell, M. J. Ochocinska, S. E. Ross, W. S. Wright, and O. A. MacDougald. 2002. Wnt signaling protects 3T3-L1 preadipocytes from apoptosis through induction of insulin-like growth factors. J. Biol. Chem. 1:1.. [DOI] [PubMed] [Google Scholar]
- 21.MacDougald, O. A., and M. D. Lane. 1995. Transcriptional regulation of gene expression during adipocyte differentiation. Annu. Rev. Biochem. 64:345-373. [DOI] [PubMed] [Google Scholar]
- 22.MacDougald, O. A., and S. Mandrup. 2002. Adipogenesis: forces that tip the scales. Trends Endocrinol. Metab. 13:5-11. [DOI] [PubMed] [Google Scholar]
- 23.Matzuk, M. M., M. J. Finegold, J. P. Mather, L. Krummen, H. Lu, and A. Bradley. 1994. Development of cancer cachexia-like syndrome and adrenal tumors in inhibin-deficient mice. Proc. Natl. Acad. Sci. USA 91:8817-8821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miyagishi, M., F. R. Hatta, M. Yoshida, E. Araya, N. Nagafuchi, A. Ishihara, S. Nakajima, and T. Fukamizu. 2000. Regulation of Lef-mediated transcription and p53-dependent pathway by associating β-catenin with CBP/p300. J. Biol. Chem. 275:35170-35175. [DOI] [PubMed] [Google Scholar]
- 25.Mizusaki, H., K. Kawabe, T. Mukai, E. Ariyoshi, M. Kasahara, H. Yoshioka, A. Swain, and K. I. Morohashi. 2003. Dax-1 gene transcription is regulated by Wnt4 in the female developing gonad. Mol. Endocrinol. 17:507-519. [DOI] [PubMed]
- 26.Morrison, R. F., and S. R. Farmer. 2000. Hormonal signaling and transcriptional control of adipocyte differentiation. J. Nutr. 130:3116S-3121S. [DOI] [PubMed]
- 27.Pedersen, T. A., E. Kowenz-Leutz, A. Leutz, and C. Nerlov. 2001. Cooperation between C/EBPα TATA binding protein/TFIIB and SWI/SNF recruiting domains is required for adipocyte differentiation. Genes Dev. 15:3208-3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Piedra, J., M. D. Castano, J. Miravet, S. Dunach, and M. de Herreros. 2001. Regulation of β-catenin structure and activity by tyrosine phosphorylation. J. Biol. Chem. 276:20436-20442. [DOI] [PubMed] [Google Scholar]
- 29.Polakis, P. 2000. Wnt signaling and cancer. Genes Dev. 14:1837-1851. [PubMed] [Google Scholar]
- 30.Poy, F., M. Lepourcelet, R. A. Shivdasani, and M. J. Eck. 2001. Structure of a human Tcf-4-β-catenin complex. Nature 8:1053-1057. [DOI] [PubMed] [Google Scholar]
- 31.Preece, A. 1972. A manual for histologic technicians. Little Brown, Boston, Mass.
- 32.Rangwala, S. M., and M. A. Lazar. 2000. Transcriptional control of adipogenesis. Annu. Rev. Nutr. 20:535-559. [DOI] [PubMed] [Google Scholar]
- 33.Roose, J., G. Huls, M. van Beest, P. Moerer, K. van der Horn, R. Goldschmeding, T. Logtenberg, and H. Clevers. 1999. Synergy between tumor suppressor APC and the β-catenin-Tcf4 target Tcf1. Science 285:1923-1926. [DOI] [PubMed] [Google Scholar]
- 34.Rosen, E. D., and B. M. Spiegelman. 2000. Molecular regulation of adipogenesis. Annu. Rev. Cell. Dev. Biol. 16:145-171. [DOI] [PubMed] [Google Scholar]
- 35.Ross, S. E., R. L. Erickson, I. Gerin, P. M. DeRose, L. Bajnok, K. A. Longo, D. E. Misek, R. Kuick, S. M. Hanash, K. B. Atkins, S. M. Andresen, H. I. Nebb, L. Madsen, K. Kristiansen, and O. A. MacDougald. 2002. Microarray analyses during adipogenesis: understanding the effects of Wnt signaling on adipogenesis and the roles of liver X receptor α in adipocyte metabolism. Mol. Cell. Biol. 22:5989-5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ross, S. E., R. L. Erickson, N. Hemati, and O. A. MacDougald. 1999. Glycogen synthase kinase 3 is an insulin-regulated C/EBPα kinase. Mol. Cell. Biol. 19:8433-8441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ross, S. E., N. Hemati, K. A. Longo, C. N. Bennett, P. C. Lucas, R. L. Erickson, and O. A. MacDougald. 2000. Inhibition of adipogenesis by Wnt signaling. Science 289:950-953. [DOI] [PubMed] [Google Scholar]
- 38.Schaufele, F., J. F. Enwright 3rd, X. Wang, C. Teoh, R. Srihari, R. Erickson, O. A. MacDougald, and R. N. Day. 2001. CCAAT/enhancer binding protein alpha assembles essential cooperating factors in common subnuclear domains. Mol. Endocrinol. 15:1665-1676. [DOI] [PubMed] [Google Scholar]
- 39.Sharma, M., W. W. Chuang, and Z. Sun. 2002. Phosphatidylinositol 3-kinase/Akt stimulates androgen pathway through glycogen synthase kinase 3β inhibition and nuclear β-catenin accumulation. J. Biol. Chem. 277:30935-30941. [DOI] [PubMed]
- 40.Shtutman, M., J. Zhurinsky, I. Simcha, C. Albanese, M. D'Amico, R. Pestell, and A. Ben-Ze'ev. 1999. The cyclin D1 gene is a target of the β-catenin/LEF-1 pathway. Proc. Natl. Acad. Sci. USA 96:5522-5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shulman, J. M., N. Perrimon, and J. D. Axelrod. 1998. Frizzled signaling and the developmental control of cell polarity. Trends Genet. 14:452-458. [DOI] [PubMed] [Google Scholar]
- 42.Student, A. K., R. Y. Hsu, and M. D. Lane. 1980. Induction of fatty acid synthetase synthesis in differentiating 3T3-L1 preadipocytes. J. Biol. Chem. 255:4745-4750. [PubMed] [Google Scholar]
- 43.Sun, Y., F. T. Kolligs, M. O. Hottiger, R. Mosavin, E. R. Fearon, and G. J. Nabel. 2000. Regulation of beta-catenin transformation by the p300 transcriptional coactivator. Proc. Natl. Acad. Sci. USA 97:12613-12618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tetsu, O., and F. McCormick. 1999. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature 398:422-426. [DOI] [PubMed] [Google Scholar]
- 45.van de Wetering, M., W. de Lau, and H. Clevers. 2002. WNT signaling and lymphocyte development. Cell 109(Suppl.):S13-S19. [DOI] [PubMed] [Google Scholar]
- 46.van Noort, M., and H. Clevers. 2002. TCF transcription factors, mediators of Wnt-signaling in development and cancer. Dev. Biol. 244:1-8. [DOI] [PubMed] [Google Scholar]
- 47.Wolf, D., M. Rodova, E. A. Miska, J. P. Calvet, and T. Kouzarides. 2002. Acetylation of beta-catenin by CREB-binding protein (CBP). J. Biol. Chem. 277:25562-25567. [DOI] [PubMed] [Google Scholar]
- 48.Yang, F., X. Li, M. Sharma, C. Y. Sasaki, D. L. Longo, B. Lim, and Z. Sun. 2002. Linking beta-catenin to androgen-signaling pathway. J. Biol. Chem. 277:11336-11344. [DOI] [PubMed] [Google Scholar]