Abstract
Rgt1 is a glucose-responsive transcription factor that binds to the promoters of several HXT genes encoding glucose transporters in Saccharomyces cerevisiae and regulates their expression in response to glucose. Rgt1 contains a Zn2Cys6 binuclear cluster responsible for DNA binding. Most proteins that contain this sequence motif bind as dimers to regularly spaced pairs of the sequence CGG. However, there are no CGG pairs with regular spacing in promoters of genes regulated by Rgt1, suggesting that Rgt1 binds as a monomer to CGG or to another sequence. We identified the Rgt1 consensus binding site sequence 5′-CGGANNA-3′, multiple copies of which are present in all HXT promoters regulated by Rgt1. Rgt1 binds in vivo to multiple sites in the HXT3 promoter in a nonadditive, synergistic manner, leading to synergistic repression of HXT3 transcription. We show that glucose inhibits the DNA-binding ability of Rgt1, thereby relieving repression of HXT gene expression. This regulation of Rgt1 DNA-binding activity is caused by its glucose-induced phosphorylation: the hyperphosphorylated Rgt1 present in cells growing on high levels of glucose does not bind DNA in vivo or in vitro; dephosphorylation of this form of Rgt1 in vitro restores its DNA-binding ability. Furthermore, an altered Rgt1 that functions as a constitutive repressor remains hypophosphorylated when glucose is added to cells and binds DNA under these conditions. These results suggest that glucose regulates the DNA-binding ability of Rgt1 by inducing its phosphorylation.
The yeast Saccharomyces cerevisiae carefully regulates the utilization of glucose, its favorite carbon source. The first step of glucose utilization—its transport across the cell membrane—is tightly regulated: the expression of many HXT genes that encode glucose transporters is induced by glucose (27). Rgt1 is a transcription factor responsible for regulating expression of the HXT genes in response to glucose (26). In the absence of glucose, Rgt1 binds to HXT promoters and recruits the transcriptional corepressor complex Ssn6-Tup1, which establishes a repressive chromatin domain (9) and/or directly inhibits RNA polymerase II function by interacting with the mediator complex (36). A glucose signal generated by the Snf3 and Rgt2 glucose sensors is transmitted to Rgt1 through the SCFGrr1 ubiquitin-conjugating enzyme, inhibiting transcriptional repression by Rgt1 and leading to derepression of HXT gene expression (27). How this glucose signal alters Rgt1 function is not known. Here, we present evidence that glucose regulates the DNA-binding ability of Rgt1 by inducing its phosphorylation.
The DNA-binding domain of Rgt1 consists of a Zn2Cys6 zinc cluster, a DNA-binding domain found in several fungal proteins, such as Gal4, Leu3, and Hap1 (1, 29, 33). Most proteins that contain this sequence motif bind as dimers to two 5′-CGG-3′ sequences, with each monomer contacting one CGG sequence. The binding sites of the different zinc cluster-containing proteins differ in the orientation of the CGG sequences and in the number of nucleotides separating them. For example, in the binding sites of Gal4, Ppr1, and Put3, the pair of CGG sequences is inverted (5′-CGG-CCG-3′) and separated by 11, 6, and 10 bp, respectively (21, 22, 31). Leu3 binds to an everted pair of CGG sequences (5′-CCG-CGG-3′) that are separated by 4 bp (15). Each of these proteins homodimerizes via an interaction of a leucine zipper-like coiled-coil domain located C-terminal to the zinc cluster. The length of the linker sequence that connects the zinc cluster and the coiled-coil motif dictates the orientation and separation of the CGG sequences recognized by a zinc cluster protein. The linker of Gal4 has an extended conformation to accommodate the 11 bp between CGG sequences; Ppr1 has a more compact linker that causes it to bind to CGG sequences separated by only 6 bp (21, 22). Thus, dimerization is a major determinant for sequence-specific binding of this class of proteins, though some zinc cluster proteins act as monomers (e.g., AlcR of Aspergillus nidulans [10]); others are part of heterodimers (e.g., Oaf1/Pip2 [3]).
Rgt1 appears to be devoid of a coiled-coil dimerization domain, and the promoters of genes regulated by Rgt1 contain many CGG sequences, but none appear to be paired. These observations suggest that Rgt1 binds to DNA as a monomer (26). We sought to define the Rgt1 binding site with the hope of learning how Rgt1 specifically recognizes HXT gene promoters. We found that Rgt1 binds to a surprisingly simple sequence but requires several such sequences to regulate gene expression efficiently.
MATERIALS AND METHODS
Production of Rgt1 protein for DNA-binding assays.
The 564-bp RGT1 DNA fragment encoding the first 188 amino acids, encompassing the Zn2Cys6 DNA-binding domain, was PCR amplified and fused to the glutathione-S-transferase (GST) gene in the pGEX-5x-3 plasmid (Pharmacia) to yield pBM4338. The primers used for the PCR are OM2415 (5′-TCGTGGGATCCCCAACGAGCTGAACACTGTTTCGACTAACTCCAG-3′) and OM2417 (5′-ACGAGTCGACCCAAAACTGCTGTTGGCCAATATTACCCGCGGGC-3′), which were flanked by BamHI and SalI sites as indicated by underlining. GST-Rgt1 was expressed from the T7 promoter in Escherichia coli BL21(DE3), and the fusion proteins were purified with glutathione agarose (Sigma) according to instructions from the manufacturer. To eliminate the possible bias of dimerization of the GST moiety (34), the Rgt1 fragment was liberated by treatment with factor Xa (Pierce).
Footprinting analysis and EMSA.
Probe DNA (200 to 250 bp) was PCR amplified using sequences upstream of HXT1, HXT2, and HXT3 as templates (the sequences of the primer sets for PCR are available upon request). For end labeling of the PCR fragments, they were cut with EcoRI or HindIII (sites incorporated in the forward and reverse primers, respectively), treated with α-32P (Amersham) and the Klenow fragment of DNA polymerase (Promega), and gel purified. For the DNA-binding assay, Rgt1 was incubated with 32P-labeled DNA fragments (2 × 104 cpm) in 20 μl of TGZD buffer (20 mM Tris-HCl, pH. 8.0, 75 mM KCl, 10 μM ZnCl2, 5% glycerol) containing 1 μg of poly(dI-dC) · poly(dI-dC) at room temperature for 20 min. For the electrophoretic mobility shift assay (EMSA), the reaction mixtures were directly loaded in 5% nondenaturing polyacrylamide gels. For DNase I or dimethylsulfate (DMS) protection experiments, Rgt1-DNA complexes were incubated with 2 U of DNase I (Promega) at 25°C for 2 min or with 0.5% (vol/vol; final concentration) DMS (Sigma) at 20°C for 1 min, respectively. DNase I digests were extracted with phenol and precipitated with ethanol. The methylated bases were cleaved by boiling DNA in a solution of 10% (vol/vol) piperidine and 10 mM EDTA at 90°C for 30 min. For UV photofootprinting, performed as described previously (2), the reaction mixtures were irradiated with six 254-nm-wavelength germicidal UV lamps at a distance of 22 cm for 30 s to 2 min. The DNA was precipitated with ethanol and then incubated with 20 U of T4 endonuclease V (Epicentre) at 37°C for 1 h. DNA samples from all footprinting reactions were resolved by electrophoresis through 8% polyacrylamide gels containing 8 M urea.
Cell growth and assaying function of Rgt1 binding sites.
To monitor the behavior of Rgt1 under repressing and inducing conditions, cells expressing Rgt1 were grown to mid-log phase (optical density at 600 nm, 1.0) in minimal medium containing 2% galactose and 0.05% glucose. The cells were washed with water, divided into three fractions, and further incubated in minimal medium containing 2% galactose, 2% raffinose (equivalent to ∼0.2% glucose), or 4% glucose for 1.5 h.
Gene expression reporter plasmids containing Rgt1 binding sites were prepared by cloning various Rgt1 sites between the LEU2 upstream activation sequence and the TATA box of HIS3 fused to lacZ (pBM2832). One or two Rgt1 binding sites were prepared by annealing complementary oligonucleotides containing Rgt1 sites; three or more sites were prepared by PCR, in which both oligonucleotides and PCR primers were flanked with BamHI and PstI sites (Table 1). The annealed or PCR products were inserted into the same restriction sites upstream of lacZ in pBM2832. Mutations in Rgt1 binding sites were generated by recombinational gap repair. Briefly, two PCR fragments were generated, the first with a mutation in the Rgt1 binding site (CGG to TTT or CTT) and sequences identical to the vector at the 5′ end (the BamHI side of pBM2832) and the second with sequences identical to the vector at the 3′ end (the PstI side of pBM2832) and identical to the first PCR fragment at the 5′ end. The two PCR products were cotransformed with pBM2832 cut with BamHI and PstI into yeast YM4127 to select for repair of the gapped vector. Wild-type (YM4127) and Δrgt1 (YM4509) yeasts were transformed with these plasmids, and β-galactosidase levels were determined in three independent transformants as previously described (35).
TABLE 1.
Plasmids used in this study
| Plasmid | Rgt1 site | Cloning method | Primersa |
|---|---|---|---|
| pBM4305 | A | Annealing | OM2919 and OM2920 |
| pBM4306 | B | Annealing | OM2921 and OM2922 |
| pBM4307 | C | Annealing | OM2927 and OM2928 |
| pBM4308 | D | Annealing | OM2957 and OM2958 |
| pBM4309 | A to B | Annealing | OM2923 and OM2924 |
| pBM4310 | C to D | Annealing | OM2959 and OM2960 |
| pBM4314 | E | Annealing | OM2971 and OM2972 |
| pBM4315 | G | Annealing | OM2977 and OM2978 |
| pBM4316 | B to C | PCR | OM2742 and OM2745 |
| pBM4317 | A to D | PCR | OM2741 and OM2744 |
| pBM4318 | A to G | PCR | OM3014 and OM3015 |
| pBM4319 | D to G | PCR | OM3013 and OM3014 |
| pBM4320 | F to G | Annealing | OM3019 and OM3020 |
| pBM4321 | F | Annealing | OM3017 and OM3018 |
| pBM4322 | A to C | PCR | OM3015 and OM2745 |
| pBM4323 | B to D | PCR | OM2742 and OM2744 |
| pBM4324 | D to E | Annealing | OM3030 and OM3031 |
| pBM4325 | Clusters I + II | PCR | OM3014 and OM3027 |
| pBM4326 | Cluster II | PCR | OM3027 and OM3028 |
| pBM4327 | C to E | PCR | OM3032 and OM2743 |
| pBM4328 | D to F | PCR | OM3033 and OM3035 |
| pBM4329 | E to G | PCR | OM3034 and OM3014 |
| pBM4330 | ΔB in A-D | PCR | OM2407-OM2412 and OM2408-OM2409 |
| pBM4331 | ΔC2 in A-D | PCR | OM2410-OM2412 and OM2411-OM2409 |
| pBM4332 | ΔE in D-G | PCR | OM3064-OM2412 and OM3063-OM2409 |
| pBM4333 | ΔF in D-G | PCR | OM2647-OM2412 and OM2646-OM2409 |
| pBM4335 | 4 Rgt1 sites | Annealing | OM3325 and OM3326 |
| pBM4337 | HXT3 promoter | PCR | OM2436 and OM3014 |
Sequences of primers are available upon request.
Immunoprecipitation.
Extracts of yeast cells expressing the LexA-Rgt1 fusion protein, expressed under the ADH1 promoter (in pBM3307), were prepared by vortexing cells with acid-washed glass beads (0.5-mm diameter) in NP-40 lysis buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% NP-40) containing phosphatase inhibitors (10 mM Na-pyrophosphate, 200 μM Na-orthovanadate, 50 mM Na-flouride) at 4°C for 10 min. The cell lysates (2 mg) were incubated with anti-LexA mouse monoclonal antibody (Santa Cruz) at 4°C for 3 h and further incubated with protein G-conjugated agarose beads (Santa Cruz) for 1 h. After the beads were washed with NP-40 lysis buffer containing 1 M NaCl, proteins were eluted by boiling the beads in sodium dodecyl sulfate (SDS) sample buffer for 5 min and were resolved in SDS-polyacrylamide gels. Western blot analysis was carried out as described previously (8) using LexA antibody (Santa Cruz) and an enhanced chemiluminescence system (Pierce). Immunoprecipitated Rgt1 was dephosphorylated by incubation with 10 U of calf intestinal alkaline phosphatase (CIP) (Roche) at 37°C for 30 min.
Production of Rgt1 antibody.
A peptide with the last 25 amino acids of Rgt1 (N-LENVALENFVSIGWKLLDDSELGWY-C) was coupled with maleimide-activated keyhole limpet hemocyanin (Pierce) and injected by Babco into rabbits three times with 1-month intervening periods. Polyclonal anti-Rgt1 antibody was purified using protein A beads from total antiserum (14).
ChIP.
Chromatin immunoprecipitation (ChIP) assays were carried out essentially as described previously (30). Briefly, yeast cells grown to mid-log phase were incubated with formaldehyde (1% final concentration) for 15 to 20 min at room temperature, after which glycine was added to a final concentration of 125 mM to quench the cross-linking reaction for 5 min. The cells were disrupted by vortexing them with glass beads in lysis buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 1% Triton X-100, 0.1% Na-deoxycholate). The lysate was sonicated (ultrasonic cell disruptor with a microtip) five times with 10-s pulses. The genomic DNA fragments, which averaged 200 to 500 bp in length, were immunoprecipitated with rabbit polyclonal anti-Rgt1 antibody prepared as described above. The immunoprecipitated DNA and 1/100 of the input DNA were used as templates in a 25-cycle PCR. For quantitative analysis, PCR was conducted with reaction mixtures containing 0.5 μCi of [α-32P]dATP. The PCR products were resolved in 6% polyacrylamide gels, and autoradiograms were scanned and quantified with ImageQuaNT software (Molecular Dynamics). The sequences of primer pairs used for ChIP of the HXT gene promoters were as follows: HXT1, 5′-ATATAATTCCCCCCTCCTGAAG-3′ (OM3109) and 5′-TGATTCTACGTTTTTGCAAGC-3′ (OM3111); HXT2, 5′-GAGAGGAAAAACCAAAATATA-3′ (OM3197) and 5′-ATCTAGTTCCCGAAGAAAACGCG-3′ (OM3198); and HXT3, 5′-CGGATACCTACTATTATCCCC-3′ (OM2642) and 5′-AAAATTCTCCACGAAGCTTTC-3′ (OM2643). The sequences of another set of PCR primers for ChIP (see Fig. 5) were 5′-CCGCCTCTCCCCGCGCGTTGGCCGATTCATTCCC-3′ (OM3128) and 5′-ATCACATTACTTTATATAATGTATAATTCATTA-3′ (OM3129).
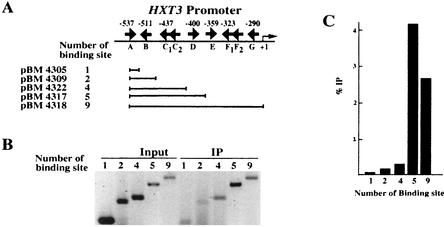
FIG. 5.
Rgt1 is synergistically recruited to multiple binding sites in vivo. (A) The same plasmids used for the repression assay shown in Fig. 4 were used for a ChIP assay. The arrows indicate the locations and orientations of Rgt1 binding sites. (B) Chromatins prepared from cells (YM4127) containing the reporter plasmids grown under repressing conditions were immunoprecipitated with anti-Rgt1 antibody. Rgt1 binding sites in the immunoprecipitated DNA (IP) were PCR amplified using a primer set (OM3128 and OM3129) containing [α-32P]dATP and analyzed in a 6% polyacrylamide gel. (C) The amounts of the input DNA and immunoprecipitated DNA were measured by a PhosphorImager, quantified by the ImageQuaNT program, and presented as the ratio of immunoprecipitated counts to input counts. Fig. 8C, box, shows the control of ChIP using anti-Rgt1 antibody.
IDBA.
Rgt1 proteins exist in very small amounts in a cell and are very difficult to purify, although they are overexpressed under the control of a strong promoter, thereby limiting any thorough in vitro investigations due to insufficient proteins. To overcome this problem, we have described a novel method, the immobilized-DNA-binding assay (IDBA), to assay DNA binding in vitro easily and rapidly with full-length proteins purified directly from yeast cell extract.
The DNA-binding activity of immunoprecipitated full-length LexA-Rgt1 was measured in vitro by incubating it with a DNA fragment (60 bp) containing four Rgt1 binding sites (Table 1), followed by elution and quantification of the bound DNA. LexA-Rgt1 was immunoprecipitated from crude yeast cell extracts with anti-LexA antisera as described above and washed with NP-40 lysis buffer containing 1 M NaCl. The beads decorated with LexA-Rgt1 were resuspended in 30 μl of TGZD buffer (75 mM KCl) containing 32P-labeled DNA fragments (2 × 104 cpm; prepared by annealing oligonucleotide OM3325 with OM3326, followed by end labeling with γ-32P) and 1 μg of poly(dI-dC) · poly(dI-dC). After incubation at room temperature for 30 min, the beads were washed twice with TGZD buffer containing 0.25 M NaCl. The DNA bound to Rgt1 was eluted with Tris-EDTA buffer containing 1 M NaCl, followed by elution of LexA-Rgt1 by boiling the beads in SDS sample buffer at 90°C for 5 min. The eluted DNA and LexA-Rgt1 were resolved by electrophoresis through polyacrylamide gels and visualized by autoradiography and Western blotting, respectively.
Imaging of GFP-Rgt1.
The green fluorescent protein (GFP)-Rgt1 fusion plasmid (pBM3911) was constructed by inserting the Rgt1 fragment from pBM3183 into the EcoRI and SalI sites of pUG36 (25). Cells containing the GFP-Rgt1 fusion plasmid were grown under the same conditions as for the β-galactosidase assay described above. The cells were stained with DAPI (4,6-diamidino-2-phenylindole) and visualized on a Bmax-60F microscope (Olympus, Lake Success, N.Y.) with the 41014 filter set (for detecting GFP) (Chroma Technology, Brattleboro, Vt.) as described previously (8).
RESULTS
Analysis of Rgt1 binding sites in the HXT promoters.
To identify the sequences to which Rgt1 binds, we performed DNase I and DMS footprinting assays on the promoters of HXT1, HXT2, and HXT3 (data not shown). We mapped 31 Rgt1 binding sites in ∼3.5 kb of HXT promoters (Fig. 1A). Nearly all of the binding sites include the sequence 5′-CGGANNA-3′, which we consider to be the consensus Rgt1 binding site; the few remaining sites have 5′-AGG-3′ or 5′-TGG-3′ instead of 5′-CGG-3′ (Fig. 1B). Each HXT promoter has multiple Rgt1 sites (Fig. 1A): the HXT3 promoter contains 15 sites, 9 of them between −560 and −280 (relative to the first ATG codon), with 20 to 70 bp separating the sites; the HXT2 promoter contains only 3 Rgt1 binding sites; 13 Rgt1 binding sites in the HXT1 promoter are arrayed in several clusters. The different architectures of the clusters of Rgt1 binding sites in the HXT promoters raise the possibility that Rgt1 might deploy its DNA-binding domain differently in the different HXT promoters.
FIG. 1.
Identification of Rgt1 binding sites upstream of HXT genes. (A) Diagram of arrangement of sequences upstream of HXT3, HXT1, and HXT2 that are protected from DNase I digestion and DMS methylation. Arrows, locations and orientations of Rgt1 binding sites. (B) Alignment of the sequences footprinted by Rgt1. The number of bases from the central G residue of the 5′-CGG-3′ triplet to the ATG codon are in parentheses. The sequence 5′-CGGANNA-3′ is the Rgt1 consensus binding site.
Probing interactions between Rgt1 and its binding sites in vitro.
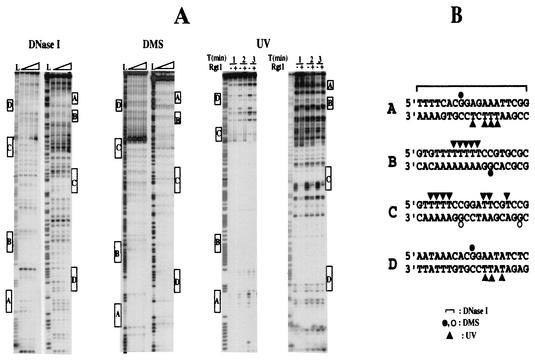
To probe the contacts Rgt1 makes with its binding site, three different probes—DNase I, DMS, and UV—were used in footprinting assays (Fig. 2A). DNase I footprinting indicates that Rgt1 covers 15 to 18 bp around the Rgt1 consensus sequence. Rgt1 protects from methylation by DMS only the central G residue of its binding site. Rgt1 makes an intense photofootprint on the T residues (underlined) in each binding site on the strand opposite the CGG sequence (5′-TTTTCCG-3′) (Fig. 2B). The strong DNase I-hypersensitive site in the C2 binding site may be due to Rgt1-induced DNA bending, since poly(A) tracts can cause DNA bending (23) and some Zn cluster proteins significantly bend DNA (16).
FIG. 2.
Probing the interaction between Rgt1 and its binding sites. (A) Footprinting assays using DNase I, DMS, and UV as probes were carried out with a DNA fragment of the HXT3 promoter containing five Rgt1 binding sites (−372 to −572 [Fig. 3A]) as described in Materials and Methods. For DNase I and DMS protections, a 32P-labeled DNA fragment (2 × 104 cpm) was incubated with different amounts of Rgt1 (30, 60, and 120 ng, from the third gel lane from the left). The first (L) and second gel lanes are the A+G ladder and the control without Rgt1, respectively. For UV photofootprinting, Rgt1 (100 ng) was incubated (+) with the same amounts of DNA used for DNase I and DMS protections, and then DNA-protein complexes were irradiated with UV for the time indicated above each lane. The Rgt1 binding sites are indicated by boxes. (B) Summary of interactions between Rgt1 and its binding sites observed from three different footprinting assays. DMS protection assays show that Rgt1 weakly contacts CGG at C1 and C2 (open circles).
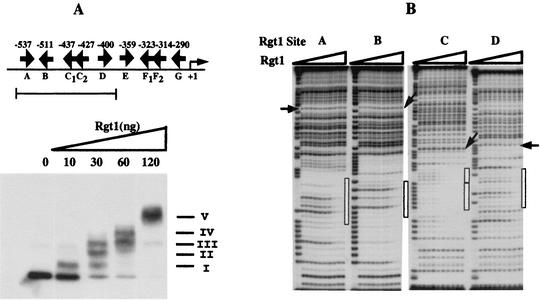
An EMSA using as a probe a radiolabeled DNA fragment of the HXT3 promoter containing five Rgt1 binding sites (site A to site D) revealed five discrete Rgt1-DNA complexes (Fig. 3A). Significantly, there is no rapid transition from free to fully bound DNA. The dissociation constants (Kd) of Rgt1 for individual binding sites appear not to be significantly different and are relatively high (∼10−7 M, assuming all of the protein is active for DNA binding) (Fig. 3B). These results suggest that the Rgt1 DNA-binding domain does not bind to DNA cooperatively in vitro.
FIG. 3.
Binding of Rgt1 to the HXT3 promoter. (A) EMSA was carried out with a DNA fragment containing five Rgt1 binding sites. A constant amount of 32P-labeled DNA fragment (3 × 104 cpm) was incubated with increasing amounts of Rgt1 as indicated above the gel lanes. Arrows, locations and orientations of Rgt1 binding sites. Roman numerals denote Rgt1-DNA complexes. (B) The five Rgt1 sites were separated and used as probes for DNase I footprinting assays. After incubation of a 32P-labeled DNA fragment (2 × 105 cpm) with increasing amounts of Rgt1 (1, 3, 6, 10, 30, 60, 100, 300, and 600 ng per lane, from the third gel lane from the left) for 20 min, the DNA-protein complexes were digested with 2 U of DNase I for 1 min. The first and second gel lanes are the A+G ladder and the control (without Rgt1), respectively. Site C consists of two Rgt1 binding sites, C1 and C2, 3 bp apart. The binding affinities of Rgt1 were estimated by comparing the intensities (measured on a PhosphorImager by the ImageQuaNT program) of a reference band (indicated by arrows) and a band within the Rgt1 footprint (indicated by boxes).
Synergistic transcription repression by Rgt1 at multiple binding sites.
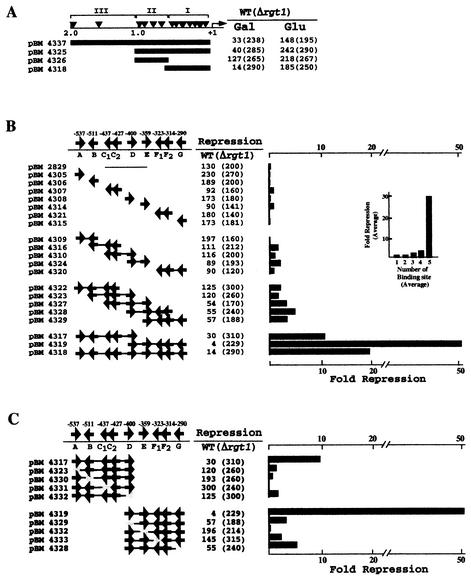
To determine the functionality in vivo of the Rgt1 binding sites identified by footprinting, the upstream region of HXT3 was divided into three sections (clusters I, II, and III) and inserted into the promoter of the CYC1-lacZ reporter gene. Clusters I and II (pBM4325) direct sevenfold repression of β-galactosidase expression, the same as for the whole 2-kb upstream region (pBM4337) (Fig. 4A). Cluster I (pBM4318) directs 20-fold repression; only 2-fold repression is effected by cluster II (pBM4326).
FIG. 4.
Multiple Rgt1 binding sites mediate synergistic repression. (A) Three different segments of the HXT3 promoter were fused to a HIS3-lacZ reporter gene (pBM2832) and introduced into yeast wild-type (WT) (YM4127) and Δrgt1 (YM4509) strains. Expression of the reporter gene was measured in cells grown on minimal medium containing 2% galactose (Gal) to mid-log phase and then switched to glucose (4%) (Glu) for 1.5 h. (B) Single or multiple copies of Rgt1 sites in cluster I were inserted in the reporter plasmid (pBM2832) assayed for the function of Rgt1 binding sites as described for panel A. These Rgt1 binding sites were generated by annealing complementary oligonucleotides containing one or two Rgt1 sites or by amplifying more than three Rgt1 sites. (C) Mutations in Rgt1 binding sites were generated by recombinational gap repair. Repression is the ratio of β-galactosidase activity in the WT (YM4127) to that in Δrgt1 (YM4509). Arrows (B and C), locations and orientations of Rgt1 binding sites.
Since Rgt1 binding sites in a promoter are not functionally equivalent (Fig. 4A), we wondered how many sites are required for Rgt1-mediated transcriptional repression and if they operate in an additive or cooperative fashion. Different numbers of Rgt1 sites in cluster I were inserted into the promoter of the HIS3-lacZ reporter (pBM2832). One Rgt1 binding site is insufficient for transcriptional repression; two, three, or four sites cause two- to fivefold repression (Fig. 4B). However, five Rgt1 binding sites (pBM4317 and pBM4319) provide significantly more repression: 10- and 57-fold, respectively. Inactivation of any one of the five Rgt1 binding sites reduces repression to two- to threefold (Fig. 4C). Thus, multiple Rgt1 binding sites (five in this case) are necessary for a significant amount of repression. This is reminiscent of AlcR, which binds to single sites in vitro but functions through multiple sites in vivo (28).
Rgt1 binds its target sites cooperatively in vivo.
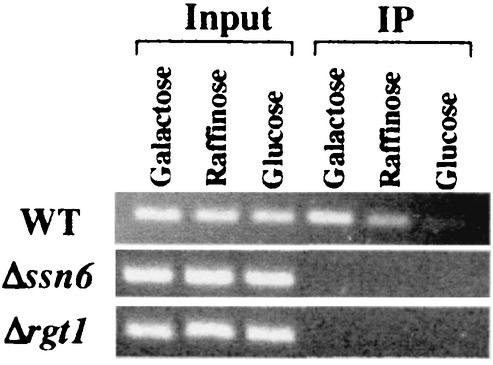
Although Rgt1 is able to bind to a single binding site in vitro (Fig. 3B), a single site does not cause transcriptional repression in vivo (Fig. 4B). Therefore, Rgt1 was tested for its DNA binding in vivo using a ChIP assay (Fig. 5). Rgt1 binding to promoters containing one (pBM4305) or two (pBM4309) binding sites is barely detectable, but Rgt1 binding to promoters with five binding sites (pBM4317) is obvious: they are ∼20- and 40-fold more abundant in Rgt1 immunoprecipitates than promoters with two or one Rgt1 binding site, respectively. This result suggests that Rgt1 binds synergistically to multiple binding sites in vitro, congruent with in vivo results (Fig. 4). The Ssn6-Tup1 corepressor may play a role in this synergistic binding of Rgt1, because anti-Ssn6 antisera can be used in a ChIP experiment to detect Rgt1 DNA binding to HXT promoters (Fig. 6).
FIG. 6.
Rgt1 binds with Ssn6-Tup1 on an HXT promoter. Chromatin prepared from yeast cells (WT, YM4127; Δssn6, YM4554; Δrgt1, YM4509) grown on YP containing different carbon sources as indicated above each lane was immunoprecipitated using anti-Ssn6 antibody (Santa Cruz). The HXT3 promoter in the immunoprecipitated DNA (IP) was amplified in a PCR, resolved in an agarose gel (2%), and visualized by ethidium bromide staining.
Glucose regulates Rgt1 DNA-binding function.
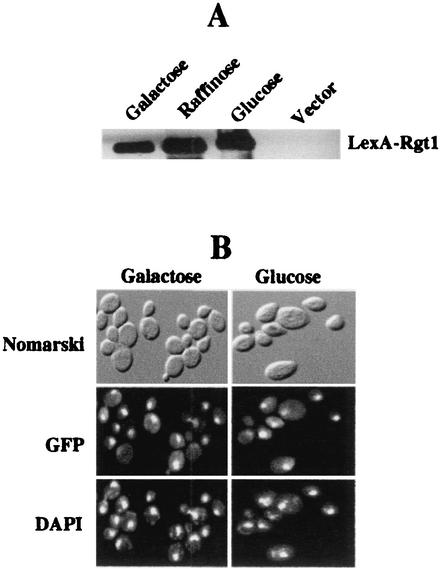
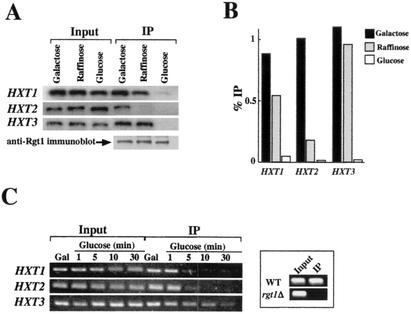
We attempted to determine which activities of Rgt1 are regulated by glucose. Rgt1 is not degraded after the addition of glucose to yeast cells (Fig. 7A), nor is its subcellular localization affected by glucose (Fig. 7B). The DNA-binding ability of Rgt1 is regulated by glucose: it binds to the HXT promoters in cells grown in the absence of glucose or on raffinose (equivalent to a low level [∼0.2%] of glucose), but binding is greatly reduced in cells grown with a high level (4%) of glucose (Fig. 8A and B). This regulation of Rgt1 DNA-binding activity is recapitulated in an in vitro DNA-binding assay: Rgt1 in extracts of cells grown in the absence of glucose (i.e., on galactose) or on raffinose (approximating low glucose) binds to DNA, but Rgt1 from cells grown with high levels of glucose has a low affinity for DNA (Fig. 9A). Rgt1 from cells grown on raffinose seems to have an intermediate affinity for its binding sites in vitro, consistent with the observation that a low level of glucose appears to reduce the affinity of Rgt1 for the HXT2 promoter in vivo (Fig. 8A and B). This is congruent with the fact that expression of HXT2 is induced only by low levels of glucose (27). Rgt1 is dissociated from the HXT3 promoter within 10 min after the addition of a high level of glucose to cells growing on galactose (Fig. 8C). Thus, glucose inhibits the DNA-binding activity of Rgt1.
FIG. 7.
Glucose does not regulate Rgt1 levels or nuclear localization. (A) Yeast cells (YM4509) expressing LexA-Rgt1 (expressed from pBM3307) were grown on minimal medium containing either 2% galactose or 2% raffinose (equivalent to low levels of glucose) or 4% glucose. Yeast cells (YM4509) expressing only LexA (pBM2662) were grown on minimal medium containing 2% galactose (Vector). LexA-Rgt1 was immunoprecipitated and subjected to Western blot analysis using anti-LexA antibody as a probe. (B) Yeast cells (YM4127) expressing GFP-Rgt1 (expressed from pBM3911) were grown to mid-log phase under repressing conditions (2% galactose), the carbon source was switched to 4% glucose, and growth was continued for 1.5 h. The cells were harvested and stained with DAPI before and after the carbon source was switched and then were imaged for DAPI and GFP fluorescence or by Nomarski optics.
FIG. 8.
Association of Rgt1 with the HXT promoters was determined by ChIP in cells (YM4127) grown on media containing different carbon sources (2% galactose, 2% raffinose, and 4% glucose). (A and B) ChIP and immunoblot analyses of Rgt1 using anti-Rgt1 antibody were performed as described in the legends to Fig. 5 and 7A, respectively. The HXT promoters in the immunoprecipitated DNA (IP) were PCR amplified (A), quantified by the ImageQuaNT program, and presented as the ratio of immunoprecipitated counts to input counts (B). (C) Time course of dissociation of Rgt1 from the HXT promoters. Yeast cells (YM4127) grown on galactose (2%) (Gal) were harvested after glucose (4%) was added at the time points indicated above each lane and subjected to ChIP. After the HXT promoters in the immunoprecipitated DNA were amplified by PCR, the products were analyzed as described in the legend to Fig. 6. Chromatin prepared from the Δrgt1 (YM4509) strain was used as the control for ChIP using anti-Rgt1 antibody (box). WT, wild type.
FIG. 9.
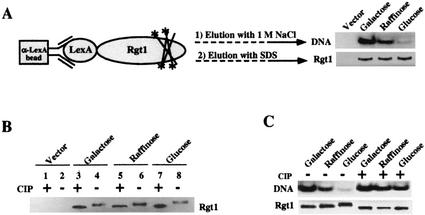
Glucose-induced phosphorylation inhibits DNA-binding activity of Rgt1 in vitro. (A) IDBA. LexA-Rgt1 was immunoprecipitated from yeast extracts as described in the legend to Fig. 7A and incubated with 32P-labeled DNA containing four Rgt1 binding sites (a repeat of Rgt1 binding sites C1 and C2). Approximately 85% of the DNA was bound to anti-LexA-Rgt1 beads. The bound DNA was eluted with 1 M NaCl. The Rgt1 bound to anti-LexA beads was subsequently eluted by boiling the beads in SDS buffer. The eluted DNA and Rgt1 were resolved in polyacrylamide gels and visualized by autoradiography and Western blotting (using anti-LexA antibody), respectively. (B) Rgt1 is phosphorylated. Rgt1 was immunoprecipitated as described above and incubated with (+) or without (−) 10 U of CIP at 37°C for 30 min and then subjected to Western blotting. (C) Glucose-induced phosphorylation of Rgt1 inhibits its DNA-binding activity. IDBA (DNA) and Western blotting (Rgt1) were performed on phosphorylated and dephosphorylated (CIP-treated) Rgt1 as for panels A and B.
Phosphorylation of Rgt1 inhibits its DNA-binding activity.
Rgt1 from cells grown on different carbon sources exhibits different mobilities in SDS-polyacrylamide gel electrophoresis: Rgt1 has the highest mobility in cells grown in the absence of glucose (Fig. 9B, lane 4) and the lowest mobility in cells grown with a high level of glucose (Fig. 9B, lane 8); Rgt1 from cells grown on raffinose (approximating low glucose) has intermediate mobility (Fig. 9B, lane 6). Evidence that these mobility differences are due to differential phosphorylation of Rgt1 is that phosphatase (CIP) treatment increased the mobilities of all forms of the protein (Fig. 8B, lanes 3, 5, and 7). Dephosphorylation of the hyperphosphorylated (high-glucose) form of Rgt1 restores its in vitro DNA-binding ability (Fig. 9C). These results support the idea that glucose-induced phosphorylation inhibits the DNA-binding activity of Rgt1.
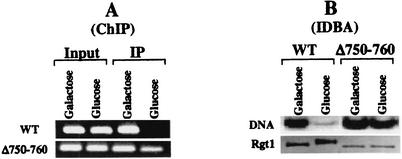
Further evidence that phosphorylation of Rgt1 inhibits its DNA-binding activity comes from analysis of an Rgt1 variant missing 10 amino acids in its central region (Δ750-760). This altered Rgt1 functions as a constitutive repressor (it represses gene expression even in the presence of high levels of glucose [J. Polish, unpublished data]). Unlike full-length Rgt1, it does not become hyperphosphorylated in cells grown with high levels of glucose (Fig. 10B, bottom) and it binds to DNA under these conditions, both in vivo (Fig. 10A, bottom) and in vitro (Fig. 10B, top). Thus, the phosphorylation state of Rgt1 is correlated with its DNA-binding activity.
FIG. 10.
Constitutive DNA binding by Rgt1 leads to constitutive repression. (A) Chromatin prepared from cells expressing wild type (WT) Rgt1 (pBM3580) and an Rgt1 derivative missing 10 amino acids (Δ750-760; pBM4058) was immunoprecipitated using anti-Rgt1 antibody, and the HXT3 promoter in the immunoprecipitated DNA (IP) was detected by PCR as described in the legend to Fig. 8C. (B) DNA binding of Rgt1 in vitro. IDBA (DNA) and Western blotting (Rgt1) were performed on the two forms of Rgt1 as described in the legend to Fig. 9.
DISCUSSION
Rgt1 is at the end of a signal transduction pathway that causes expression of many HXT genes to be induced by glucose. We addressed two central issues concerning the function of Rgt1: how it recognizes its target sites and how its repression function is inactivated.
Rgt1 binds to 5′-CGGANNA-3′.
We determined the sequence of the DNA-binding site of Rgt1 (Fig. 1). Inspection of the sequences of the 31 Rgt1 binding sites we mapped revealed a consensus sequence of 5′-CGGANNA-3′. This simple site is found in many promoters in the yeast genome, so Rgt1 must have a way to limit its binding to a subset of these promoters. One way this could be achieved would be by synergistic binding to multiple sites. Indeed, at least five binding sites are necessary for Rgt1 to bind to the HXT3 promoter (Fig. 5) and for a significant amount of transcriptional repression (Fig. 4). Promoters with this many Rgt1 binding sites are rare: there are 2,630 promoters in the yeast genome with at least one 5′-CGGANNA-3′ sequence, but only 10 of these promoters have five of these sequences; 3 of these genes (HXT3, MIG2, and MIG3) are known to be regulated by Rgt1. Several promoters regulated by Rgt1 possess <5 Rgt1 binding sites. For example, the HXT2 promoter has only three binding sites. Perhaps another protein directs Rgt1 to these promoters.
Interestingly, Rgt1 protects from methylation by DMS only the central G residue of the CGG triplet (Fig. 2). This is in contrast to other Zn cluster proteins, which make strong contacts with two or more G residues in both strands of DNA (33). DMS footprinting (20) and nuclear magnetic resonance structural analysis of AlcR (6) revealed that the protein contacts two G residues (underlined) of the CGG triplet (5′-CGG-3′). Ppr1, a dimeric Zn cluster protein with almost the same sequence of the conserved recognition helix (5′-CRLKKIKC-3′) as Rgt1 (5′-CRKKKIKC-3′), directly interacts with the two G residues of the CGG triplet (22). In any case, Rgt1 seems to function differently at different promoters, perhaps due to different architectures of the Rgt1 binding sites.
Cooperative DNA binding of Rgt1 at multiple binding sites confers synergistic repression.
Many transcription factors bind to multiple binding sites and regulate transcription in a nonadditive fashion. This results from protein-protein interactions of two types: multimerization of the DNA-binding protein (19, 32) and its interactions with the transcription machinery (36). Cooperative DNA binding through protein-protein interaction at multiple sites has been well described for both prokaryotic (5) and eukaryotic (4) transcription factors, but this is unexpected behavior for monomeric DNA-binding proteins. Several lines of evidence support the idea that Rgt1 binds as a monomer to multiple binding sites. First, no dimerization domain is apparent in Rgt1. Second, the Rgt1 binding site does not contain repeats (inverted, everted, or direct) of the CGG sequence (Fig. 1). Third, no regular spacing between Rgt1 binding sites seems necessary for Rgt1 binding (Fig. 1 and unpublished data from helix-phasing experiments using Rgt1 binding sites with different spacings). Fourth, efficient DNA binding and repression by Rgt1 occur only on multiple binding sites (Fig. 4 and 5). How Rgt1 binds synergistically to the multiple binding sites remains unclear. Within a functional unit, however, each individual site appears to be functionally equivalent to the others: a deletion of any single site within a functional unit significantly diminished transcriptional repression and binding of Rgt1 (Fig. 4C). This suggests that Rgt1 is efficiently recruited by multiple binding sites, causing it to mediate synergistic repression of transcription. This could be due to aggregation of Rgt1 on multiple binding sites, due to interactions of Rgt1 with itself or with the transcription machinery. The Ssn6-Tup1 corepressor could play a role, because it interacts with the mediator complex of RNA polymerase (13, 36) and with Rgt1 (Fig. 6).
Transcription derepression caused by the inactivation of Rgt1 DNA binding.
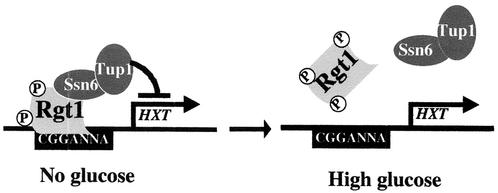
Glucose induction (derepression) of HXT gene expression is achieved by inactivation of the DNA-binding activity of Rgt1. Some other signal transduction pathways work in this manner (17), but others work by causing degradation of repressors (18) or changing their subcellular localization (8). We were surprised by this result, because Rgt1 activates transcription (at least of HXT1) in the presence of high levels of glucose (26). Perhaps Rgt1 does this by a “hit-and-run” mechanism (7, 11, 12) or by activating expression of another transcriptional activator that activates HXT1 expression. The affinity of Rgt1 for HXT promoters depends on the amount of glucose available, with the highest affinity in cells grown in the absence of glucose, modest affinity in cells grown with low levels of glucose, and very low affinity in cells grown with high levels of glucose (Fig. 8). The different DNA-binding affinities are correlated with the degree of Rgt1 phosphorylation: hyperphosphorylated Rgt1 has a much lower affinity for DNA than hypophosphorylated Rgt1 (Fig. 9). (Similar findings were recently reported by Mosley et al. [24]). An Rgt1 derivative (Δ750-760) that functions as a constitutive repressor does not become hyperphosphorylated in the presence of glucose and binds DNA in this condition (Fig. 10). The three serine residues in this region of Rgt1 (at positions 753, 755, and 758) could be the sites of phosphorylation that affect its DNA-binding activity, or this region of Rgt1 could influence the phosphorylation states of residues elsewhere in the protein. In any case, glucose-induced phosphorylation of Rgt1 seems to inactivate its DNA-binding activity, thereby relieving repression of HXT gene expression (Fig. 11).
FIG. 11.
Model for glucose induction of HXT gene expression. Hypophosphorylated Rgt1 is efficiently recruited and forms a stable repression complex with the Ssn6-Tup1 corepressor complex at multiple Rgt1 binding sites in the HXT promoters, leading to the repression of HXT gene expression. Rgt1 is hyperphosphorylated in response to glucose, causing its rapid dissociation from the HXT promoters, leading to induction of gene expression.
Acknowledgments
We thank Ted Hansen for providing Rgt1 C-terminal peptide and John Majors for his insight, suggestions, and criticism.
This work was supported by NIH grant GM-54725.
REFERENCES
- 1.Akache, B., K. Wu, and B. Turcotte. 2001. Phenotypic analysis of genes encoding yeast zinc cluster proteins. Nucleic Acids Res. 29:2181-2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Axelrod, J. D., M. S. Reagan, and J. Majors. 1993. GAL4 disrupts a repressing nucleosome during activation of GAL1 transcription in vivo. Genes Dev. 7:857-869. [DOI] [PubMed] [Google Scholar]
- 3.Baumgartner, U., B. Hamilton, M. Piskacek, H. Ruis, and H. Rottensteiner. 1999. Functional analysis of the Zn2Cys6 transcription factors Oaf1p and Pip2p: different roles in fatty acid induction of β-oxidation in Saccharomyces cerevisiae. J. Biol. Chem. 274:22208-22216. [DOI] [PubMed] [Google Scholar]
- 4.Burz, D. S., R. Rivera-Pomar, H. Jäckle, and S. D. Hanes. 1998. Cooperative DNA-binding by Biocoid provides a mechanism for threshold-dependent gene activation in the Drosophila embryo. EMBO J. 17:5998-6009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carlson, N. G., and J. W. Little. 1993. Highly cooperative DNA binding by Coliphage HK022 repressor. J. Mol. Biol. 230:1108-1130. [DOI] [PubMed] [Google Scholar]
- 6.Cerdan, R., B. Cahuzac, B. Felenbok, and E. Guittet. 2000. NMR solution structure of AlcR (1-60) provides insight in the unusual DNA-binding properties of this zinc binuclear cluster protein. J. Mol. Biol. 295:729-736. [DOI] [PubMed] [Google Scholar]
- 7.Cosma, M. P., T. Tanaka, and K. Nasmyth. 1999. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell 97:299-311. [DOI] [PubMed] [Google Scholar]
- 8.De Vit, M. J., J. A. Waddle, and M. Johnston. 1997. Regulated nuclear translocation of the Mig1 glucose repressor. Mol. Biol. Cell 8:1603-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edmondson, D. G., M. M. Smith, and S. Y. Roth. 1996. Repression domain of the yeast global repressor TUP1 interacts directly with histones H3 and H4. Genes Dev. 10:1247-1259. [DOI] [PubMed] [Google Scholar]
- 10.Felenbok, B., M. Flipphi, and I. Nikolaev. 2001. Ethanol catabolism in Aspergillus nidulans: a model system for studying gene regulation. Prog. Nucleic Acids Res. Mol. Biol. 69:149-204. [DOI] [PubMed] [Google Scholar]
- 11.Fletcher, T. M., N. Xiao, G. Mautino, C. T. Baumann, R. Wolford, B. S. Warren, and G. L. Hager. 2002. ATP-dependent mobilization of the glucocorticoid receptor during chromatin remodeling. Mol. Cell. Biol. 22:3255-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frolova, E., M. Johnston, and J. Majors. 1999. Binding of the glucose-dependent Mig1p repressor to the GAL1 and GAL4 promoters in vivo: regulation by glucose and chromatin structure. Nucleic Acids Res. 27:1350-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gromoller, A., and N. Lehming. 2000. Srb7p is a physical and physiological target of Tup1p. EMBO J. 19:6845-6852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harlow, E., and D. Lane (ed.). 1999. Using antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 15.Hellauer, K., M.-H. Rochon, and B. Turcotte. 1996. A novel DNA-binding motif for yeast zinc cluster proteins: the Leu3p and Pdr3p transcriptional activators recognize everted repeats. Mol. Cell. Biol. 16:6096-6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoffmann, P., and A. Schepartz. 1997. Evidence for induced DNA bending by the yeast zinc cluster protein PUT3. Bio. Med. Chem. Lett. 7:2049-2054. [Google Scholar]
- 17.Huang, M., Z. Zhou, and S. J. Elledge. 1998. The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell 94:595-605. [DOI] [PubMed] [Google Scholar]
- 18.Johnson, P. R., R. Swanson, L. Rakhilina, and M. Hochstrasser. 1998. Degradation signal masking by heterodimerization of MATα2 and Mata1 blocks their mutual destruction by the ubiquitin-proteasome pathway. Cell 94:217-227. [DOI] [PubMed] [Google Scholar]
- 19.Kim, B., and J. W. Little. 1992. Dimerization of a specific DNA-binding protein on the DNA. Science 255:203-206. [DOI] [PubMed] [Google Scholar]
- 20.Lenouvel, F., I. Nikolaev, and B. Felenbok. 1997. In vitro recognition of specific DNA targets by AlcR, a Zinc binuclear cluster activator different from the other proteins of this class. J. Biol. Chem. 272:15521-15526. [DOI] [PubMed] [Google Scholar]
- 21.Marmorstein, R., M. Carey, M. Ptashne, and S. C. Harrison. 1992. DNA recognition by GAL4: structure of a protein-DNA complex. Nature 356:408-414. [DOI] [PubMed] [Google Scholar]
- 22.Marmorstein, R., and S. C. Harrison. 1994. Crystal structure of a PPR1-DNA complex: DNA recognition by proteins containing a Zn2Cys6 binuclear cluster. Genes Dev. 8:2504-2512. [DOI] [PubMed] [Google Scholar]
- 23.McConnell, K. J., and D. L. Beveridge. 2001. Molecular dynamics simulations of B′-DNA: sequence effects on A-tract-induced bending and flexibility. J. Mol. Biol. 314:23-40. [DOI] [PubMed] [Google Scholar]
- 24.Mosley, A. L., J. Lakshmanan, B. K. Aryal, and S. Özcan. 2003. Glucose-mediated phosphorylation converts the transcription factor Rgt1 from a repressor to an activator. J. Biol. Chem. 278:10322-10327. [DOI] [PubMed] [Google Scholar]
- 25.Niedenthal, R. K., L. Riles, M. Johnston, and J. H. Hegemann. 1996. Green fluorescent protein as a marker for gene expression and subcellular localization in budding yeast. Yeast 12:773-786. [DOI] [PubMed] [Google Scholar]
- 26.Özcan, S., T. Leong, and M. Johnston. 1996. Rgt1p of Saccharomyces cerevisiae, a key regulator of glucose-induced genes, is both an activator and a repressor of transcription. Mol. Cell. Biol. 16:6419-6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Özcan, S., and M. Johnston. 1999. Function and regulation of yeast hexose transporters. Micribiol. Mol. Biol. Rev. 63:554-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Panozzo, C., V. Capuano, S. Fillinger, and B. Felenbok. 1997. The zinc binuclear cluster activator AlcR is able to bind to single sites but requires multiple repeated sites for synergistic activation of the alcA gene in Aspergillus nidulans. J. Biol. Chem. 272:22859-22865. [DOI] [PubMed] [Google Scholar]
- 29.Schjerling, P., and S. Holmberg. 1996. Comparative amino acid sequence analysis of the C6 zinc cluster family of transcriptional regulators. Nucleic Acids Res. 24:4599-4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strahl-Bolsinger, S., A. Hecht, K. Luo, and M. Grunstein. 1997. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 11:83-93. [DOI] [PubMed] [Google Scholar]
- 31.Swaminathan, K., P. Flynn, R. R. Reece, and R. Marmorstein. 1997. Crystal structure of a PUT3-DNA complex reveals a novel mechanism for DNA recognition by a protein containing a Zn2Cys6 binuclear cluster. Nat. Struct. Biol. 4:751-759. [DOI] [PubMed] [Google Scholar]
- 32.Tanaka, M. 1996. Modulation of promoter occupancy by cooperative DNA-binding and activation-domain function is a major determinant of transcriptional regulation by activators in vivo. Proc. Natl. Acad. Sci. USA 93:4311-4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Todd, R. B., and A. Andrianopoulos. 1997. Evolution of a fungal regulatory gene family: the Zn(II)2Cys6 binuclear cluster DNA-binding motif. Fungal Genet. Biol. 21:388-405. [DOI] [PubMed] [Google Scholar]
- 34.Walker, J., P. Crowley, A. D. Moreman, and J. Barrett. 1993. Biochemical properties of cloned glutathione. Mol. Biochem. Parasitol. 61:255-264. [DOI] [PubMed] [Google Scholar]
- 35.Yocum, R. R., S. Hanley, J. R. West, and M. Ptashne. 1984. Use of LacZ fusions to delimit regulatory elements of the inducible divergent Gal1-Gal10 promoter site in Saccharomyces cerevisiae. Mol. Cell. Biol. 4:1985-1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zaman, Z., A. Z. Ansari, S. S. Koh, R. A. Young, and M. Ptashne. 2001. Interaction of a transcriptional repressor with the RNA polymerase II holoenzyme plays a crucial role in repression. Proc. Natl. Acad. Sci. USA 98:2550-2554. [DOI] [PMC free article] [PubMed] [Google Scholar]