Abstract
The yeast protein Fis1p has been shown to participate in mitochondrial fission mediated by the dynamin-related protein Dnm1p. In mammalian cells, the dynamin-like protein DLP1/Drp1 functions as a mitochondrial fission protein, but the mechanisms by which DLP1/Drp1 and the mitochondrial membrane interact during the fission process are undefined. In this study, we have tested the role of a mammalian homologue of Fis1p, hFis1, and provided new and mechanistic information about the control of mitochondrial fission in mammalian cells. Through differential tagging and deletion experiments, we demonstrate that the intact C-terminal structure of hFis1 is essential for mitochondrial localization, whereas the N-terminal region of hFis1 is necessary for mitochondrial fission. Remarkably, an increased level of cellular hFis1 strongly promotes mitochondrial fission, resulting in an accumulation of fragmented mitochondria. Conversely, cell microinjection of hFis1 antibodies or treatment with hFis1 antisense oligonucleotides induces an elongated and collapsed mitochondrial morphology. Further, fluorescence resonance energy transfer and coimmunoprecipitation studies demonstrate that hFis1 interacts with DLP1. These results suggest that hFis1 participates in mitochondrial fission through an interaction that recruits DLP1 from the cytosol. We propose that hFis1 is a limiting factor in mitochondrial fission and that the number of hFis1 molecules on the mitochondrial surface determines fission frequency.
Mitochondria in mammalian cells are dynamic structures displaying frequent fission, fusion, and translocation (1). These dynamic processes are believed to ensure an appropriate distribution of mitochondria during cell proliferation and provide sufficient energy to a localized cytoplasmic region. It is now known that fusion and fission of mitochondria from yeast to human are balanced to maintain normal tubular mitochondria through the antagonistic action of two distinct large GTPases (6, 9, 16, 17, 19, 20, 24). An imbalance between these two events results in excessive fragmentation or tubulation of mitochondria. One of these GTPases is a dynamin-like enzyme called Dnm1p in yeast cells or DLP1/Drp1 in mammalian cells. Studies with yeast and mammalian cells indicated that Dnm1p/DLP1/Drp1 plays a role in mitochondrial fission (16, 17, 20, 23, 24). In a previous study, DLP1/Drp1 has been found to deform biological membranes in a GTP-dependent manner (32).
Although DLP1 is involved in controlling mitochondrial morphology, its distribution to mitochondria in mammalian cells is modest. Both morphological and biochemical data have shown that the majority of DLP1 is cytosolic while subpopulations localize to mitochondria and other cellular organelles such as the endoplasmic reticulum, microtubules, and peroxisomes (10, 13, 14, 22, 31). Although DLP1 is known to function as a mitochondrial fission enzyme, overexpression of DLP1 within cells does not increase mitochondrial fission (17), indicating that there are other limiting factors in the mitochondrial fission process. These factors may recruit cytosolic DLP1 to the mitochondrial surface and/or activate the DLP1 GTPase, leading to mitochondrial fission. Yeast genetic screening has identified two proteins, Mdv1p and Fis1p, which are both predicted to function together with Dnm1p in mitochondrial fission (2, 4, 15, 25). Yeast cells defective in these proteins display interconnected net-like mitochondria, which is identical to the Dnm1p-defective phenotype. Morphological studies found that Dnm1p and Mdv1p colocalized on mitochondrial tubules as punctate spots, and in the absence of Fis1p, the size and number of these Dnm1p-containing structures were grossly altered (15, 25). It was also found that the normal distribution of Mdv1p was perturbed in the absence of Dnm1p and/or Fis1p (15, 25). These results suggest that the three proteins interact either directly or indirectly to mediate mitochondrial fission through a multistep pathway (15, 25). Although a cognate homologue of Mdv1p does not exist in higher eukaryotes, it is possible that a structurally similar protein may participate as a functional homologue in the mitochondrial fission process.
As mammalian DLP1 distributes predominantly to the cytosol, it seemed important to identify a protein that might recruit DLP1 to mitochondria in a regulated manner. Fis1p is a likely candidate for this role based on its membrane topology and proposed function. Fis1p is a small, 17-kDa protein and is predicted to be anchored in the outer mitochondrial membrane with its N-terminal region exposed to the cytosol and a short C-terminal tail protruding into the mitochondrial intermembrane space (15). It has been proposed that Fis1p is required for the proper assembly and activation of the fission-mediating complex for yeast mitochondrial division (15, 25). Furthermore, homologues of Fis1p have been found in many higher eukaryotes, including human (15). Here, we have cloned a human homologue of Fis1p, hFis1, and obtained mechanistic information regarding the role of hFis1 in mitochondrial fission. We demonstrated that the N-terminal region of hFis1 is required for mitochondrial fission whereas the C-terminal tail is necessary for mitochondrial localization of hFis1. More importantly, we found that an elevated level of cellular hFis1 drastically increases mitochondrial fission frequency, resulting in fragmented mitochondria, while disruption of hFis1 function causes inhibition of mitochondrial fission. Further, hFis1 and DLP1 form a complex in cells and are able to interact with each other directly in vitro. These findings both support and expand the earlier observations made for yeast and provide evidence that hFis1 regulates mitochondrial fission through a protein-protein interaction that recruits DLP1 from the cytosol to the mitochondrial surface. These results suggest that hFis1 is a limiting factor in mitochondrial fission and that the number of hFis1 molecules on the mitochondrial surface determines fission frequency.
MATERIALS AND METHODS
Cell culture, DNAs, and transfection.
The cell lines Clone 9 (ATCC CRL-1439), HeLa, COS-7, and BHK-21 were used for all experiments. Cells were maintained at 37°C with 5% CO2 in Ham's F-12K medium for Clone 9 cells and in Dulbecco's modified Eagle's medium for HeLa, COS-7, and BHK-21 cells, supplemented with 10% fetal bovine serum, 100 U of penicillin per ml, and 100 μg of streptomycin per ml. The cDNA for hFis1 was cloned by reverse transcription-PCR with total human RNA as the template. PCR primers were synthesized according to the sequence deposited in GenBank (accession number AF151893). All subcloning into different vectors was carried out by standard molecular biology techniques with PCR. The absence of mutations in nucleotide sequences after PCR was verified by DNA sequencing. Stable cells carrying green fluorescent protein (GFP) or red fluorescent protein (RFP) in their mitochondrial matrix were isolated by transfecting pmitoGFP or pmitoRFP and selecting with G418. pmitoGFP and pmitoRFP were constructed by fusing the mitochondrial transit sequence of human isovaleryl coenzyme A dehydrogenase to the N terminus of GFP (pEGFP-N1; Clontech, Inc.) or RFP (pDsRed1-N1; Clontech, Inc.), respectively. Plasmids for recombinant DLP1 and hFis1 were constructed in the pQE-80 series vector (Qiagen, Inc.). For cell transfection, DNA constructs were purified by using plasmid purification columns (Qiagen, Inc.). Cells were plated for 16 to 24 h before transfection on glass coverslips in 35-mm-diameter tissue culture dishes. Transfections were performed by using Lipofectamine (Invitrogen, Inc.) or GeneJammer (Stratagene, Inc.) according to the manufacturer's instructions or by microinjecting DNA into the nucleus as described previously (17). Transfected cells were allowed to recover for 16 to 24 h before being processed for either immunofluorescence or electron microscopy.
Immunofluorescence and microscopy.
Indirect immunofluorescence was performed as described previously (8, 31). Briefly, cells were fixed, permeabilized, and then incubated in a blocking buffer containing 5% goat serum for 1 h at 37°C. Mouse monoclonal anti-Myc antibodies (Zymed, Inc.) and either Alexa 488- or 594-conjugated goat anti-mouse immunoglobulin G (Molecular Probes, Inc.) were used for primary and secondary antibodies, respectively. Permeabilization, blocking, and antibody incubation were omitted when the immunodetection was unnecessary. After appropriate rinsings, coverslips were mounted in ProLong antifade reagent (Molecular Probes, Inc.) on glass slides and cells were viewed with an Axiovert 35 epifluorescence microscope (Carl Zeiss) equipped with a 100× objective (Zeiss Plan-Neofluar; numerical aperture, 1.30). Fluorescence images were acquired with an OrcaII cooled charge-coupled device (Hamamatsu Photonics K.K.) driven by IPLab imaging software (Scanalytics, Inc.). For confocal microscopy, images were acquired in a 0.5-μm optical section with a Zeiss LSM510 confocal microscope.
Electron microscopy.
Cells were plated on etched grid coverslips with numbered imprints (Bellco Biotechnology). Cells within one square on the coverslip were microinjected with Myc-hFis1 or control DNA constructs. After 24 h, cells were rinsed with 37°C phosphate-buffered saline and submerged sequentially in primary fixative (100 mM Na-PO4 [pH 7.2], 50 mM sucrose, 3.0% glutaraldehyde) for 1 h, in 1% osmium tetroxide for 30 min, and in 1% uranyl acetate for 30 min. The fixed cells were then dehydrated in a graded series of ethanol solutions and embedded in Quetol 651 (Ted Pella, Inc.). The block face of the resin was then trimmed to leave a square box that contained only injected cells and processed as described previously (7). Samples were viewed and photographed with a JEOL 1200 electron microscope.
Inhibition of endogenous hFis1.
Microinjections were performed as described previously (17). Anti-hFis1 antibodies raised against the peptide EDLLKFEKKFQSEKAAGSVSKSTQFEY were affinity purified on a column with the immobilized immunogen peptide and then dialyzed against microinjection buffer (10 mM KH2PO4 [pH 7.2], 75 mM KCl) and concentrated to a final concentration of 5 to 10 mg/ml. Rhodamine-conjugated dextran (molecular weight, 3,000; Molecular Probes, Inc.) was added to the antibody solution as an injection marker. Solutions for control injections were microinjection buffer with fluorescent markers. Injected cells were allowed to recover for 2 to 4 h, fixed, and processed for immunofluorescence. For antisense experiments, a 22-mer antisense oligonucleotide that spans positions −4 to 18 (5′-GTTCAGCACGGCCTCCATGGCC-3′) was prepared with phosphorothioate bridges. A sense oligonucleotide (5′-ACGAGCTGGTGTCTGTGGAGGA-3′) was used as a control. Oligonucleotides were introduced into cells by using Oligofectamine reagent (Invitrogen, Inc.) according to the manufacturer's instructions. After 24- to 72-h treatments, cells were processed for immunoblotting or immunofluorescence.
FRET technique.
COS-7 cells were transfected with Myc-hFis1 and processed for indirect immunofluorescence. The donor quenching method was used for fluorescence resonance energy transfer (FRET) imaging (12). This method measures release of donor quenching after acceptor photobleaching. For this, secondary antibodies conjugated to the fluorescent cyanine dyes Cy3 and Cy5 (Jackson ImmunoResearch Laboratories, Inc.) were used as the donor and acceptor pairs, respectively. An LSM510 confocal microscope (Carl Zeiss) with a 100× oil immersion Plan-Apochromat lens with a numerical aperture of 1.4 was used for image acquisition. DLP1-Cy3 was excited with a 543-nm HeNe laser at 10% power, and emission was detected after passage through a 560- to 615-nm band-pass filter. Myc-hFis-Cy5 was excited with a 633-nm HeNe laser at 10% power, and emission was detected after passage through a 650-nm long-pass filter. Images were acquired at 1,024 by 1,024 resolution at a 12-bit pixel depth in a 1.0-μm optical section. Photobleaching of the acceptor dye Cy5 was performed with a 633-nm HeNe laser at 100% power for 250 iterations on a box of 200 by 50 pixels. Images were acquired in a specific order, i.e., an image of Cy5 alone (Cy5pre) and an image of Cy3 alone (Cy3pre), followed by photobleaching of Cy5, another image of Cy3 alone (Cy3post), and an image of Cy5 (Cy5post) to show complete photobleaching of the acceptor molecule. Fluorescence profiles and numerical data for Cy3pre and Cy3post were obtained by line scanning, and FRET intensity was expressed by the difference in fluorescence (Cy3post minus Cy3pre) as arbitrary units. For a positive control, DLP1 was labeled with both Cy3 and Cy5 and analyzed for FRET. Either DLP1 spots showing negative FRET or DLP1 and dynamin labeled with Cy3 and Cy5, respectively, were used as a negative control.
Purification of recombinant hFis1 and DLP1.
hFis1ΔTM/C and DLP1 cDNAs were cloned into the pQE vector (Qiagen, Inc.) to overexpress His6-tagged proteins. The His6-tagged protein was purified with ProBond resin (Invitrogen, Inc.) according to the manufacturer's instruction. Briefly, M15 bacteria (Qiagen, Inc.) carrying a plasmid for His6-tagged DLP1 were induced with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 16 to 18 h at room temperature to overproduce DLP1. Soluble bacterial extracts were obtained by sonication followed by centrifugation at 15,000 × g for 20 min. Bacterial extracts were incubated with Ni+-resin (ProBond; Invitrogen, Inc.) for 30 min at 4°C with gentle rotation to isolate proteins. After the resin was rinsed, His6-protein was eluted with 500 mM imidazole.
Detection of the interaction between hFis1 and DLP1 by immunoprecipitation.
To test for a DLP1-hFis1 interaction in intact cells, BHK-21 cells were transfected with the Myc-hFis1 plasmid and whole cells were subjected to chemical cross-linking at 24 h posttransfection. A cleavable, homobifunctional cross-linker [dithiobis(succinimidylpropionate) (DSP)] (Pierce Chemical Co.) was diluted to a 1.0 mM final concentration in phosphate-buffered saline and added to cultured cells. After incubation for 1 h at room temperature, cross-linking was stopped by the addition of Tris (pH 7.7) to a 10 mM final concentration. Cells were lysed with radioimmunoprecipitation assay buffer (50 mM Tris [pH 8.0], 150 mM NaCl, 0.1% sodium dodecyl sulfate, 0.5% deoxycholic acid, and 1.0% Triton X-100), and the immunoprecipitation was performed with anti-DLP1 or anti-hFis1 antibodies. To test for a direct interaction between hFis1 and DLP1, purified proteins were concentrated and dialyzed against buffer A (20 mM HEPES [pH 7.4], 100 mM KCl, 2 mM MgCl). A 10 μM concentration of hFis1 and the same concentration of purified DLP1 or dynamin were combined in buffer A and left for 20 min at room temperature. DSP was added to the protein mixture and left for 30 min. Cross-linking was stopped by the addition of 10 mM Tris (pH 7.7). Immunoprecipitation was done with anti-DLP1 (DLP-Mid) or antidynamin (Dyn2-II) antibodies and protein A-Sepharose (Sigma Chemical Co.). Samples were washed three times in buffer A and boiled in sodium dodecyl sulfate sample buffer containing 5% 2-mercaptoethanol to cleave the cross-linking. Samples were run on 15% acrylamide gels and subjected to Western blot analysis with anti-DLP1, anti-Myc (Zymed, Inc.), or anti-RGS-His mouse monoclonal antibody (Qiagen, Inc.).
GTP hydrolysis assay.
GTPase assays were carried out by using thin-layer chromatography. Recombinant DLP1 was diluted to 3 μM in PEM buffer {100 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)] [pH 6.9], 1 mM MgSO4, 2 mM EGTA} plus 0.25 mg of bovine serum albumin per ml in the absence or presence of 3 μM recombinant hFis1 and incubated for 15 min at room temperature. The reaction mixtures were then placed on ice while 500 μM cold GTP and 5 μCi of [α-32P]GTP (ICN Biomedicals) were added. After addition of GTP, the reaction mixtures were incubated at room temperature, and 1-μl aliquots were removed at 0, 5, 10, 20, 30, 40, and 50 min and spotted on polyethyleneimine cellulose thin-layer chromatography plates (Sigma Chemical Co.). The thin-layer chromatography plates were resolved in 1.0 M LiCl2, dried, and exposed to film. The ratio of GDP to total nucleotide (GDP plus GTP) at each time point was determined by densitometry.
RESULTS
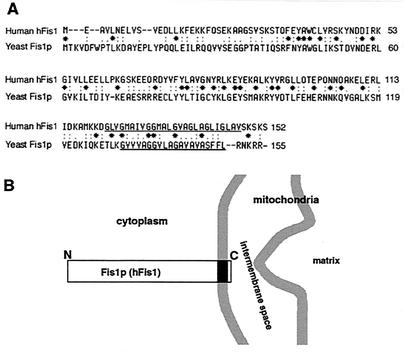
To test for a role of a Fis1p-like protein in mammalian mitochondrial division, we cloned a human homologue of yeast Fis1p by reverse transcription-PCR based on the nucleotide sequence deposited in GenBank (accession number AF151893). The predicted human homologue of Fis1p, which we call hFis1, is a 17-kDa protein of 152 amino acids. It has 25% amino acid identity and 63% similarity to its yeast counterpart (Fig. 1A). The C-terminal region of hFis1 contains a 26-amino-acid stretch of predicted transmembrane domain (amino acids 122 to 147), leaving a short, 5-amino-acid tail protruding from the membrane (Fig. 1B). The rat Fis1 amino acid sequence is now available (GenBank accession number XM_213746.1), and it has 97% identity and 99% similarity to hFis1.
FIG. 1.
Identification of human hFis1, a homologue of the yeast mitochondrial outer membrane protein Fis1p. (A) Sequence alignment for human hFis1 and yeast Fis1p. The two proteins share 25% identity and 63% similarity in amino acid sequences. The C-terminal regions of both proteins contain a stretch with a putative transmembrane domain (underlines). (B) Fis1p (hFis1) is a transmembrane protein anchored at the outer mitochondrial membrane with its N-terminal region exposed to the cytoplasm and a short C-terminal tail in the intermembrane space.
Mitochondrial localization of hFis1 requires an intact C terminus.
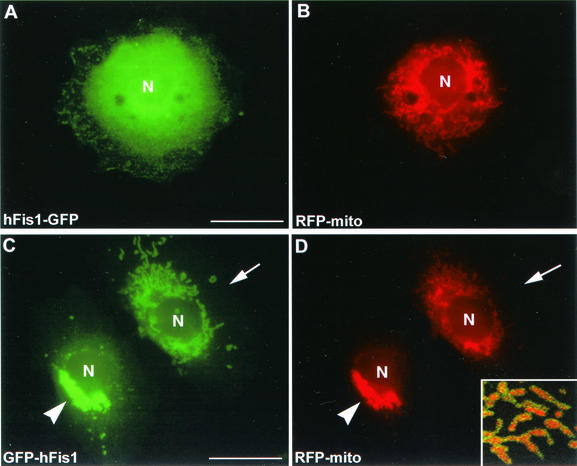
To test whether hFis1 is a mitochondrial protein like the yeast Fis1p, we made two hFis1 constructs tagged with GFP at either the C or N terminus of hFis1. These constructs were transfected into HeLa cells or Clone 9 cells (normal rat liver cells), which stably expressed RFP in the mitochondrial matrix. When the hFis1 tagged with GFP at its C-terminal end (hFis1-GFP) was expressed, a diffuse cytosolic distribution was observed (Fig. 2A). In some cells, however, hFis1-GFP partially localized to mitochondria in addition to the cytosolic distribution (results not shown). It is possible that the large GFP tag (26 kDa) on the small hFis1 (17 kDa) disrupted mitochondrial localization. We tested this possibility by tagging the protein with a short Myc epitope at the C terminus of hFis1 (hFis1-Myc) and observed a diffuse distribution similar to that of hFis1-GFP (results not shown). This cytosolic localization of hFis1-GFP and hFis1-Myc suggested that tagging at the C terminus interfered with the correct localization of hFis1 to mitochondria. We next tested whether the intact C-terminal sequence of hFis1 is necessary for the correct localization to mitochondria by expressing hFis1 tagged with GFP at the N terminus (GFP-hFis1). We found that GFP-hFis1 consistently localized to mitochondria (Fig. 2C and D). Confocal microscopy revealed that the GFP fluorescence of hFis1 surrounds the matrix RFP, suggesting that hFis1 is localized to the mitochondrial outer membrane as shown for yeast Fis1p (4, 15). GFP-hFis1 appeared punctate or as small patches on the outer surface of mitochondrial tubules (Fig. 2D, inset). Among transiently transfected cells, however, we frequently found that cells expressing high levels of GFP-hFis1 exhibited a dramatic change in mitochondrial morphology. In these cells, mitochondria became aggregated around the nucleus, appearing as a clumped mass (Fig. 2C and D). It is possible that overexpression of GFP-hFis1 on the mitochondrial surface inhibits the DLP1-mediated mitochondrial fission and that ongoing fusion induces the formation of clumped mitochondria. However, the mitochondrial morphology induced by DLP1 inhibition is mainly elongated and entangled tubules (17, 24), morphologically distinct from these mitochondrial clumps. We reasoned that the mitochondrial clumps induced by GFP-hFis1 overexpression are due to uncontrolled aggregation or fusion, possibly mediated by GFP tags on the mitochondrial surface (see Discussion).
FIG. 2.
GFP-tagged hFis1 proteins distribute differently in cells depending on the position of the tag. Clone 9 cells expressing an RFP-labeled mitochondrial protein (RFP-mito) as a marker were transfected with hFis1 protein tagged with GFP at either the C terminus (hFis1-GFP) (A and B) or the N terminus (GFP-hFis1) (C and D). The distribution of hFis1-GFP was diffuse (A) and did not show mitochondrial localization (B), suggesting that the C-terminal tagging inhibited mitochondrial localization of hFis1. In contrast, expressed GFP-hFis1 (C) distributed exclusively to mitochondria (D) (arrows). Overexpression of GFP-hFis1 often induced formation of mitochondrial clumps (arrowheads). An enlarged confocal image of mitochondria shows GFP-hFis1 on the surface of mitochondria, surrounding RFP in the matrix (inset in panel D). GFP-hFis1 localized to the mitochondrial surface as small patches and punctate spots. N, nucleus. Bar, 20 μm.
Increased expression of functional hFis1 promotes mitochondrial fission.
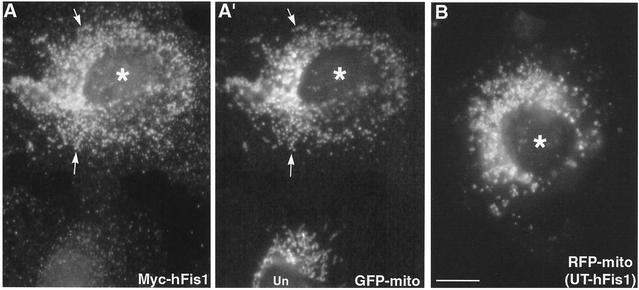
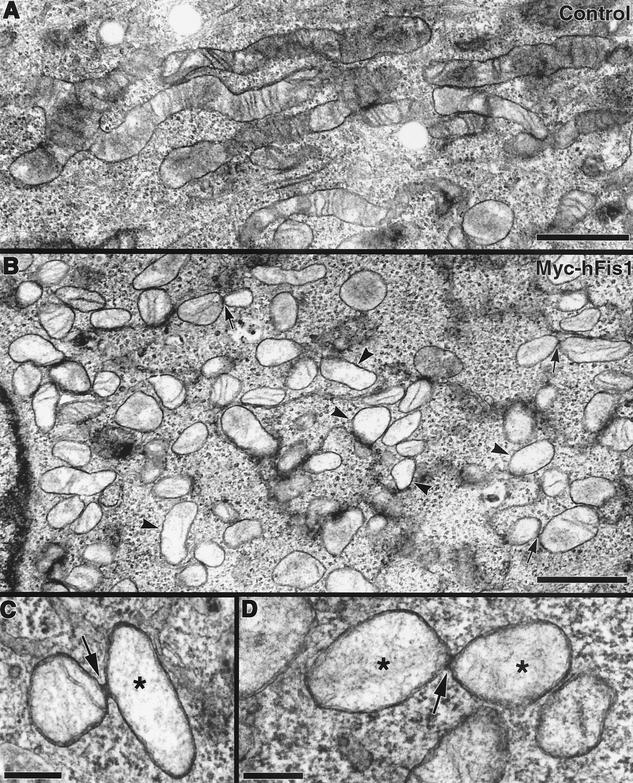
Mitochondrial deformation, presumably aggregation or fusion, induced by overexpression of GFP-hFis1 suggested that the large GFP tag may interfere with the proper function of hFis1. Thus, to examine the cellular function of hFis1, we expressed full-length hFis1 tagged with the short, 10-amino-acid Myc epitope at the N terminus (Myc-hFis1). Remarkably, cells expressing Myc-hFis1 exhibited completely fragmented mitochondria, while the expressed Myc-hFis1 was localized to mitochondria as judged by immunofluorescence with the Myc antibody (Fig. 3A and A′). All of the cells expressing Myc-hFis1, at both low and high levels, contained extremely short rods or fine puncta as mitochondria (Fig. 3A′). We also expressed untagged hFis1 to demonstrate that the Myc tag had no effect on normal hFis1 function. We transfected the untagged hFis1 by microinjection of cDNA and found an essentially identical mitochondrial phenotype (Fig. 3B). This indicated that an elevated level of hFis1 drastically increased mitochondrial fission. To examine mitochondrial fragmentation at the ultrastructural level, we performed electron microscopy of Myc-hFis1-expressing cells. As shown in Fig. 4B, consistent with light microscopy, the Myc-hFis1-expressing cells contained predominantly small spherical mitochondria, compared to the mainly normal tubular mitochondria in untransfected cells (Fig. 4A). In addition, we frequently found two mitochondria connected by a narrow neck in these cells (Fig. 4B to D). Although it is unclear whether they were undergoing fission or fusion, we presumed that these were mitochondria caught actively dividing, as these small mitochondrial spheres were the result of increased mitochondrial fission and were not found in untransfected cells. Interestingly, the matrices of many small mitochondria appeared to be less electron dense than those of control mitochondria. In addition, some of the fragmented mitochondria also lost cristae structures (Fig. 4B), suggesting that mitochondrial fragments induced by hFis1 expression may be functionally compromised (see Discussion).
FIG. 3.
Expression of Myc-hFis1 or untagged hFis1 induces fragmentation of mitochondria. Clone 9 cells carrying GFP-labeled mitochondria (GFP-mito) were transfected with Myc-hFis1 and subjected to indirect immunofluorescence with antibodies to Myc. Cells expressing Myc-hFis1 (A) (asterisk) contained completely fragmented mitochondria (A′) to which Myc-hFis1 localized (arrows). Note the normal mitochondria in the untransfected cell in panel A′ (Un). (B) Clone 9 cells carrying RFP-labeled mitochondria (RFP-mito) were microinjected with untagged, full-length hFis1 DNA (UT-hFis1) along with GFP vector as a transfection marker. Mitochondria became fragmented in cells expressing untagged hFis1. Bar, 10 μm.
FIG. 4.
Electron microscopy of Myc-hFis1-expressing cells confirms mitochondrial fragmentation. (A) Transmission electron microscopy image of an untransfected HeLa cell showing normal tubular mitochondria with cristae. (B) HeLa cells expressing Myc-hFis1 contain numerous small mitochondria throughout the cytoplasm. Mitochondria in these cells have a hollow appearance, with a less electron-dense matrix and markedly reduced cristae (arrowheads). Connected mitochondria were occasionally observed in these cells (arrows). (C and D) Mitochondria connected by a narrow neck, presumably undergoing fission. Electron-dense necks are apparent at the point of constriction (arrows). Again, note the loss of cristae and lightly stained matrices in these mitochondria (asterisks). Bars, 1.0 μm (A and B) and 200 nm (C and D).
Because we did not observe mitochondrial fragmentation in cells expressing hFis1-GFP or hFis1-Myc, which distribute to the cytosol (Fig. 2A and B), it is likely that the mitochondrial localization of hFis1 is a prerequisite for mitochondrial fission. We confirmed this prediction by expressing hFis1 constructs lacking either the 5-amino-acid tail (Myc-hFis1-ΔC) or both the transmembrane region and the tail (Myc-hFis1-ΔTM/C). These truncated proteins failed to localize to mitochondria and showed a diffuse cytosolic distribution. Mitochondria were normal in these transfected cells, without showing fragmentation. However, we found that some cells displayed collapsed mitochondria similar to those seen in DLP1-defective cells (see Fig. 6A and A′). Although quantitation was difficult due to the different protein expression levels during transient expression, cells expressing high levels of these truncations tended to show the collapsed mitochondrial phenotype more prevalently. A similar phenotype was found for cells expressing hFis1-GFP or hFis1-Myc upon more careful observation (results not shown). These findings suggested that hFis1 interacts with DLP1 and that overexpressed hFis1 mutants in the cytosol may sequester DLP1 away from mitochondria and inhibit mitochondrial fission. Conversely, the increased number of functional hFis1 molecules within the mitochondrial membrane recruits excess DLP1 to mitochondria, leading to mitochondrial fragmentation.
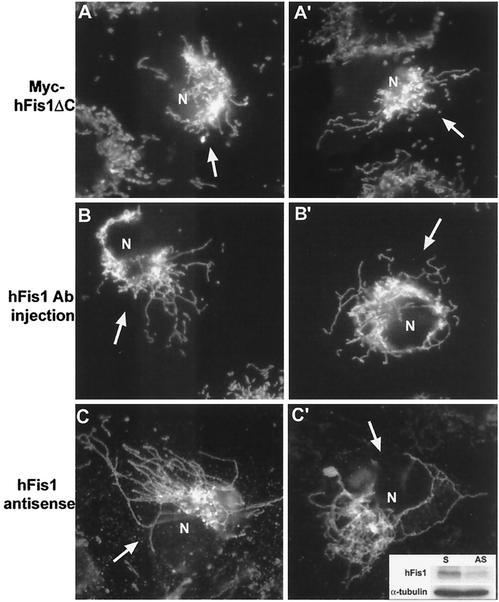
FIG. 6.
Disruption of hFis1 function alters mitochondrial shape and distribution. (A and A′) Clone 9 cells harboring GFP-mitochondria were transfected with Myc-hFis1ΔC. Mitochondria in the cell overexpressing Myc-hFis1ΔC (arrows) displayed a collapsed phenotype. (B and B′) Anti-hFis1 antibodies were microinjected into Clone 9 cells harboring GFP-mitochondria to inhibit hFis1 function. By 2 h, mitochondria in antibody-injected cells (arrows) became elongated and collapsed around the nucleus. (C and C′) Antisense oligonucleotides were introduced into Clone 9 cells to reduce the expression of hFis1. At 48 h posttransfection, cells were either fixed for immunofluorescence or lysed for immunoblotting. A 50% reduction of hFis1 expression was observed when normalized to tubulin by immunoblotting (inset). Anti-cytochrome c staining showed that cells (arrows) displayed a fission-defective mitochondrial morphology. N, nucleus; S, sense; AS, antisense.
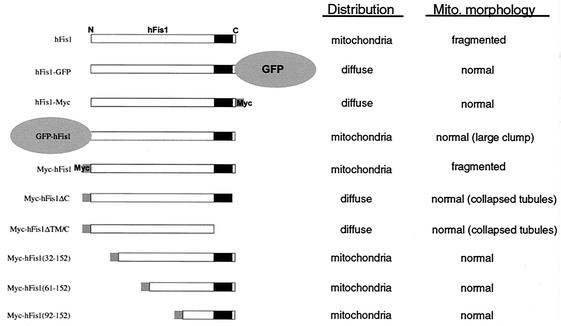
A partial inhibition of mitochondrial fission by cytosolic hFis1 suggests that the N-terminal region of hFis1 interacts with DLP1. To further elaborate the role of the N-terminal region of hFis1, we expressed three N-terminally truncated hFis1 constructs [Myc-hFis1(32-152), Myc-hFis1(61-152), and Myc-hFis1(92-152)] in Clone 9 cells harboring GFP-mitochondria. All three truncated hFis1 constructs localized to mitochondria without causing mitochondrial fragmentation (Fig. 5). The mitochondrial morphology in these cells was normal, although mitochondria in some cells appeared to be unhealthy, showing several large swollen spheres probably due to gross overexpression of abnormal mitochondrial proteins. These results suggested that the first 31 amino acids of hFis1 contain crucial information for mitochondrial fission, possibly regulating DLP1 function. These findings also indicated that while the intact C-terminal structure of hFis1 is essential for mitochondrial localization, the vast majority of the N-terminal sequence (up to the first 91 amino acids) is not required for the correct distribution of hFis1. A summary of the constructs, their localizations, and their mitochondrial phenotypes is shown in Fig. 5.
FIG. 5.
Summary of cellular distributions and alterations in mitochondrial (Mito.) morphology of different constructs used in this study. GFP or Myc was used as an epitope tag at either the N or C terminus of hFis1. Ten different hFis1 constructs were used in this study, including five truncation mutants with mutations in either the C or N terminus. hFis1 proteins modified at the C terminus distributed to the cytosol, whereas those with N-terminal modifications maintained a mitochondrial localization.
hFis1 functions in mitochondrial fission.
To expand upon our observations that altered hFis1 expression affects mitochondrial morphology, we attempted to disrupt the normal function of hFis1 by either antibody microinjection or antisense oligonucleotides. Upon microinjection of purified antibodies against hFis1, mitochondria became elongated and collapsed around the nucleus (Fig. 6B and B′). This mitochondrial phenotype was remarkably similar to that observed in DLP1-defective cells (17, 24) and cells overexpressing high levels of C-terminally truncated hFis1 (Fig. 6A and A′). Control cells injected with buffer alone did not show any change in mitochondrial morphology (data not shown). In addition to antibody injection, cells were also treated with antisense oligonucleotides to suppress hFis1 expression. Although we could not discern cells with reduced hFis1 expression (endogenous hFis1 was not detectable by our antibodies, probably because its level in cells is below the detection limit), we were able to find a reduction of hFis1 in the antisense oligonucleotide-treated cell population by immunoblotting (Fig. 6C′, inset). The reduction in hFis1 levels was 30 to 50% compared to control cells by 48 h of treatment. Approximately 20% of the antisense oligonucleotide-treated cells displayed drastic changes in mitochondrial morphology similar to those seen with antibody injection (Fig. 6C and C′), while an additional 20 to 30% of the cells showed various degrees of fission-defective mitochondrial morphology. The mitochondrial phenotypic changes found in these and antibody-injected cells are consistent with the notion that hFis1 functions in mitochondrial fission.
Inhibition of DLP1 function reduces mitochondrial fragmentation induced by hFis1 overexpression.
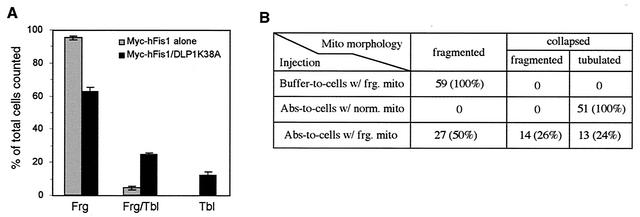
To provide a functional test for hFis1 playing a role in DLP1-mediated mitochondrial fission, we coexpressed Myc-hFis1 with DLP1-K38A, a dominant-negative DLP1 mutant. We predicted that expression of the DLP1 mutant, which normally blocks mitochondrial fission (17), would inhibit mitochondrial fragmentation promoted by Myc-hFis1 overexpression. Consistent with this prediction, we found that expression of DLP1-K38A reduced mitochondrial fragmentation induced by hFis1. When Myc-hFis1 was expressed alone, 95% of Myc-hFis1-expressing cells contained fragmented mitochondria, with 5% having a mixture of tubular and fragmented mitochondria. No cells showed tubular mitochondria exclusively (Fig. 7A, gray bars). However, when Myc-hFis1 was coexpressed with DLP1-K38A, cells with fragmented mitochondria were reduced to 63%, while cells containing tubular mitochondria increased by over sevenfold (37% total; 12% with tubular mitochondria and 25% with a mixture of tubular and fragmented mitochondria) (Fig. 7A, black bars). These results suggest that mitochondrial fragmentation induced by hFis1 expression is suppressed through inhibition of DLP1 function, supporting the finding with yeast that Fis1p and Dnm1p function together in the same mitochondrial fission pathway (15). Although mitochondrial fragmentation in cells coexpressing Myc-hFis1 and DLP1-K38A was reduced, this suppression was incomplete, showing that more than half of the cells contained fragmented mitochondria. This partial suppression suggests that the dominant-negative effect of DLP1-K38A may be incomplete in cells coexpressing Myc-hFis1 and DLP1-K38A. Possibly, the activity of endogenous wild-type DLP1 in DLP1-K38A-expressing cells is sufficient to mediate mitochondrial fragmentation facilitated by hFis1 overexpression. To test whether we could further restore tubular mitochondria from fragmentation, anti-DLP1 antibodies were microinjected into cells expressing Myc-hFis1. A substantial recovery of tubular mitochondria would be expected in the antibody-injected cells, as these DLP1 antibodies inhibit the endogenous DLP1 function (17). When DLP1 antibodies were introduced into Myc-hFis1-expressing cells, 50% of the injected cells exhibited mitochondria collapsed around the nucleus. Among cells with collapsed mitochondria, however, tubular mitochondria were evident in half of the cells while the remaining cells appeared to contain fragmented mitochondria (Fig. 7B). These results are similar to those obtained from coexpression of hFis1 and DLP1-K38A, showing a partial recovery of tubular mitochondria. The reason for the partial recovery is not clear, but it is likely that the extensively fragmented mitochondria in Myc-hFis1-expressing cells are fusion incompetent. Consistent with the fusion incompetence of fragmented mitochondria in hFis1-expressing cells, mitochondrial fragments caused by mutation in yeast Mgm1 have been shown to be incompetent for fusion (29).
FIG. 7.
Inhibition of DLP1 function reduces mitochondrial fragmentation induced by Myc-hFis1 expression. (A) Clone 9 cells with GFP-labeled mitochondria were cotransfected with Myc-hFis1 and DLP1-K38A. Coexpression was detected by immunofluorescence staining with anti-Myc and anti-DLP1 antibodies. Mitochondrial morphology was scored by counting 150 to 200 coexpressing cells on three different coverslips. The number of cells containing either fragmented mitochondria (Frg), a mixture of fragmented and tubular mitochondria (Frg/Tbl), or tubular mitochondria (Tbl) was expressed as percentage of total counted cells. Cells expressing Myc-hFis1 alone are shown as gray bars, and Myc-hFis1- and DLP1-K38A-coexpressing cells are shown as solid black bars. Tubular mitochondria increased in the coexpressing cells, indicating that inhibition of DLP1 function suppressed mitochondrial fragmentation induced by hFis1 expression. Error bars indicate standard deviations. (B) Clone 9 cells carrying RFP-labeled mitochondria were injected with Myc-hFis1 DNA to fragment mitochondria. Fifteen hours later, DLP1-N antibodies were injected into cells displaying fragmented mitochondria to inhibit DLP1 (Abs-to-cells w/frg. mito). Twenty-four percent of these injected cells formed tubulated mitochondria from the fragmented state, indicating a suppression of mitochondrial fragmentation by inhibition of DLP1 function. In the negative control, buffer was injected into cells with fragmented mitochondria (Buffer-to-cells w/frg. mito) and had no effect on mitochondrial fragmentation. For a positive control, DLP1 antibodies were injected into cells with normal mitochondria (Abs-to-cells w/norm. mito), and mitochondria in all injected cells became tubulated and collapsed around the nucleus.
hFis1 and DLP1 form a complex in cells and interact with each other in vitro.
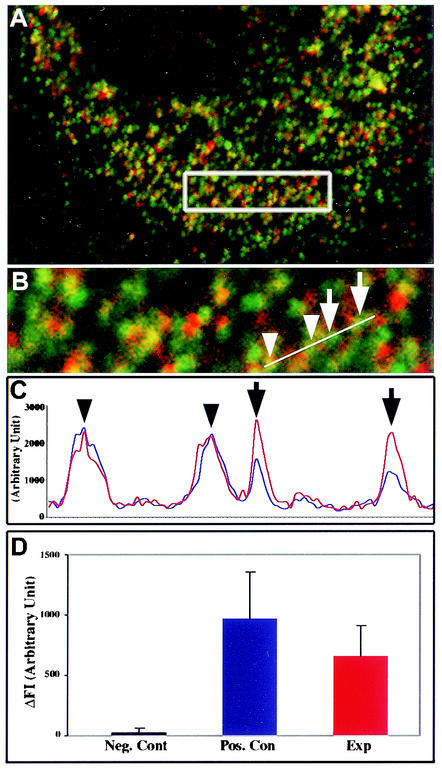
Promotion of DLP1-mediated mitochondrial fission by expression of Myc-hFis1 suggested that hFis1 facilitates targeting of cytosolic DLP1 to the mitochondrial surface, where DLP1 mediates the fission reaction. Alternatively, hFis1 may enhance the GTPase activity of DLP1, thus promoting the fission reaction. These predictions suggest that hFis1 may interact with DLP1. To test whether these two proteins interact with each other, we utilized FRET technology, which is widely used to demonstrate protein-protein interactions in the confines of intact cells (12, 27). As we found that GFP-tagged hFis1 did not behave normally (Fig. 2), DLP1 and Myc-hFis1 were labeled with Cy3 and Cy5 secondary antibodies, respectively. Indirect immunofluorescence with secondary antibodies has been successful for FRET techniques (3, 11). Myc-hFis1 localized to fragmented mitochondrial puncta as shown earlier in Fig. 3 (red channel) (Fig. 8A and B). A population of DLP1 spots associated with mitochondria displaying yellow color, while others distributed to other cytoplasmic locations. After photobleaching of the acceptor fluorophore (Myc-hFis1-Cy5), an increased fluorescence of donor molecules (DLP1-Cy3) was detected, indicating that the two proteins were in close proximity (Fig. 8C). More than 20 cells transfected with Myc-hFis1 were analyzed and consistently produced a positive FRET. The FRET intensity between hFis1 and DLP1 was significant, showing approximately 70% of the level of a positive control in which DLP1 was labeled with both Cy3 and Cy5 (Fig. 8D). However, among the DLP1 puncta colocalized with hFis1, a small number (5 to 12%) of DLP1 spots displayed a large positive FRET, while others showed lower or no FRET. This selective FRET provides an important control for nonspecific FRET and also indicates a transient interaction between the two proteins. The use of indirect immunofluorescence methods for FRET experiments has a potential limitation because the distance of two primary-secondary antibody complexes can be extended as far as 68 nm (3). However, FRET drops to the sixth power of the separation, and virtually no FRET occurs beyond a 14-nm distance (12). Our calculated FRET efficiency was 34.7% in the experimental set compared to 42.5% in the positive control, indicating very close proximity between two fluorophores. Considering the random orientation of the antibodies to each other and the strong positive FRET in our experiments, hFis1 and DLP1 are much closer to each other than the possible 82-nm separation. Although convincing, these data do not preclude the association of hFis1 and DLP1 within a complex that includes additional proteins. Regardless of whether they interact directly or indirectly, these data demonstrated that the two proteins are in very close proximity to each other.
FIG. 8.
FRET between hFis1 and DLP1. (A) DLP1 and Myc-hFis1 were labeled with secondary antibodies conjugated to Cy3 (green channel) and Cy5 (red channel) as the donor and acceptor pair, respectively, in COS-7 cells transfected with Myc-hFis1. (B) Four DLP1 spots (arrows and arrowheads) were analyzed for FRET by the donor quenching method (see Materials and Methods). (C) Donor fluorescence (Cy3-DLP1) was measured before (blue line) and after (red line) photobleaching of the acceptor molecule (Cy5) in the boxed region in panel A. Cy3 fluorescence of two DLP1 spots (arrows) significantly increased after the acceptor photobleaching, while the other DLP1 spots (arrowheads) showed no change. (D) FRET intensity was obtained by averaging the difference of fluorescence values (ΔFI [Cy3post minus Cy3pre]) of DLP1 spots showing positive FRET (red bar). Thirty to forty DLP1 spots from negative and positive controls and the experiment (Exp) were subjected to the analysis. DLP1 and dynamin labeled with Cy3 and Cy5, respectively, were used as a negative control. For a positive control, DLP1 was labeled with both Cy3 and Cy5 and analyzed for FRET.
To support the in vivo FRET data, coimmunoprecipitation experiments were performed from cell lysates. Our initial attempt to detect this interaction by coimmunoprecipitation was inconclusive, suggesting that the interaction might be weak and/or transient as predicted from our FRET results. To detect a potentially transient and weak interaction, we increased the cellular hFis1 level by transfecting BHK-21 cells with Myc-hFis1 and treated them with the membrane-permeant cross-linker DSP. Cells were solubilized, and the homogenates were subjected to immunoprecipitation. As shown in Fig. 9A, anti-DLP1 antibodies immunoprecipitated Myc-hFis1 from lysates of cross-linked cells. A reciprocal immunoprecipitation with anti-hFis1 antibodies also pulled down DLP1. As the levels of Myc-hFis1 and DLP1 coimmunoprecipitated are not extensive, it is likely that this interaction is transient as predicted by the FRET studies. Note that coimmunoprecipitated Myc-hFis1 and DLP1 bands in Fig. 9A were overexposed for better display and thus are not quantitative. Along with the FRET data, the coimmunoprecipitation of hFis1 and DLP1 indicates that these two proteins are in the same complex. However, it is possible that this interaction may involve other components such as a mammalian Mdv1p-like protein, as suggested in yeast studies (26).
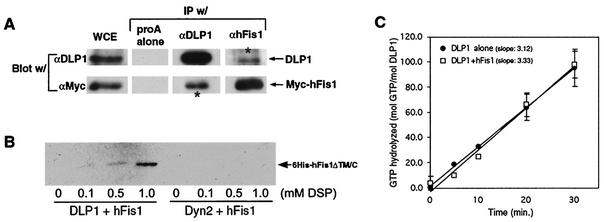
FIG. 9.
hFis1 and DLP1 interact with each other. (A) BHK-21 cells were transfected with Myc-hFis1 and cross-linked by DSP. The cell lysate (whole-cell extract [WCE]) was subjected to immunoprecipitation (IP) by anti-DLP1 (αDLP1) or anti-hFis1 antibodies. Immunoprecipitated proteins were analyzed by immunoblotting with anti-DLP1 and anti-Myc antibodies. Immune complexes isolated by DLP1 or hFis1 antibodies contained Myc-hFis1 or DLP1, respectively (asterisks), indicating that the two proteins were in the same complex. (B) Purified recombinant His6-tagged hFis1ΔTM/C was incubated with either DLP1 or conventional dynamin (Dyn2) in the presence of increasing concentrations of the cross-linker DSP. DLP1 or Dyn2 was isolated by immunoprecipitation with anti-DLP1 or antidynamin antibodies, respectively. Coimmunoprecipitation of hFis1 was assessed with anti-His antibodies. His6-hFis1ΔTM/C was detected in the DLP1 immunoprecipitate in the presence of DSP, showing the most in 1.0 mM DSP. No hFis1 was present in the dynamin immunoprecipitate even in 1.0 mM DSP. (C) The GTPase activity of DLP1 was assayed by thin-layer chromatography. Hydrolyzed GTP was calculated by the ratios between GDP and total nucleotide (GTP plus GDP) and plotted for reaction periods. The rate of GTP hydrolysis is expressed as the slope of average trendlines in the graph. DLP1 alone has a rate of 3.1 mol of GTP/min/mol of DLP1, and addition of equal moles of purified hFis1 to DLP1 did not significantly affect the GTPase activity of DLP1, showing 3.3 mol of GTP/min/mol of DLP1. Error bars indicate standard deviations.
To further test the possibility that hFis1 interacts with DLP1 directly, we used purified recombinant proteins and again employed the cross-linking reagent to stabilize a possible transient interaction. His6-tagged DLP1 and hFis1 were expressed and purified by using a bacterial system. For the ease of purification, we used a truncated form of hFis1 in which the transmembrane domain and 5-amino-acid C-terminal tail were deleted, as the hFis1-DLP1 interaction is likely through the N-terminal region of hFis1. After incubation of the two purified proteins, DLP1 was isolated from the reaction mixture by immunoprecipitation and examined if hFis1 was coprecipitated with DLP1. As shown in Fig. 9B, hFis1 was isolated consistently along with DLP1 in the presence of the cross-linker, indicating that hFis1 and DLP1 interact with each other in vitro. In a control experiment, purified conventional dynamin (Dyn2) was used to show that random cross-linking between two proteins did not contribute to the hFis1-DLP1 association. Dynamin did not associate with hFis1, indicating that hFis1 specifically interacts with DLP1.
Because DLP1 and hFis1 appear to interact with each other, we tested whether this interaction could alter the GTPase activity of DLP1. The same recombinant proteins were used in GTP hydrolysis assays to test whether hFis1 functions as an enzymatic cofactor for DLP1. We found that addition of hFis1 did not yield any significant effect on GTP hydrolysis by DLP1 (Fig. 9C). Without hFis1, DLP1 hydrolyzed GTP at an average rate of 3.1 mol of GTP/min/mol of DLP1. Addition of an equal molar concentration of purified hFis1 to the reaction mixture resulted in no significant change in GTP hydrolysis (Fig. 7B). Further attempts with different molar ratios between DLP1 and hFis1 did not influence GTP hydrolysis by DLP1 (results not shown). However, we cannot rule out that other factors might have contributed to the inability of hFis1 to affect GTPase activity. These include a possible involvement of other proteins, the lack of a transmembrane domain in hFis1, potential incorrect folding or aggregation of hFis1, and the lack of membranes in the assay. Regardless, these results suggest that hFis1 alone does not enhance the GTPase activity of DLP1 in a significant fashion.
DISCUSSION
In this study, we have cloned a human homologue of yeast Fis1p, hFis1, and examined its function in mitochondrial morphology maintenance. We found that cells expressing an increased level of hFis1 contained completely fragmented mitochondria, suggesting that mitochondrial fission was promoted. This phenomenon was not observed in previous studies and provided useful information that the frequency of mitochondrial fission may be controlled by the number of hFis1 molecules on the mitochondrial surface. We were also able to detect an interaction between DLP1 and hFis1 by coimmunoprecipitation and FRET experiments. Further, DLP1 and hFis1 were able to interact directly, as the two purified proteins associated together in vitro. However, this interaction did not appear to change the GTPase activity of DLP1. These results suggest that hFis1 participates in mitochondrial fission by recruiting DLP1 from the cytosol.
Structural requirements of hFis1 for mitochondrial localization and fission.
Our data obtained by using epitope tags and truncations of hFis1 provide useful information regarding the structural requirement of hFis1. Studies with yeast Fis1p indicate that the N-terminal region of hFis1 is exposed to the cytosol and that the short C-terminal tail protrudes into the intermembrane space of mitochondria (15). We have found that tagging with either GFP or Myc at the C terminus or truncating the 5-amino-acid C-terminal tail of hFis1 disrupted mitochondrial localization, resulting in a diffuse distribution of hFis1 (Fig. 2 and 5). These results suggest that the intact C-terminal structure of hFis1 is essential for proper distribution of this protein. Further, while either the GFP or Myc tag at the N terminus did not interfere with the mitochondrial localization of hFis1, these N-terminally tagged proteins did affect mitochondrial morphology in different ways (Fig. 2 and 3). Myc-hFis1 promoted mitochondrial fission, whereas GFP-hFis1 appeared to be nonfunctional, failing to promote mitochondrial fission. In cells expressing Myc-hFis1, mitochondria were completely fragmented, and it was confirmed by expressing untagged hFis1 that this phenotype was the functional consequence of the increased amount of hFis1 and was not due to an artifact from the Myc tag (Fig. 3).
In some cells overexpressing GFP-hFis1, mitochondria were drastically deformed into large aggregated clumps (Fig. 2C and D). It is possible that these mitochondrial clumps are the result of mitochondrial fusion due to the inhibition of DLP1-mediated fission by nonfunctional hFis1. However, this clumped mitochondrial morphology is different from the one induced by a direct inhibition of DLP1 function. When DLP1 was inhibited, mitochondria became elongated and entangled around the nucleus, sometimes showing a net-like morphology (17, 23, 24). Electron microscopy of cells overexpressing GFP-hFis1 showed that mitochondria were aggregated laterally to form clusters of discrete mitochondria, suggesting that these mitochondria were not fused (results not shown). It is known that GFP molecules dimerize in a low-ionic-strength environment (30). It is possible that overexpressed GFP-hFis1 molecules on mitochondrial surfaces cause uncontrolled lateral interaction of mitochondria by dimerization of GFP moieties. This nonfunctionality of GFP-hFis1 is likely caused by the large GFP tag at the N terminus, because the small Myc tag did not interfere with the function of hFis1. The importance of the N-terminal region in hFis1 function was further supported by studies of N-terminal truncations (Fig. 5). We found that deletion of the first 31 amino acids of hFis1 abolished the ability of hFis1 to promote mitochondrial fission, suggesting that this region contains a functional motif necessary to mediate mitochondrial fission. However, while the N-terminal region of hFis1 is required for mitochondrial fission, the vast majority of this domain is not needed for the mitochondrial localization, because the truncation of the N-terminal 90 amino acids still distributed to mitochondria. Taken together, the data indicate that the region mediating mitochondrial fission resides within the first 31 amino acids of hFis1 whereas mitochondrial localization requires the transmembrane domain and an intact C-terminal tail structure.
Role of hFis1 in mitochondrial fission.
Overexpression of functional hFis1 promotes mitochondrial fission, resulting in fragmented mitochondria (Fig. 3). It has been shown by genetic and morphological analyses that yeast Fis1p participates in the Dnm1p-mediated mitochondrial fission pathway (4, 15, 25). Possible functions of Fis1p in mitochondrial fission have been proposed: (i) Fis1p regulates proper assembly of Dnm1p-containing complexes on the mitochondrial membrane, or (ii) Fis1p activates the fission activity of Dnm1p complexes in the later stage. Our results are consistent with these proposed roles of Fis1p, since mitochondrial fragmentation was induced upon overexpression of hFis1. In mammalian cells, DLP1 is more abundant in the cytoplasm than on mitochondria at steady state (31). This suggests that the association of DLP1 with mitochondria is transient and likely to be highly regulated. Our in vivo and in vitro coimmunoprecipitation and FRET experiments clearly demonstrate an interaction between hFis1 and DLP1 (Fig. 8 and 9). However, this interaction appears to be unstable, as no interaction is detected without cross-linking, indicating that this interaction is indeed extremely transient. It has been proposed that GTP-bound DLP1 associates with membrane (e.g., mitochondria) and, upon release of the nucleotide, DLP1 dissociates from the membrane (32). Further, like dynamin, DLP1 is likely to have a low affinity for GTP and a high rate of GTP hydrolysis and release (28), predicting a transient association of DLP1 with membranes.
If a DLP1 association with the mitochondrial membrane involves hFis1, it is possible that DLP1 binds to hFis1 when GTP is bound. However, when GTP or GTPγS was added in our binding experiments, the hFis1-DLP1 interaction was not affected (results not shown), suggesting that this interaction is not nucleotide dependent. However, it cannot be ruled out that hFis1 and specific lipid molecules cooperatively recruit DLP1 to the mitochondrial surface in a nucleotide-dependent manner or that yet-unidentified components are also involved. Recently, it was reported that yeast Mdv1p, another protein that participates in Dnm1p-mediated mitochondrial fission, functions as a molecular adaptor that links Dnm1p and Fis1p (26). It is possible that an Mdv1p-like protein in mammalian cells is required to facilitate the hFis1-DLP1 interaction. Because DLP1 is able to bind directly to membranes, as purified DLP1 binds and tubulates synthetic liposomes (32), it is likely that hFis1 mediates a highly regulated and specific targeting of DLP1 to the mitochondrial surface.
We propose that hFis1 is a limiting factor in mitochondrial fission and that the number of hFis1 molecules on the mitochondrial surface determines fission frequency. Several lines of evidence support this hypothesis. First, only a slight increase in the expression of hFis1 induces fragmentation of mitochondria. Second, DLP1 is already abundant in the cytosol, and further overexpression of DLP1 does not result in fragmented mitochondria (17). Third, increased levels of hFis1 in cells expressing a dominant-negative DLP1 (DLP1-K38A) still fragment mitochondria, indicating that the endogenous DLP1 can mediate mitochondrial fission in response to the increased hFis1. Finally, overexpression of N-terminal truncations of hFis1 did not inhibit mitochondrial fission, suggesting that endogenous hFis1 levels are sufficient to carry out normal fission to maintain normal mitochondrial morphology. Therefore, hFis1 is likely to function as a DLP1-recruiting molecule. Detection of the interaction between DLP1 and hFis1 supports a targeting role for hFis1 (Fig. 8 and 9). Results for yeast also support that Fis1p recruits Dnm1p to mitochondria (15): first, Dnm1p distribution to mitochondria was reduced in fis1Δ mutants (>8 puncta versus 1 to 3 puncta), and second, in immunogold labeling for Dnm1p, fis1Δ mutants had a majority of Dnm1p-gold particles in the cytoplasm (92% in cytoplasm and 8% in mitochondria) whereas wild type cells showed a completely reversed distribution (7.5% in cytoplasm and 90.4% in mitochondria). It has been suggested that Fis1p functions in yeast during the late stage of the fission reaction by activating the fission activity of Dnm1p complexes on the mitochondrial surface. Although the hFis1-DLP1 interaction did not affect the GTPase activity of DLP1 in our GTP hydrolysis assay (Fig. 9), it is possible that hFis1 activates the fission reaction in a different way (e.g., through changes in the membrane binding affinity, composition, or structure of the DLP1 complex). Given the high sensitivity of mitochondrial fission to the cellular hFis1 level, the expression of hFis1 may be tightly regulated to prevent excessive fragmentation of mitochondria.
It is interesting that the recovery of tubular mitochondria by inhibition of DLP1 function was incomplete in hFis1-expressing cells (Fig. 7). This indicates that excessively fragmented mitochondria are fusion incompetent. Formation of fusion-incompetent mitochondrial fragments was observed in yeast cells harboring mgm1 or fzo1 mutations. In these cells, mitochondria lost their DNA and respiration capacity (5, 9, 18, 21, 29), indicating functional defects of fragmented mitochondria. Our electron microscopy results showing a decreased electron density of mitochondrial matrices and loss of cristae in Myc-hFis1-expressing cells also suggest a functional compromise of fragmented mitochondria (Fig. 4). These results support the premise that the proper control of mitochondrial morphology is essential for mitochondrial function.
Acknowledgments
We thank Bing Huang for helping with electron microscope sample preparation and James Orth and Heather Thompson for critical reading.
This work was supported by grants from the National Institute of Diabetes and Digestive and Kidney Diseases to Y. Yoon (DK 02648) and to M. A. McNiven (DK 44650)
REFERENCES
- 1.Bereiter-Hahn, J., and M. Voth. 1994. Dynamics of mitochondria in living cells: shape changes, dislocation, fusion, and fission of mitochondria. Microsc. Res. Technol. 27:198-219. [DOI] [PubMed] [Google Scholar]
- 2.Cerveny, K. L., J. M. McCaffery, and R. E. Jensen. 2001. Division of mitochondria requires a novel DNM1-interacting protein, Net2p. Mol. Biol. Cell 12:309-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dictenberg, J. B., W. Zimmerman, C. A. Sparks, A. Young, C. Vidair, Y. Zheng, W. Carrington, F. S. Fay, and S. J. Doxsey. 1998. Pericentrin and gamma-tubulin form a protein complex and are organized into a novel lattice at the centrosome. J. Cell Biol. 141:163-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fekkes, P., K. A. Shepard, and M. P. Yaffe. 2000. Gag3p, an outer membrane protein required for fission of mitochondrial tubules. J. Cell Biol. 151:333-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guan, K., L. Farh, T. K. Marshall, and R. J. Deschenes. 1993. Normal mitochondrial structure and genome maintenance in yeast requires the dynamin-like product of the MGM1 gene. Curr. Genet. 24:141-148. [DOI] [PubMed] [Google Scholar]
- 6.Hales, K. G., and M. T. Fuller. 1997. Developmentally regulated mitochondrial fusion mediated by a conserved, novel, predicted GTPase. Cell 90:121-129. [DOI] [PubMed] [Google Scholar]
- 7.Henley, J. R., E. W. A. Krueger, B. J. Oswald, and M. A. McNiven. 1998. Dynamin-mediated internalization of caveolae. J. Cell Biol. 141:85-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henley, J. R., and M. A. McNiven. 1996. Association of a dynamin-like protein with the Golgi apparatus in mammalian cells. J. Cell Biol. 133:761-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hermann, G. J., J. W. Thatcher, J. P. Mills, K. G. Hales, M. T. Fuller, J. Nunnari, and J. M. Shaw. 1998. Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p. J. Cell Biol. 143:359-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Imoto, M., I. Tachibana, and R. Urrutia. 1998. Identification and functional characterization of a novel human protein highly related to the yeast dynamin-like GTPase Vps1p. J. Cell Sci. 111:1341-1349. [DOI] [PubMed] [Google Scholar]
- 11.Kam, Z., T. Volberg, and B. Geiger. 1995. Mapping of adherens junction components using microscopic resonance energy transfer imaging. J. Cell Sci. 108:1051-1062. [DOI] [PubMed] [Google Scholar]
- 12.Kenworthy, A. K. 2001. Imaging protein-protein interactions using fluorescence resonance energy transfer microscopy. Methods 24:289-296. [DOI] [PubMed] [Google Scholar]
- 13.Koch, A., M. Thiemann, M. Grabenbauer, Y. Yoon, M. A. McNiven, and M. Schrader. 2003. Dynamin-like protein 1 is involved in peroxisomal fission. J. Biol. Chem. 278:8597-8605. [DOI] [PubMed] [Google Scholar]
- 14.Li, X., and S. J. Gould. 2003. The dynamin-like GTPase DLP1 is essential for peroxisome division and is recruited to peroxisomes in part by PEX11. J. Biol. Chem., 278:17012-17020. [DOI] [PubMed]
- 15.Mozdy, A. D., J. M. McCaffery, and J. M. Shaw. 2000. Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J. Cell Biol. 151:367-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Otsuga, D., B. R. Keegan, E. Brisch, J. W. Thatcher, G. J. Hermann, W. Bleazard, and J. M. Shaw. 1998. The dynamin-related GTPase, Dnm1p, controls mitochondrial morphology in yeast. J. Cell Biol. 143:333-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pitts, K. R., Y. Yoon, E. W. Krueger, and M. A. McNiven. 1999. The dynamin-like protein DLP1 is essential for normal distribution and morphology of the endoplasmic reticulum and mitochondria in mammalian cells. Mol. Biol. Cell. 10:4403-4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rapaport, D., M. Brunner, W. Neupert, and B. Westermann. 1998. Fzo1p is a mitochondrial outer membrane protein essential for the biogenesis of functional mitochondria in Saccharomyces cerevisiae. J. Biol. Chem. 273:20150-20155. [DOI] [PubMed] [Google Scholar]
- 19.Santel, A., and M. T. Fuller. 2001. Control of mitochondrial morphology by a human mitofusin. J. Cell Sci. 114:867-874. [DOI] [PubMed] [Google Scholar]
- 20.Sesaki, H., and R. E. Jensen. 1999. Division versus fusion: Dnm1p and Fzo1p antagonistically regulate mitochondrial shape. J. Cell Biol. 147:699-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shepard, K. A., and M. P. Yaffe. 1999. The yeast dynamin-like protein, Mgm1p, functions on the mitochondrial outer membrane to mediate mitochondrial inheritance. J. Cell Biol. 144:711-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shin, H. W., C. Shinotsuka, S. Torii, K. Murakami, and K. Nakayama. 1997. Identification and subcellular localization of a novel mammalian dynamin-related protein homologous to yeast Vps1p and Dnm1p. J. Biochem. 122:525-530. [DOI] [PubMed] [Google Scholar]
- 23.Smirnova, E., L. Griparic, D. L. Shurland, and A. M. van der Bliek. 2001. Dynamin-related protein drp1 is required for mitochondrial division in mammalian cells. Mol. Biol. Cell 12:2245-2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smirnova, E., D. L. Shurland, S. N. Ryazantsev, and A. M. van der Bliek. 1998. A human dynamin-related protein controls the distribution of mitochondria. J. Cell Biol. 143:351-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tieu, Q., and J. Nunnari. 2000. Mdv1p is a WD repeat protein that interacts with the dynamin-related GTPase, Dnm1p, to trigger mitochondrial division. J. Cell Biol. 151:353-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tieu, Q., V. Okreglak, K. Naylor, and J. Nunnari. 2002. The WD repeat protein, Mdv1p, functions as a molecular adaptor by interacting with Dnm1p and Fis1p during mitochondrial fission. J. Cell Biol. 158:445-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Truong, K., and M. Ikura. 2001. The use of FRET imaging microscopy to detect protein-protein interactions and protein conformational changes in vivo. Curr. Opin. Struct. Biol. 11:573-578. [DOI] [PubMed] [Google Scholar]
- 28.Warnock, D. E., and S. L. Schmid. 1996. Dynamin GTPase, a force-generating molecular switch. Bioessays 18:885-893. [DOI] [PubMed] [Google Scholar]
- 29.Wong, E. D., J. A. Wagner, S. W. Gorsich, J. M. McCaffery, J. M. Shaw, and J. Nunnari. 2000. The dynamin-related GTPase, Mgm1p, is an intermembrane space protein required for maintenance of fusion competent mitochondria. J. Cell Biol. 151:341-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang, F., L. G. Moss, and G. N. Phillips, Jr. 1996. The molecular structure of green fluorescent protein. Nat. Biotechnol. 14:1246-1251. [DOI] [PubMed] [Google Scholar]
- 31.Yoon, Y., K. R. Pitts, S. Dahan, and M. A. McNiven. 1998. A novel dynamin-like protein associates with cytoplasmic vesicles and tubules of the endoplasmic reticulum in mammalian cells. J. Cell Biol. 140:779-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoon, Y., K. R. Pitts, and M. A. McNiven. 2001. Mammalian dynamin-like protein DLP1 tubulates membranes. Mol. Biol. Cell 12:2894-2905. [DOI] [PMC free article] [PubMed] [Google Scholar]