Abstract
The Mre11-Rad50-Nbs1(Xrs2) complex and the Ku70-Ku80 heterodimer are thought to compete with each other for binding to DNA ends. To investigate the mechanism underlying this competition, we analyzed both DNA damage sensitivity and telomere overhangs in Schizosaccharomyces pombe rad50-d, rad50-d pku70-d, rad50-d exo1-d, and pku70-d rad50-d exo1-d cells. We found that rad50 exo1 double mutants are more methyl methanesulfonate (MMS) sensitive than the respective single mutants. The MMS sensitivity of rad50-d cells was suppressed by concomitant deletion of pku70+. However, the MMS sensitivity of the rad50 exo1 double mutant was not suppressed by the deletion of pku70+. The G-rich overhang at telomere ends in taz1-d cells disappeared upon deletion of rad50+, but the overhang reappeared following concomitant deletion of pku70+. Our data suggest that the Rad50 complex can process DSB ends and telomere ends in the presence of the Ku heterodimer. However, the Ku heterodimer inhibits processing of DSB ends and telomere ends by alternative nucleases in the absence of the Rad50-Rad32 protein complex. While we have identified Exo1 as the alternative nuclease targeting DNA break sites, the identity of the nuclease acting on the telomere ends remains elusive.
While a DNA double-strand break (DSB) within a chromosome must be repaired to prevent cell death, a chromosome end is not recognized as DNA damage and is thus protected from the action of repair enzymes. It was therefore surprising when it was shown that several DNA repair proteins, including Mre11 and Ku, are involved in both DNA DSB repair and telomere maintenance (23, 24). Telomeres, the natural DNA ends of eukaryotic chromosomes (9), are stable and do not fuse with other chromosome ends. So that telomeres can be treated as specialized DNA structures and not as DNA damage, they are composed of repetitive DNA elements and associated with specialized proteins including human TRF1 and TRF2, Schizosaccharomyces pombe Taz1p or Saccharomyces cerevisiae Rap1 (4, 10, 11, 31, 62). Disruption of telomere architecture caused by the deletion of S. pombe taz1+, for example, leads to massive telomere elongation and Ku-dependent end-to-end fusions (11, 20).
In eukaryotic cells, DSBs are mainly repaired either by homologous recombination (HR) or nonhomologous end joining (NHEJ) (12-14, 23, 24, 27). In the yeast S. cerevisiae, DNA DSBs are predominantly repaired by HR, which requires genes of the Rad52 epistasis group (50). The first step of HR is single-stranded DNA (ssDNA) end resection in a 5′ to 3′ direction to form long 3′ single-stranded tails (53). Although the Mre11-Rad50-Xrs2 complex is thought to participate in this step (29), Mre11 exhibits exonuclease activity of the opposite polarity under in vitro conditions, namely 3′ to 5′ exonuclease activity against both ssDNA and double-stranded DNA (dsDNA) (21, 40, 51, 61). Mre11 also displays ssDNA endonuclease activity. Consistent with this observation, mutants in the Mre11 nuclease motif are not as sensitive to ionized radiation (IR) as would be expected for mutants in an enzyme required for end processing (40). Furthermore, the observation that overexpression of exonuclease I (Exo1) partially suppress the DNA damage sensitivity of Mre11 mutants (30, 41, 58) suggests that Exo1 acts redundantly with Mre11 in end processing.
In contrast to the situation in yeast, the major mechanism for the repair of radiation-induced DSBs in higher eukaryotes is NHEJ. The Ku70-Ku80 heterodimer, DNA-PKcs, and a DNA ligase IV-Xrcc4 complex are all required for this process (19, 22, 63). NHEJ is not a major mechanism of DNA repair in S. cerevisiae, but yeast Ku70, Ku80, and Lig4 are essential for the repair of plasmid DSBs following the transformation of the linearized plasmid into cells (38, 60). In S. cerevisiae, the IR sensitivity of mre11 null mutants is partially suppressed by loss of YKU70 (7). Furthermore, the rate of 5′ to 3′ degradation of HO-induced DSBs is decreased by deletion of MRE11 and increased by deletion of YKU70. These data have led to the development of a model in which the Ku pathway competes with 5′ to 3′ exonuclease at DNA ends (29). However, it is unclear which nuclease is competing with Yku70. In contrast to IR sensitivity, the methyl methanesulfonate (MMS) sensitivity of mre11 null mutants is not suppressed by the loss of YKU70 (38). The yku70 single mutant is not MMS sensitive, which indicates that Ku heterodimers play no role in the repair of MMS-induced DNA damage (52, 60).
Ku has also been characterized in the fission yeast S. pombe. The Ku heterodimer is required for NHEJ of transformed linear plasmids and for the maintenance of correct telomere length (3, 32, 39). In contrast to S. cerevisiae Yku70, fission yeast Ku70 does not accumulate in telomeric foci. Moreover, deletion of the fission yeast gene encoding Ku70 (i.e., pku70+) does not overcome telomere silencing, which indicates that the function of Ku at telomere ends is not fully conserved between fission and budding yeasts (6, 32, 35). S. pombe Rad50 and Rad32 (a homolog of S. cerevisiae Mre11) are both involved in telomere length maintenance and DNA repair (25, 32, 56, 65, 66), but, in contrast to their homologues in S. cerevisiae, they are not required for the delay to S-phase progression that occurs upon treatment with hydroxyurea (HU) or MMS (25, 33). Although the cellular function of the Rad32-Rad50 protein complex is still not understood, recent data suggest that, during DNA replication, the complex is required to cleave hairpin structures, which would be produced from palindromic sequences during DNA synthesis of the lagging strand (18).
The Rad50 complex is required for both HR and NHEJ in S. cerevisiae (23), making it difficult to study the exact roles of the Rad50 complex and Ku heterodimer in HR repair separately from their effects on NHEJ. However, it is possible to study the role of the S. pombe Rad50 complex in HR repair because the S. pombe Rad50 complex is not required for NHEJ (32). We report here an investigation of the different roles of the Rad50-Rad32 protein complex and the Ku heterodimer at DSB ends generated in response to DNA damage and at telomere ends in taz1-d cells. Our data suggest that both types of DNA ends are mainly processed in a manner dependent on the Rad32-Rad50 complex. We also provide evidence suggesting that, in the absence of the Rad32-Rad50 complex (which appears to play the primary role), the Ku70/80 heterodimer has to be removed from either type of DNA end to provide access for alternative end-processing pathways. Our data strongly indicate that exonuclease 1 (Exo1) is one of these alternative nucleases. Interestingly, Exo1 seems to act only on DNA ends generated by DNA damage and not on telomeric DNA ends, which are targeted by another as yet unidentified nuclease.
MATERIALS AND METHODS
Growth medium.
S. pombe cells were grown in YPAD medium (1% yeast extract, 2% polypeptone, 2% glucose, 20 μg of adenine per ml), YEA medium (0.5% yeast extract, 3% glucose, 20 μg of adenine per ml) or Edinburgh minimal medium with required supplements (42).
Strain construction.
The rad50+ gene was disrupted by replacing the region between the first HindIII restriction site and second HindIII site (nucleotide positions 529 to 1158, relative to initiation codon) with either the ura4+ or the LEU2+ gene (Table 1). Standard methods were used to create the disrupted constructs, and linear fragments were transformed into a wild-type strain (JY741) (42). rad32-d, trt1-d and taz1-d cells were constructed by insertion of the ura4+ or LEU2+ cassette into the HindIII, BglII, and PstI sites, respectively. pku70-d, pku80-d and lig4-d cells were constructed by insertion of the LEU2+ cassette into the EcoRV, EcoRV and NcoI sites, respectively. To construct pku70::leu2::ade6+ cells, the ade6+cassette was inserted into the EcoRV site in the LEU2 gene in the pku70::LEU2 disruption fragment. rad50 taz1 and rad50 pku70 double mutants were constructed by transformation of taz1 and pku70 mutants with the rad50 disruption fragment. The rad32 taz1 double mutant was constructed by transforming rad32-d cells with a taz1 disruption fragment. rhp51-d cells were constructed by insertion of the ura4+ cassette in the NheI site of the rhp51+ gene. To tag Rad32 with the Myc epitope at the C terminus, we amplified the rad32+ open reading frame by PCR with a primer set of Rad32T (5′-GCATACCCGGGATCATCTAAAATTTCGTCATCC-3′) and Rad32B (5′-GCATACCCGGGATCATCTAAAATTTCGTCATCC-3′), with wild-type genomic DNA used as a template. The SmaI cut PCR fragment was then cloned into SmaI cut pFA6a-13Myc-kanMX6, which contained 13 copies of the Myc epitope and a kanMX6 marker. pFA6a-13Myc-kanMX6 was provided by J. R. Pringle (University of North Carolina) (1). The resulting plasmid was linearized with BsaBI and used for transformation. Other double and triple mutants were constructed by genetic crossing.
TABLE 1.
S. pombe strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| 119 | h− smt-0 leu1-32 ura4-D18 his3-D1 arg3-D1 | Lab stock |
| 142 | h+ leu1-32 ura4-D18 ade6-M210 rad32::ura4+ | This work |
| 302 | smt-0 leu1-32 ura4-D18 his3-D1 arg3-D1 exo1-1::ura4+ | This work |
| 315 | smt-0 leu1-32 ura4-D18 his3-D1 arg3-D1 pku70::LEU2 rad32::ura4+ | This work |
| 319 | smt-0 leu1-32 ura4-D18 his3-D1 arg3-D1 pku70::LEU2 | This work |
| 324 | smt-0 leu1-32 ura4-D18 his3-D1 arg3-D1 rad32::ura4+ | This work |
| 394 | smt-0 leu1-32 ura4-D18 his3-D1 arg3-D1 pku70::LEU2 exo1-1::ura4+ | This work |
| 403 | smt-0 leu1-32 ura4-D18 his3-D1 arg3-D1 exo1-1::ura4+ rad32::ura4+ | This work |
| 407 | smt-0 leu1-32 ura4-D18 his3-D1 arg3-D1 exo1-1::ura4+ rad32::ura4+ pku70::LEU2 | This work |
| 435 | h+ TAP-rad50 ura4-D18 ade6-704 leu1-32 | Lab stock |
| 700 | h+ leu1-32 ura4-D18 | Lab stock |
| 701 | smt-0 ura4-D18 rad50::kanMX6 | Lab stock |
| 702 | h+ leu1-32 ura4-D18 exo1::ura4+ | Lab stock |
| 703 | h+ ura4-D18 exo1::ura4+ rad50::kanMX6 | This work |
| 704 | h+ leu1-32 ura4-D18 ade6-M216 rhp51::ura4+ | Lab stock |
| KT00c | h− leu1-32 ura4-D18 ade6-M216 rhp51::ura4+ | This work |
| KT00g | h− leu1-32 ura4-D18 ade6-M216 exo1::ura4+ | This work |
| KT001 | h− leu1-32 ura4-D18 ade6-M216 taz1::ura4+ | This work |
| KT002 | h− leu1-32 ura4-D18 ade6-M216 rad50::ura4+ | This work |
| KT007 | h− leu1-32 ura4-D18 ade6-M216 trt1::ura4+ | This work |
| KT0106M1 | h− leu1-32 rad32D25A taz1::LEU2 | This work |
| KT0106M2 | h− leu1-32 rad32D25A taz1::LEU2 | This work |
| KT016g5 | h− leu1-32 ura4-D18 ade6 taz1::LEU2 rad32::ura4+ exo1::ura4+ pku70::leu2::ade6+ | This work |
| KT02g | h− leu1-32 ura4-D18 ade6-M216 exo1::ura4+ rad50::LEU2 | This work |
| KT021 | h− leu1-32 ura4-D18 ade6-M216 rad50::LEU2 taz1::ura4+ | This work |
| KT0215 | h− leu1-32 ura4-D18 ade6 rad50::LEU2 taz1::ura4+ pku70::leu2::ade6+ | This work |
| KT090 | h− leu1-32 ura4-D18 ade6-M216 pku80::LEU2 | This work |
| KT1a0 | h+ leu1-32 ura4-D18 ade6-M210 lig4::LEU2 | This work |
| KT1a2 | h+ leu1-32 ura4-D18 ade6-M210 lig4::LEU2 rad50::ura4+ | This work |
| KT10c5 | h+ leu1-32 ura4-D18 ade6 rhp51::ura4+ pku70::leu2::ade6+ | This work |
| KT110 | h+ leu1-32 ura4-D18 ade6-M210 taz1::LEU2 | This work |
| KT116 | h+ leu1-32 ura4-D18 ade6-M210 taz1::LEU2 rad32::ura4+ | This work |
| KT1165 | h+ leu1-32 ura4-D18 ade6 taz1::LEU2 rad32::ura4+ pku70::leu2::ade6+ | This work |
| KT117 | h+ leu1-32 ura4-D18 ade6-M210 taz1::LEU2 trt1::ura4+ | This work |
| KT12g5 | h+ leu1-32 ura4-D18 ade6- exo1::ura4+ rad50::LEU2 pku70::leu2::ade6+ | This work |
| KT120 | h+ leu1-32 ura4-D18 ade6-M216 rad50::LEU2 | This work |
| KT121g5 | h+ leu1-32 ura4-D18 ade6 rad50::LEU2 taz1::ura4+ exo1::ura4+ pku70::leu2::ade6+ | This work |
| KT192 | h+ leu1-32 ura4-D18 ade6-M216 pku80::LEU2 rad50::ura4+ | This work |
| KT152 | h+ leu1-32 ura4-D18 ade6-M216 rad50::ura4+ pku70::LEU2 | This work |
| KT156 | h+ leu1-32 ura4-D18 ade6-M216 rad32::ura4+ pku70::LEU2 | This work |
| KTt2T6M | h+ leu1-32 ura4-D18 ade6-704 TAP-rad50 rad32:13Myc:kanMX6 | This work |
| KTt2T6MM1 | h− leu1-32 TAP-rad50 rad32D25A:13Myc:kanMX6 | This work |
| KTt6M | h+ leu1-32 ura4-D18 ade6-M210 rad32:13Myc:kanMX6 | This work |
| KTt6MM1 | h− leu1-32 rad32D25A:13Myc:kanMX6 | This work |
| JY741 | h− leu1-32 ura4-D18 ade6-M216 | M. Yamamoto |
| pku70L | h− leu1-32 ura4-D18 ade6-M216 pku70::LEU2 | This work |
| pku70A | h+ leu1-32 ura4-D18 ade6-M216 pku70::leu2::ade6+ | This work |
In-gel hybridization.
In-gel hybridization analysis was performed according to the protocol previously published (16), i.e., by using a G-rich probe (5′-GATCG GGTTACAAGGTTACGTGGTTACACG-3′) and a C-rich probe (5′-CGTGTAACCACGTAACCTTGTAACCCGATC-3′). A plasmid containing the telomere repeat sequence derived from pNSU70 (46) was used as a dsDNA and ssDNA control. For the dsDNA control, about 25 ng of ApaI-digested plasmid containing 300-bp-long telomere DNA was loaded. For the ssDNA control, the same amount of heat-denatured ApaI-digested plasmid was loaded. One microgram of genomic DNA was digested with EcoRI and electrophoresed on a 0.5% agarose gel in 0.5× TAE buffer (40 mM Tris-acetate [pH 8.0], 1 mM EDTA) with 0.01 mg of ethidium bromide per ml. The gel was vacuum dried at 45°C until it became thin and warm (about 45 min). Single-stranded telomeric DNA probe was labeled with [γ-32P]ATP (Amersham Pharmacia Biotech) by using T4 polynucleotide kinase. The gel was prehybridized in hybridization buffer at 37°C for 15 min, and then 10 pmol of probe was added and the incubation continued overnight at 37°C. The gel was washed with primary wash buffer at 37°C twice for 10 min and then washed with secondary wash buffer at room temperature three times for 5 min (AlkPhos Direct; Amersham Pharmacia Biotech). The gel was dried on the Whatman paper and exposed to X-ray film for about 2 days. To detect the double-stranded telomere DNA, the gel was treated with denaturing solution (0.5 M NaOH, 150 mM NaCl) for 25 min at room temperature and was treated with neutralizing solution (0.5 M Tris-HCl [pH 8.0], 150 mM NaCl) and reprobed with the same probe by in-gel hybridization.
DNA damage sensitivity assay.
Clonogenic cell survival after MMS treatment was determined as described previously (38). Logarithmically growing cells were plated directly onto solid medium containing 0.002% MMS. Colonies formed on the control and MMS-containing plates were counted after 4 days of incubation at 30°C, and the surviving fraction was calculated. For the spot assay, 4 μl of 10-fold dilutions of log-phase cells (0.5 × 107 cells/ml) was spotted onto a YEA (2% agar) plate or YEA plate containing the indicated concentration of MMS. For IR survival, logarithmically growing cells were irradiated by using a 137Cs source at a dose rate of 12.5 Gy/min. For UV survival, a germicidal lamp (FUNA-UV-LINKER, FS-800) was used at a dose rate of 50 to 200 J/m2/min. Irradiated and unirradiated cells were plated on YPAD medium plates and incubated at 30°C for 4 days. All experiments were repeated at least twice.
Indirect immunofluorescence microscopy.
Indirect immunofluorescence microscopy was performed according to the protocol previously described (8) with the following change: anti-hRad51 (Santa Cruz H-92) was diluted 1:100. To determine the percentage of cells showing nuclear foci, we visually scored 1,000 cells for each sample.
Immunoprecipitation.
For immunoprecipitation, immunoglobulin G (IgG)-conjugated magnetic beads were produced with Tosylactivated Dynabeads M-280 (DYNAL) and mouse IgG according to the manufacturer's instructions. Twenty microliters of Dynabeads was added to 12 mg of total proteins in 400 μl of buffer (50 mM HEPES/KOH [pH 7.5], 140 mM NaCl, 300 mM (NH4)2SO4, 1 mM EDTA, 1% (vol/vol) Triton X-100, 0.1% (wt/vol) sodium deoxycholate, 0.01% (wt/vol) bovine serum albumin, protease inhibitor cocktail (Roche), 1 mM phenylmethylsulfonyl fluoride, and 1 mM dithiothreitol). This mixture was incubated for 2 h at 4°C. After extensive washing, the beads were suspended in 50 μl of sodium dodecyl sulfate sample buffer. Ten microliters of the suspension was analyzed by Western blotting. The anti-Myc-Tag 9B11 antibody (Cell Signaling) and anti-protein A antibody (Sigma) were used for detection of proteins.
Chromatin immunoprecipitation.
The chromatin immunoprecipitation (ChIP) assay described by Takahashi et al. (55) was adopted with modification. Cells grown in 100 ml of YPAD culture at 30°C were fixed with formaldehyde. For immunoprecipitation, anti-Myc-Tag 9B11 antibody (Cell Signaling) and protein G-coated dynabeads (DYNAL) were used. Immunoprecipitated DNA was extracted and suspended in TE buffer (10 mM Tris-HCl, 1 mM EDTA). PCRs used the following primers to amplify the telomeric DNA (top, 5′-CGGCTGACGGGTGGGGCCCAATA-3′; bottom, 5′-GTGTGGAATTGAGTATGGTGAA-3′) or the ade6+ DNA (top, 5′-AGGTATAACGACAACAAACGTTGC-3′; bottom, 5′-CAAGGCATCAGTGTTAATATGCTC-3′).
RESULTS
The DNA damage sensitivity of rad50-d cells is suppressed by deletion of pku70+.
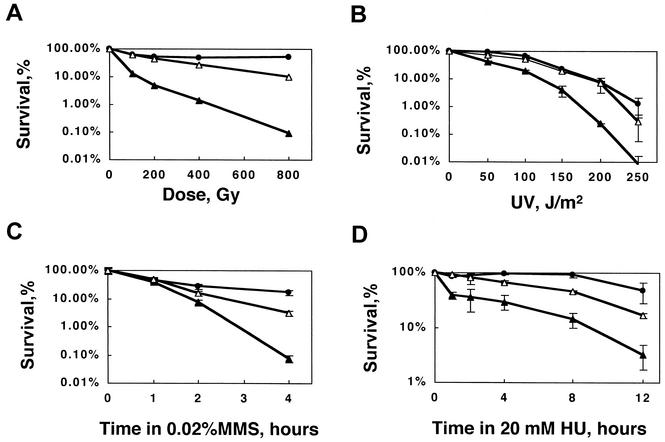
The Rad50-Mre11-Nbs1(Xrs2) complex and the Ku heterodimer are thought to compete with each other for binding to DSB ends (29). To investigate the mechanism underlying this competition, we analyzed the DNA damage sensitivities of rad50-d cells and rad50 pku70 double mutants in S. pombe. We found that the γ-ray sensitivity of rad50-d cells was suppressed by deletion of pku70+ (Fig. 1A). By analogy to the mechanism proposed for S. cerevisiae, in which the IR sensitivity of mre11 null mutants is partially suppressed by the loss of YKU70 (7), our results suggest that deletion of pku70+ in S. pombe cells lacking a functional Rad50-Rad32 protein complex improves the efficiency of HR repair by enhancing the ability to process DSB ends. In S. cerevisiae, the MMS sensitivity of mre11 mutants is not suppressed by the loss of YKU70 (38). However, we found that the MMS, UV, and HU sensitivities of rad50-d cells were suppressed by concomitant deletion of pku70+ (Fig. 1B through D). These results strongly suggest that S. pombe Ku70 plays an important role in the repair of MMS-, UV-, and HU-induced DNA damage, probably by inhibiting HR repair in the absence of the Rad50 complex. As rad50+ is epistatic to rad32+ for DNA damage sensitivity, we examined whether rad50-d cells and rad32-d cells display the same phenotype with respect to the suppression of DNA damage sensitivity. Spot assays revealed that deletion of pku70+ suppressed the sensitivity of both rad50-d and rad32-d cells to MMS and HU (data not shown).
FIG. 1.
DNA damage sensitivity of rad50-d cells is suppressed by deletion of pku70+. The sensitivities to γ rays, HU, MMS, and UV light of rad50-d cells and rad50 pku70 double mutants. The percent survival of wild-type cells, JY741 (•), rad50-d, KT002 (▴), and rad50 pku70 double mutants, KT152 (▵) is plotted against the dose of γ rays (A) or UV light (B) or is plotted against time in 0.02% MMS (C) or 20 mM HU (D). Standard deviations are shown by error bars.
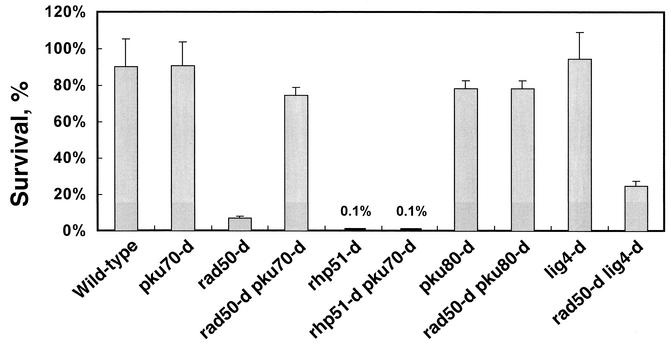
Because the function of Rad50 is thought to be in DNA damage processing, upstream of Rad51, the suppression of DNA damage sensitivity presumably reflects the enhancement of DSB end processing. In this case, suppression should be limited to the early stages of HR. We therefore asked whether the MMS sensitivity of rhp51-d cells, which are defective in later steps in HR (43), is also suppressed by the deletion of pku70+. The survival of rhp51-d cells at 0.002% MMS (0.1% ± 0.01%) was almost same as that of rhp51 pku70 double mutants (0.1% ± 0.01%) (Fig. 2). These results indicate that the suppression of DNA damage sensitivity occurs at an early stage in HR, probably before strand invasion, which requires Rad51 (Rhp51).
FIG. 2.
Suppression of DNA damage sensitivity occurs upstream of Rad51. Cell survival frequencies on 0.002% MMS-containing plates versus control plates for wild-type cells (JY741), rad50-d (KT002), rhp51-d (KT00c), rhp51-d pku70-d (KT10c5), pku80-d (KT090), rad50-d pku80-d (KT192), lig4-d (KT1a0), and rad50-d lig4-d cells (KT1a2). Standard deviations are shown by error bars (for two to four independent experiments).
Similarly, Ku70/Ku80 is thought to function at an early stage of NHEJ and Lig4 at a later stage (3, 39). We investigated whether the DNA damage sensitivity of rad50-d cells was suppressed by the deletion of either pku80+ or lig4+. The MMS sensitivity of rad50-d cells was suppressed by the concomitant deletion of pku80+ but was not significantly suppressed by the concomitant deletion of lig4+ (Fig. 2). These results indicate that the DNA binding of the Ku70/80 heterodimer plays an important role in the suppression of DNA damage sensitivity.
Evidence that Exo1 resects DNA DSB ends in the absence of Rad50 and Ku70.
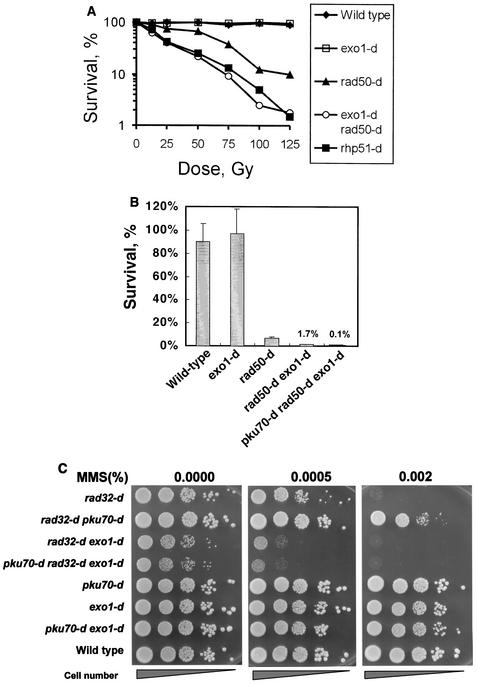
The data presented above suggest that the Ku heterodimer represses a repair function early in HR, perhaps directly at DNA ends, and the data also suggest that the derepression of this function can partially overcome the loss of Rad50. Because Mre11-Rad50 is known to encode an endonuclease, we hypothesized that the repair function that is derepressed when Ku is lost could be provided by an unknown nuclease that resects DNA DSB ends in the absence of Ku and Rad50. Exo1 is a good candidate for such a nuclease, because overexpression of EXO1 can suppress the DNA damage sensitivity of mre11 disruptants in S. cerevisiae (30, 41, 58). Thus, if S. pombe Exo1 can resect DSB ends independently of the Rad50-Rad32 protein complex, then rad50 exo1 double mutants should be more sensitive to DNA damage than the single mutants. To test this, we examined the MMS and the IR sensitivity of rad50 exo1 double mutants. As shown previously (54), exo1 single mutants were not significantly IR sensitive (Fig. 3A). However, the rad50 exo1 double mutant was significantly more IR and MMS sensitive than the single rad50 mutant (Fig. 3A and B). These results suggest that Rad50 and Exo1 can function independently. Similar results have been reported in S. cerevisiae, in which exo1 mre11 double mutants become more MMS sensitive than either of the single mutants (41, 58). Importantly, the MMS sensitivity of the rad50 mutant was suppressed by the deletion of pku70+ (Fig. 2). However, the MMS sensitivity of the rad50 exo1 double mutant was not suppressed by the deletion of pku70+ (Fig. 3B). Similar results were obtained in the spot tests when the rad32 mutant instead of the rad50 mutant was used (Fig. 3C). These results indicate that the Ku heterodimer may prevent DSB resection by Exo1 in the absence of the Rad50-Rad32 protein complex. In agreement with our hypothesis that both Rad50 and Exo1 can act in an early step in HR, the rad50 exo1 double mutant was as IR sensitive as the rhp51 single mutant.
FIG. 3.
rad50 exo1 double mutants become more sensitive to DNA damage than each single mutant. (A) The sensitivities to γ rays of wild-type cells, rad50-d, exo1-d, rad50-d exo1-d, and rhp51-d cells. The percent survival of wild-type cells (700), rad50-d (701), exo1-d (702), rad50-d exo1-d (703), and rhp51-d cells (704) is plotted against the γ ray dose. (B) Cell survival frequencies on 0.002% MMS-containing plates versus control plates for wild-type cells (JY741), exo1-d (KΤ00g), rad50-d (KT002), rad50-d exo1-d (KΤ02g), and pku70-d rad50-d exo1-d (KΤ12g5) cells. Standard deviations are shown by error bars (for two to four independent experiments). (C) The MMS sensitivity of wild-type cells (119), rad32-d (324), rad32-d pku70-d (315), rad32-d exo1-d (403), pku70-d rad32-d exo1-d (407), pku70-d (319), exo1-d (302), and exo1-d pku70-d (394) cells was assayed by the spot test. The cells were grown in YEA (0.5 × 107 cells/ml), serially diluted (1:10) with sterilized water, and spotted (4 μl of each dilution was spotted onto the MMS plates).
Rad50 and Exo1 function independently upstream of Rad51.
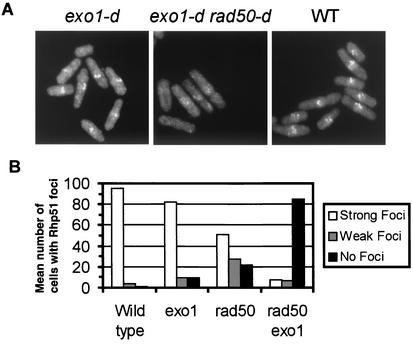
Treatment of wild-type S. pombe cells with 500 Gy of γ rays resulted in the formation of Rad51 (Rhp51) foci in almost 100% of cells within 1 h after irradiation (8). If Rad50 and Exo1 function independently and upstream of Rad51, IR-induced Rad51 focus formation should be significantly compromised in rad50 exo1 double mutants compared to the respective single mutants. Indeed, we find that rad50-d cells and exo1-d cells showed only a moderate reduction in the focal assembly of Rhp51 following IR (rad50-d, 50% of cells with foci; exo1-d, 80% of cells with foci), whereas rad50 exo1 double mutant cells were strongly impaired in Rhp51 focus formation 1 h after irradiation (rad50-d exo1-d, 8% of cells with foci) (Fig. 4). These data are consistent with a model in which Rad50-Rad32 and Exo1 process DNA DSB ends in a redundant manner upstream of Rad51.
FIG. 4.
Exo1 and rad50 function independently upstream of Rad51. (A) Indirect immunofluorescence microscopy of wild-type cells (700), rad50-d (701), exo1-d (702), and rad50-d exo1-d cells (703) at 1 h after irradiation with 500 Gy of ionizing radiation (8). A cross-reacting human anti-Rad51 antibody was used to detect Rhp51. (B) Mean percentage of various mutant cells showing Rhp51 foci at 1 h postirradiation.
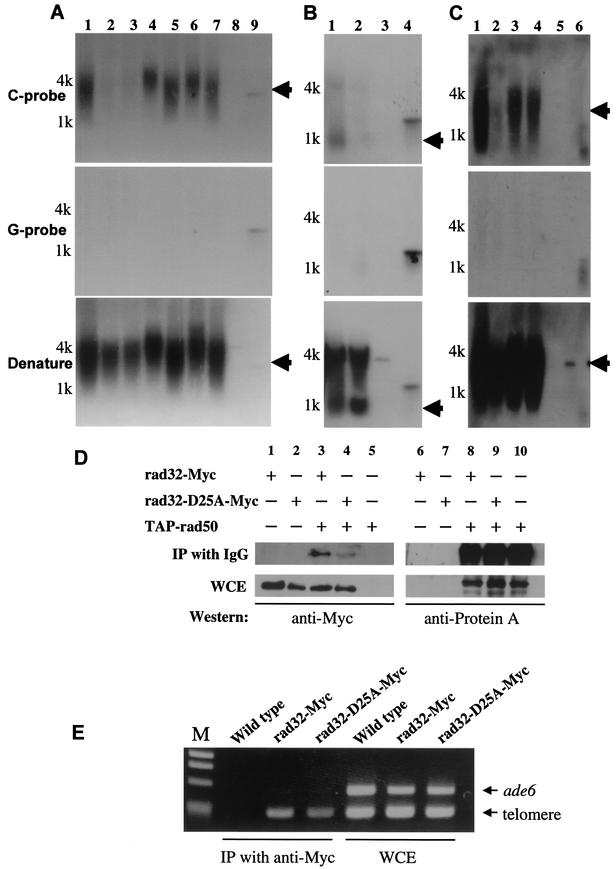
Rad50-Rad32 is involved in the production of G-strand overhang in taz1-d cells.
Our genetic data suggest that the reason that the S. pombe Rad50-Rad32 complex is required for DNA damage resistance is most probably because of its role in the processing of DSB ends. The Rad50 complex has also been implicated in the processing of telomere ends (15, 25, 49), and we therefore examined whether these interactions were reflected in the processing of these specific DNA structures. Asynchronous wild-type S. pombe cells contain only very small amounts of the G-rich overhang at telomeres (2), making it difficult to evaluate the role of Rad50-Rad32 in the processing of telomere ends in wild-type cells (Fig. 5B, top panel, lane 2). However, using an in-gel hybridization assay (16), we observed strong signals corresponding to the G-strand overhang in asynchronous taz1-d cells (Fig. 5A, top panel, lane1). We therefore constructed taz1 rad50 double mutants and taz1 rad32 double mutants and examined the extent of the single-stranded overhang at the telomeres. Intriguingly, both taz1 rad50 double mutants and taz1 rad32 double mutants lacked the G-strand overhang (Fig. 5A, top panel, lanes 2 and 3). These results suggest that the Rad50-Rad32 complex is required either for degradation of the corresponding C-rich strand in taz1-d cells or for elongation of the G-rich strand. If Rad32 and Rad50 are required for elongation of the G strand of telomeres, the elongation of telomeres themselves in taz1 rad32 double mutants would be less efficient than that in taz1 single mutants. However, the lengths of telomeres themselves in a taz1 rad32 double mutant, which was constructed by deletion of taz1+ in rad32-d cells, was identical to that seen in the taz1 single mutant (Fig. 5A, bottom panel, lanes 1 and 3), suggesting that the Rad50-Rad32 complex is not required for G-strand elongation in taz1-d cells.
FIG.5.
The roles of Rad50 and Ku70 at telomere ends in taz1-d cells. The single-stranded overhangs were detected by in-gel hybridization. (A) Lane 1, taz1-d (KT110); lane 2, rad50-d taz1-d (KT021); lane 3, rad32-d taz1-d (KT116); lane 4, pku70-d rad50-d taz1-d (KT0215); lane 5, pku70-d rad32-d taz1-d (KT1165); lane 6, pku70-d rad50-d exo1-d taz1-d (KT121g5); lane 7, pku70-d rad32-d exo1-d taz1-d (KT016g5); lane 8, dsDNA control; lane 9, ssDNA control. (B) Lane 1, taz1-d trt1-d (KT117); lane 2, wild-type cells (JY741); lane 3, dsDNA control lane 4, ssDNA control. (C) Lane 1, taz1-d (KT110); lane 2, rad32-d taz1-d. (KT116); lane 3, rad32-D25A taz1-d (KT0106M1); lane 4, rad32-D25A taz1-d (KT0106M2); lane 5, dsDNA control; lane 6, ssDNA control. Genomic DNAs were digested with EcoRI and separated by electrophoresis on a 0.5% agarose gel. Then the gel was dried and hybridized with 32P-labeled C-rich (top panel) or G-rich (middle panel) probe. To detect double-stranded telomere DNAs, the gel was treated with denaturant and reprobed with C-rich probe (bottom panel). As a control, a linearized telomeric DNA-containing plasmid was used for double-stranded telomere detection. For the single-strand control, the same linearized plasmid, which was denatured, was used (see Materials and Methods). Telomere DNA is indicated by arrows. (D) Rad32-D25A binds to Rad50 in immunoprecipitation assay. TAP-tagged Rad50 was precipitated with IgG-conjugated magnetic beads from a lysate of TAP-rad50 rad32-Myc (KTt2T6M) cells (lanes 3 and 8) and a lysate of TAP-rad50 rad32-D25A-Myc (KTt2T6MM1) cells (lanes 4 and 9), respectively. The immunoprecipitates (IP with IgG) were examined by Western blot (Western) analysis with anti-Myc antibody (anti-Myc). The protein A tag is detected with anti-protein A antibody (anti-Protein A). TAP-rad50 cells (435); lanes 5 and 10, rad32-Myc (KTt6M) cells; lanes 1 and 6, rad32-D25A-Myc (KTt6MM1) cells; lanes 2 and 7 were used as controls. The same levels of proteins were detected in the whole-cell extract (WCE). (E) Rad32-D25A is bound to telomere DNA in ChIP assay. Untagged wild-type control cells (JY741), rad32-Myc (KTt6M) cells, and rad32-D25A-Myc (KTt6MM1) cells were used. PCRs were performed on whole-cell extract (WCE) and on chromatin immunoprecipitates (IP with anti-Myc) by using primers to amplify a telomere DNA (telomere) and primers to amplify DNA from the ade6+ gene (ade6). M represents a DNA maker.
To exclude the possibility that the G-strand overhang in taz1-d cells is telomerase dependent, we also created a taz1 trt1 double mutant that lacked active telomerase. As shown previously (46, 47), taz1 trt1 double mutants lost telomeres very rapidly (Fig. 5B, bottom panel, lane 1); however, the signals corresponding to the G-rich overhang were still detected (Fig. 5B, top panel, lane 1). These results indicate that the G-rich overhang in taz1-d cells can be generated without telomerase activity, probably through degradation by the Rad50-Rad32 complex. Although degradation of the C-rich strand at the telomere ends in taz1-d cells seems to be fully Rad50-Rad32 dependent, DNA ends made by HO endonuclease are still processed in mre11 mutants in S. cerevisiae (29). This difference suggests that telomere ends may be highly protected from degradation even in the absence of the Mre11 complex.
Interestingly, as we observed for the sensitivity to DNA damage, inactivation of the Ku heterodimer could overcome the loss of Rad50-Rad32 and restore the G-strand overhang at telomere ends. In a taz1 rad50 pku70 triple mutant, a significant amount of G-rich overhang was detected (Fig. 5A, top panel, lanes 4 and 5). This again can be interpreted to indicate that an unknown nuclease activity can digest the corresponding C-rich strand to produce the G-rich overhang in the absence of the Rad32-Rad50 complex and to indicate that this nuclease activity is inhibited by the presence of Ku70. However, unlike the situation deduced from the DNA damage sensitivity analysis presented above, this nuclease activity cannot be attributed exclusively to Exo1 activity, since we still detected significant levels of G-rich overhang in the taz1 rad50 pku70 exo1 and taz1 rad32 pku70 exo1 quadruple mutants (Fig. 5A, top panel, lanes 6 and 7). These data suggest that telomere ends are processed by a different mechanism from those of DSB ends.
rad32 nuclease domain mutants possess G-strand overhang in taz1-d cells.
Our data suggest a model in which the Rad50-Rad32 protein complex is involved in end-processing at DSBs and at telomeres and further suggest that the Ku heterodimer negatively influences exonuclease I, which can act on DSBs but not on telomeric ends in the absence of Rad50-Rad32. To test whether the nuclease activity of Rad32 is indeed required for resection of telomeres, we made a rad32-D25A taz1 double mutant. Aspartate 25 in the S. pombe Rad32 protein corresponds to the catalytically important aspartate residue 8 in the P. furiosus Mre11 protein (18). This aspartate coordinates two Mn2+ atoms that are located in the active site and are required for the endo- and exonuclease activity of P. furiosus Mre11 (26). A mutation of the same protein in S. cerevisiae, Mre11 D16A, does not possess 5′ to 3′ exonuclease activity in vitro (21). S. cerevisiae mre11-D16A mutant strains exhibit MMS sensitivity, but this sensitivity is about 10-fold weaker than that of a null mutant (21). Consistent with the important role of this aspartate, rad32-D25A mutants were as MMS and HU sensitive as the rad32-d cells (data not shown). These findings are consistent with previously reported data, which showed that rad32-D25N mutants are as γ-ray sensitive as a rad32 null mutant (65).
Interestingly, taz1-d rad32-D25A double mutants contained a significant level of G-strand overhang (Fig. 5C, lanes 3 and 4) (two independent clones), indicating that the Rad32 nuclease domain is not required for degradation of the C-rich strand. Our results suggest that, at the telomere ends, the Rad50 complex does not act as a nuclease itself but probably recruits an unknown nuclease activity to telomeres. To ascertain if this nuclease is independent of Exo1, we created taz1-d exo1-d double mutants. These taz1-d exo1-d double mutants contained a significant G-rich overhang (data not shown), which suggested that the recruited nuclease activity is not exclusively due to Exo1. In S. cerevisiae, some of the Mre11 nuclease domain mutants do not form a complex with Rad50 (28). Thus, we tested the interaction between Rad32-D25A and Rad50 by coimmunoprecipitation experiments. We tagged the N terminus of Rad50 with a tandem affinity purification (TAP) tag (64) and tagged the C terminus of Rad32 with the Myc tag (1). Cells expressing both tagged proteins were lysed, and Rad50 was affinity precipitated from the soluble lysate with IgG-conjugated magnetic beads (see Materials and Methods). As expected, Rad32-Myc was coprecipitated with Rad50 (Fig. 5D). Next, we tested the interaction between Rad32-D25A and Rad50. Although the efficiency of protein binding was lower than that of wild-type Rad32, Rad32-D25A retained the ability to interact with Rad50 (Fig. 5D). We also tested the interaction between Rad32-D25A and telomere by ChIP assay. As reported previously (48), telomeric DNA was specifically amplified from Rad32-Myc immunoprecipitate (Fig. 5E). We also found that Rad32-D25A-Myc can bind to telomere ends (Fig. 5E). These two results strongly suggest that the nuclease mutant Rad32-D25A forms a complex with Rad50 on the telomere DNA. This is consistent with a model in which Rad23-D25A can recruit the unknown nuclease to telomere ends.
DISCUSSION
The roles of the Rad50-Rad32 complex and Ku70 complex at DSB ends.
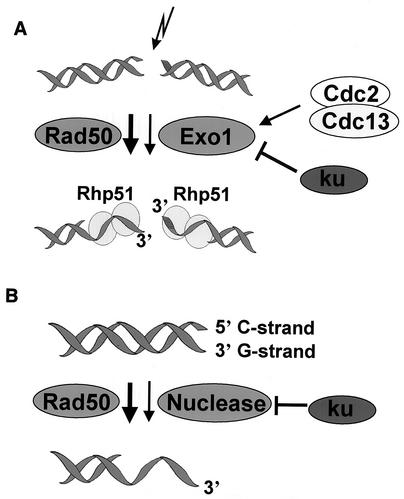
S. pombe Rad50 and Rad32 are required for efficient HR repair, but their exact roles at DSB ends remain unclear (25, 32, 56, 65, 66). Unlike for S. cerevisiae, a method to analyze the rate of degradation of DSB ends has not been developed for S. pombe. This makes it difficult to study the roles of the Rad32-Rad50 complex at DSB ends. We have demonstrated that rad50 exo1 double mutants are more IR sensitive than the respective single mutants and are as sensitive as cells lacking Rad51 (Rhp51) (Fig. 3A). Consistent with this observation, a rad50 exo1 double mutant was strongly impaired in its ability to form Rhp51 foci after irradiation (Fig. 4). These results are fully consistent with a model in which the Rad50-Rad32 complex and Exo1 can independently and redundantly act on DNA DSB ends to generate the substrates for Rhp51 filament formation (Fig. 6A).
FIG. 6.
Models. (A) Hypothetical model for DSB processing by Rad50 complex and Exo1. DSB ends are processed mainly by the Rad50-Rad32 complex. In the absence of Rad50-Rad32 complex, DNA ends can be processed by Exo1, but Ku heterodimer has to be removed from the break site to allow end processing. The Cdc13 function may act through the Exo1 pathway, in parallel to Rad50 (8). (B) Hypothetical model for degradation of C-rich strand at telomere ends in taz1-d cells. Telomere ends in taz1-d cells are processed mainly by the Rad50-Rad32 complex. Because the nuclease domain is not required for this process, we assume that the Rad50 complex recruits an unknown nuclease. In the absence of Rad50 complex, DNA ends can be processed by unknown nucleases, but Ku heterodimer has to be removed from the break site to allow end processing.
We also found that both the MMS and IR sensitivities of rad50-d cells were suppressed by concomitant deletion of pku70+, which encodes the Ku70 protein required for an early step in NHEJ (Fig. 1). However, the MMS sensitivity of the rad50 exo1 double mutants was not suppressed by deletion of pku70+ (Fig. 3B). In S. cerevisiae, it has been suggested that Ku competes with 5′ to 3′ exonucleases at DNA ends (29). However, the nuclease competing with Yku70 has not been identified. Our results strongly suggest that the nuclease competing with Ku70 at DSB ends is ExoI (Fig. 6 A). In contrast to the competition between Ku and ExoI, the Rad50 complex can process DSB ends in the presence of Ku heterodimer. pku70-d cells are not IR sensitive (32), which suggests that Ku70 does not recruit the Rad50 complex to the DNA ends. Although the biological significance of the ExoI pathway for HR repair is not clear, Ku might be removed in a controlled manner and protect DSBs from nonspecific degradation in the absence of certain activities.
Is a DNA DSB produced following exposure to UV, MMS, and HU? The sensitivities of rad50-d cells to IR, UV, MMS, and HU were all suppressed by concomitant deletion of pku70+ (Fig. 1A through D). This suggests that, in the absence of rad50+, γ rays, UV light, MMS, and HU cause the generation of similar DNA structures, probably DSBs, that may be bound by the Ku70-Ku80 heterodimer. However, the suppression of the UV sensitivity is potentially confusing because UV light produces primarily cyclobutane-pyrimidine dimers (CPDs) or 6-4 photoproducts, not DSBs. Recombination is much more important for UV survival in S. pombe than in budding yeast, as shown by the fact that rad22, rhp51, and rhp54 mutants are all significantly UV sensitive (36). To initiate homologous recombination, DNA DSBs should be generated. Interestingly, in S. pombe, CPDs and 6-4 photoproducts are not only repaired by the nuclear excision repair pathway but also by the UV damage endonuclease pathway, which facilitates homologous recombination (36). Furthermore, unrepaired UV-induced lesions are thought to become substrates for HR-based postreplication repair processes when encountered by a replication fork. Thus, DNA DSBs could be produced as a secondary lesion during the recombinogenic recovery from UV damage. A similar mechanism probably underlies the HU and MMS sensitivity of rad50-d cells. Both HU and MMS can stall the replication fork, and such stalled forks can result in DSBs, as shown in Escherichia coli (27, 37). It is now becoming clear that in many eukaryotes, including S. pombe, DNA DSBs can be produced by replication arrest and that HR is required for their repair (44, 45). In S. pombe, a Holliday junction formed at a stalled or collapsed replication fork is thought to either be reversed by Rqh1 helicase in a nonrecombinogenic pathway or resolved by a Mus81-Eme1-dependent endonuclease (potentially via a recombinogenic pathway) (5, 17). In the latter case, DSBs are suggested to be produced to initiate homologous recombination.
Given that IR, UV, MMS, and HU could all cause DSBs in S. pombe, suppression of the sensitivity of rad50-d cells to all these agents by the concomitant deletion of pku70+ could be explained by a model in which the Ku heterodimer has to be removed from the break site in the absence of Rad50 to allow end processing by alternative nucleases (Fig. 6A).
Roles of Rad32-Rad50 complex and Ku70 at the telomere ends in taz1-d cells.
Both the Rad50-Rad32 protein complex and the Ku heterodimer are involved not only in the processing of DSBs but also in telomere maintenance (25, 66). However, it is unknown how these proteins act to regulate telomere length. Asynchronous taz1-d cells contain extensive G-rich single-stranded 3′ overhangs at telomere ends (Fig. 5A, top panel, lane 1), thereby making it possible to study the roles of Rad50, Ku70, and Exo1 at these telomeres. As shown in Fig. 5B (top panel, lane 1), the generation of the G-rich overhang in taz1-d cells occurs in the absence of telomerase activity, suggesting that the G-rich overhang is generated by degradation of the C-rich strand. Our results suggest that this nuclease step is dependent on the Rad32-Rad50 complex without utilizing its nuclease activity, because the G-rich overhang in taz1-d cells disappeared upon deletion of either rad50+or rad32+ (Fig. 5A, top panel, lanes 2 and 3) but not upon mutation of the nuclease domain of Rad32 (Fig. 5C, top panel, lanes 3 and 4). To explain the physical requirement for Rad32, we propose that (in taz1 disruptants) the Rad50-Rad32 complex recruits an unknown nuclease, which contains 5′ to 3′ exonuclease activity, to the telomere (Fig. 6B). We do not know which nuclease is recruited. However, we can exclude the major activity being due to Exo1, because taz1-d exo1-d double mutants contained significant G-rich overhangs (data not shown). Although the biological significance of the Rad50-Rad32-dependent generation of G-rich overhang in taz1-d cells is not clear, Taz1 may be detached from telomeric DNA during telomere elongation. Therefore, our results may reflect the function of these proteins during telomere elongation.
Recently, it has been suggested that ExoI is required for both ssDNA generation at telomeres and the subsequent cell cycle arrest of yku70 mutants in S. cerevisiae (34). Our results in S. pombe are not consistent with this—we found no detectable role for ExoI at telomere ends. It is possible that there are significant differences between S. pombe and S. cerevisiae or that the different assays used affect competing activities in distinct ways.
The roles of Rad32-Rad50 at DSB ends.
The importance of nuclease activity in Rad32 for the processing of DSB ends remains clear. Although Mre11 has a 3′ to 5′ exonuclease and endonuclease activity in vitro (51, 57), in vivo observations suggest that Mre11 is required for the oppositely oriented (i.e., 5′ to 3′) exonuclease activity (29). There are two possible models to explain this discrepancy. (i) A long 3′ ssDNA is generated by the endonuclease activity of Rad32 combined with unidentified helicase components. (ii) An unidentified 5′ to 3′ exonuclease is recruited to DSB ends by a Rad32 (Mre11) complex. At this point it is difficult to distinguish these two possibilities, and further studies are required to resolve this discrepancy. It has been reported that the in vivo 5′ to 3′ resection of DNA ends is strongly dependent upon the successful formation of the Mre11 protein complex, perhaps along with other as yet unidentified components (28). We found that Rad32 nuclease domain mutants were significantly MMS sensitive (data not shown). However, we cannot conclude that the nuclease activity in Rad32 is required for the processing of DSB ends, because the interaction between Rad32-D25A and Rad50 were less efficient than the interaction between Rad32 and Rad50 (Fig. 5D). The reduced stability of Rad32-D25A-Rad50 complex may impair the 5′ to 3′ resection ability by affecting the unknown function of the Rad32-Rad50 complex, perhaps binding to unidentified proteins. As suggested for the roles of the Rad32-Rad50 complex at telomere ends in taz1-d cells, the main roles of the Rad50 complex might be the recruitment of unidentified nuclease or other factors to DSB ends. These factors are also suggested to exist in S. cerevisiae (28).
Our data suggest that the Rad50 complex is required for the processing of DSB ends and telomere ends in the presence of Ku heterodimer. However, Ku heterodimer inhibits processing of DSB ends and telomere ends by alternative nucleases in the absence of the Rad50-Rad32 protein complex. While we have identified Exo1 as the alternative nuclease targeting DNA break sites, the identity of the nuclease acting on the telomere ends remains elusive. The nuclease function of the Rad50-Rad32 protein complex seems not to be important for degradation of the C-rich strand at telomeres. Moreover, our data allow the speculation that cells regulate the resection of DNA ends in the absence of Rad50 complex through controlled binding of the Ku heterodimer. A similar regulation might underlie the cell-cycle-specific appearance of the G-rich overhang at telomeres.
Acknowledgments
We thank Kohta Takahashi, Shigeaki Saitoh, and Mitsuhiro Yanagida for ChIP assay protocol and Hiroyuki Araki for helping with UV-irradiation experiments, and we thank Takeshi Saito, Shinji Yasuhira, and Hiroshi Utsumi for helping with γ-ray irradiation; we also thank Masayuki Yamamoto for providing strains and John R. Pringle for providing a plasmid.
This work was supported by Grants-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports, and Culture of Japan to A.M., K.O., and M.U. Part of this work was performed under the Basic Research 21 for Breakthroughs in Info-Communications project supported by the Ministry of Public Management, Home Affairs, Posts, and Telecommunications and the Support Center for Advanced Telecommunications Technology Research Foundation (grant to M.U.). Part of this work was performed by using facilities of the Research Reactor Institute, Kyoto University.
REFERENCES
- 1.Bahler, J., J. Q. Wu, M. S. Longtine, N. G. Shah, A. McKenzie III, A. B. Steever, A. Wach, P. Philippsen, and J. R. Pringle. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14:943-945. [DOI] [PubMed] [Google Scholar]
- 2.Baumann, P., and T. R. Cech. 2001. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 292:1171-1175. [DOI] [PubMed] [Google Scholar]
- 3.Baumann, P., and T. R. Cech. 2000. Protection of telomeres by the Ku protein in fission yeast. Mol. Biol. Cell 11:3265-3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blackburn, E. H. 2001. Switching and signaling at the telomere. Cell 106:661-673. [DOI] [PubMed] [Google Scholar]
- 5.Boddy, M. N., A. Lopez-Girona, P. Shanahan, H. Interthal, W. D. Heyer, and P. Russell. 2000. Damage tolerance protein Mus81 associates with the FHA1 domain of checkpoint kinase Cds1. Mol. Cell. Biol. 20:8758-8766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boulton, S. J., and S. P. Jackson. 1998. Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 17:1819-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bressan, D. A., B. K. Baxter, and J. H. Petrini. 1999. The Mre11-Rad50-Xrs2 protein complex facilitates homologous recombination-based double-strand break repair in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:7681-7687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caspari, T., J. M. Murray, and A. M. Carr. 2002. Cdc2-cyclin B kinase activity links Crb2 and Rqh1-topoisomerase III. Genes Dev. 16:1195-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohn, M., and E. H. Blackburn. 1995. Telomerase in yeast. Science 269:396-400. [DOI] [PubMed] [Google Scholar]
- 10.Conrad, M. N., J. H. Wright, A. J. Wolf, and V. A. Zakian. 1990. RAP1 protein interacts with yeast telomeres in vivo: overproduction alters telomere structure and decreases chromosome stability. Cell 63:739-750. [DOI] [PubMed] [Google Scholar]
- 11.Cooper, J. P., E. R. Nimmo, R. C. Allshire, and T. R. Cech. 1997. Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature 385:744-747. [DOI] [PubMed] [Google Scholar]
- 12.Critchlow, S. E., and S. P. Jackson. 1998. DNA end-joining: from yeast to man. Trends Biochem. Sci. 23:394-398. [DOI] [PubMed] [Google Scholar]
- 13.Cromie, G. A., J. C. Connelly, and D. R. Leach. 2001. Recombination at double-strand breaks and DNA ends: conserved mechanisms. Mol. Cell 8:1163-1174. [DOI] [PubMed] [Google Scholar]
- 14.D'Amours, D., and S. P. Jackson. 2002. The Mre11 complex: at the crossroads of DNA repair and checkpoint signalling. Nat. Rev. Mol. Cell. Biol. 3:317-327. [DOI] [PubMed] [Google Scholar]
- 15.Diede, S. J., and D. E. Gottschling. 2001. Exonuclease activity is required for sequence addition and Cdc13p loading at a de novo telomere. Curr. Biol. 11:1336-1340. [DOI] [PubMed] [Google Scholar]
- 16.Dionne, I., and R. J. Wellinger. 1996. Cell cycle-regulated generation of single-stranded G-rich DNA in the absence of telomerase. Proc. Natl. Acad. Sci. USA 93:13902-13907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doe, C. L., J. S. Ahn, J. Dixon, and M. C. Whitby. 2002. Mus81-Eme1 and Rqh1 involvement in processing stalled and collapsed replication forks. J. Biol. Chem. 277:32753-32759. [DOI] [PubMed] [Google Scholar]
- 18.Farah, J. A., E. Hartsuiker, K. Mizuno, K. Ohta, and G. R. Smith. 2002. A 160-bp palindrome is a Rad50-Rad32-dependent mitotic recombination hotspot in Schizosaccharomyces pombe. Genetics 161:461-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Featherstone, C., and S. P. Jackson. 1999. Ku, a DNA repair protein with multiple cellular functions? Mutat. Res. 434:3-15. [DOI] [PubMed] [Google Scholar]
- 20.Ferreira, M. G., and J. P. Cooper. 2001. The fission yeast Taz1 protein protects chromosomes from Ku-dependent end-to-end fusions. Mol. Cell 7:55-63. [DOI] [PubMed] [Google Scholar]
- 21.Furuse, M., Y. Nagase, H. Tsubouchi, K. Murakami-Murofushi, T. Shibata, and K. Ohta. 1998. Distinct roles of two separable in vitro activities of yeast Mre11 in mitotic and meiotic recombination. EMBO J. 17:6412-6425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gu, Y., K. J. Seidl, G. A. Rathbun, C. Zhu, J. P. Manis, N. van der Stoep, L. Davidson, H. L. Cheng, J. M. Sekiguchi, K. Frank, P. Stanhope-Baker, M. S. Schlissel, D. B. Roth, and F. W. Alt. 1997. Growth retardation and leaky SCID phenotype of Ku70-deficient mice. Immunity 7:653-665. [DOI] [PubMed] [Google Scholar]
- 23.Haber, J. E. 1998. The many interfaces of Mre11. Cell 95:583-586. [DOI] [PubMed] [Google Scholar]
- 24.Haber, J. E. 1999. Sir-Ku-itous routes to make ends meet. Cell 97:829-832. [DOI] [PubMed] [Google Scholar]
- 25.Hartsuiker, E., E. Vaessen, A. M. Carr, and J. Kohli. 2001. Fission yeast Rad50 stimulates sister chromatid recombination and links cohesion with repair. EMBO J. 20:6660-6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hopfner, K. P., A. Karcher, L. Craig, T. T. Woo, J. P. Carney, and J. A. Tainer. 2001. Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase. Cell 105:473-485. [DOI] [PubMed] [Google Scholar]
- 27.Kowalczykowski, S. C. 2000. Initiation of genetic recombination and recombination-dependent replication. Trends Biochem. Sci. 25:156-165. [DOI] [PubMed] [Google Scholar]
- 28.Lee, S. E., D. A. Bressan, J. H. Petrini, and J. E. Haber. 2002. Complementation between N-terminal Saccharomyces cerevisiae mre11 alleles in DNA repair and telomere length maintenance. DNA Repair (Amst.) 1:27-40. [DOI] [PubMed] [Google Scholar]
- 29.Lee, S. E., J. K. Moore, A. Holmes, K. Umezu, R. D. Kolodner, and J. E. Haber. 1998. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell 94:399-409. [DOI] [PubMed] [Google Scholar]
- 30.Lewis, L. K., G. Karthikeyan, J. W. Westmoreland, and M. A. Resnick. 2002. Differential suppression of DNA repair deficiencies of yeast rad50, mre11 and xrs2 mutants by EXO1 and TLC1 (the RNA component of telomerase). Genetics 160:49-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lustig, A. J., S. Kurtz, and D. Shore. 1990. Involvement of the silencer and UAS binding protein RAP1 in regulation of telomere length. Science 250:549-553. [DOI] [PubMed] [Google Scholar]
- 32.Manolis, K. G., E. R. Nimmo, E. Hartsuiker, A. M. Carr, P. A. Jeggo, and R. C. Allshire. 2001. Novel functional requirements for non-homologous DNA end joining in Schizosaccharomyces pombe. EMBO J. 20:210-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marchetti, M. A., S. Kumar, E. Hartsuiker, M. Maftahi, A. M. Carr, G. A. Freyer, W. C. Burhans, and J. A. Huberman. 2002. A single unbranched S-phase DNA damage and replication fork blockage checkpoint pathway. Proc. Natl. Acad. Sci. USA 99:7472-7477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maringele, L., and D. Lydall. 2002. EXO1-dependent single-stranded DNA at telomeres activates subsets of DNA damage and spindle checkpoint pathways in budding yeast yku70Δ mutants. Genes Dev. 16:1919-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin, S. G., T. Laroche, N. Suka, M. Grunstein, and S. M. Gasser. 1999. Relocalization of telomeric Ku and SIR proteins in response to DNA strand breaks in yeast. Cell 97:621-633. [DOI] [PubMed] [Google Scholar]
- 36.McCready, S. J., F. Osman, and A. Yasui. 2000. Repair of UV damage in the fission yeast Schizosaccharomyces pombe. Mutat. Res. 451:197-210. [DOI] [PubMed] [Google Scholar]
- 37.Michel, B., S. D. Ehrlich, and M. Uzest. 1997. DNA double-strand breaks caused by replication arrest. EMBO J. 16:430-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Milne, G. T., S. Jin, K. B. Shannon, and D. T. Weaver. 1996. Mutations in two Ku homologs define a DNA end-joining repair pathway in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:4189-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miyoshi, T., M. Sadaie, J. Kanoh, and F. Ishikawa. 2003. Telomeric DNA ends are essential for the localization of Ku at telomeres in fission yeast. J. Biol. Chem. 278:1924-1931. [DOI] [PubMed] [Google Scholar]
- 40.Moreau, S., J. R. Ferguson, and L. S. Symington. 1999. The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol. Cell. Biol. 19:556-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moreau, S., E. A. Morgan, and L. S. Symington. 2001. Overlapping functions of the Saccharomyces cerevisiae Mre11, Exo1 and Rad27 nucleases in DNA metabolism. Genetics 159:1423-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moreno, S., A. Klar, and P. Nurse. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194:795-823. [DOI] [PubMed] [Google Scholar]
- 43.Muris, D. F., K. Vreeken, A. M. Carr, B. C. Broughton, A. R. Lehmann, P. H. Lohman, and A. Pastink. 1993. Cloning the RAD51 homologue of Schizosaccharomyces pombe. Nucleic Acids Res. 21:4586-4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muris, D. F., K. Vreeken, A. M. Carr, J. M. Murray, C. Smit, P. H. Lohman, and A. Pastink. 1996. Isolation of the Schizosaccharomyces pombe RAD54 homologue, rhp54+, a gene involved in the repair of radiation damage and replication fidelity. J. Cell Sci. 109(Pt 1):73-81. [DOI] [PubMed] [Google Scholar]
- 45.Murray, J. M., H. D. Lindsay, C. A. Munday, and A. M. Carr. 1997. Role of Schizosaccharomyces pombe RecQ homolog, recombination, and checkpoint genes in UV damage tolerance. Mol. Cell. Biol. 17:6868-6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakamura, T. M., J. P. Cooper, and T. R. Cech. 1998. Two modes of survival of fission yeast without telomerase. Science 282:493-496. [DOI] [PubMed] [Google Scholar]
- 47.Nakamura, T. M., G. B. Morin, K. B. Chapman, S. L. Weinrich, W. H. Andrews, J. Lingner, C. B. Harley, and T. R. Cech. 1997. Telomerase catalytic subunit homologs from fission yeast and human. Science 277:955-959. [DOI] [PubMed] [Google Scholar]
- 48.Nakamura, T. M., B. A. Moser, and P. Russell. 2002. Telomere binding of checkpoint sensor and DNA repair proteins contributes to maintenance of functional fission yeast telomeres. Genetics 161:1437-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nugent, C. I., G. Bosco, L. O. Ross, S. K. Evans, A. P. Salinger, J. K. Moore, J. E. Haber, and V. Lundblad. 1998. Telomere maintenance is dependent on activities required for end repair of double-strand breaks. Curr. Biol. 8:657-660. [DOI] [PubMed] [Google Scholar]
- 50.Paques, F., and J. E. Haber. 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63:349-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paull, T. T., and M. Gellert. 1998. The 3′ to 5′ exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol. Cell 1:969-979. [DOI] [PubMed] [Google Scholar]
- 52.Siede, W., A. A. Friedl, I. Dianova, F. Eckardt-Schupp, and E. C. Friedberg. 1996. The Saccharomyces cerevisiae Ku autoantigen homologue affects radiosensitivity only in the absence of homologous recombination. Genetics 142:91-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sugawara, N., and J. E. Haber. 1992. Characterization of double-strand break-induced recombination: homology requirements and single-stranded DNA formation. Mol. Cell. Biol. 12:563-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Szankasi, P., and G. R. Smith. 1995. A role for exonuclease I from S. pombe in mutation avoidance and mismatch correction. Science 267:1166-1169. [DOI] [PubMed] [Google Scholar]
- 55.Takahashi, K., S. Saitoh, and M. Yanagida. 2000. Application of the chromatin immunoprecipitation method to identify in vivo protein-DNA associations in fission yeast. Sci. STKE 2000:PL1.. [DOI] [PubMed]
- 56.Tavassoli, M., M. Shayeghi, A. Nasim, and F. Z. Watts. 1995. Cloning and characterisation of the Schizosaccharomyces pombe rad32 gene: a gene required for repair of double strand breaks and recombination. Nucleic Acids Res. 23:383-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Trujillo, K. M., S. S. Yuan, E. Y. Lee, and P. Sung. 1998. Nuclease activities in a complex of human recombination and DNA repair factors Rad50, Mre11, and p95. J. Biol. Chem. 273:21447-21450. [DOI] [PubMed] [Google Scholar]
- 58.Tsubouchi, H., and H. Ogawa. 2000. Exo1 roles for repair of DNA double-strand breaks and meiotic crossing over in Saccharomyces cerevisiae. Mol. Biol. Cell 11:2221-2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tsukamoto, Y., J. Kato, and H. Ikeda. 1996. Effects of mutations of RAD50, RAD51, RAD52, and related genes on illegitimate recombination in Saccharomyces cerevisiae. Genetics 142:383-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tsukamoto, Y., J. Kato, and H. Ikeda. 1996. Hdf1, a yeast Ku-protein homologue, is involved in illegitimate recombination, but not in homologous recombination. Nucleic Acids Res. 24:2067-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Usui, T., T. Ohta, H. Oshiumi, J. Tomizawa, H. Ogawa, and T. Ogawa. 1998. Complex formation and functional versatility of Mre11 of budding yeast in recombination. Cell 95:705-716. [DOI] [PubMed] [Google Scholar]
- 62.van Steensel, B., and T. de Lange. 1997. Control of telomere length by the human telomeric protein TRF1. Nature 385:740-743. [DOI] [PubMed] [Google Scholar]
- 63.Vogel, H., D. S. Lim, G. Karsenty, M. Finegold, and P. Hasty. 1999. Deletion of Ku86 causes early onset of senescence in mice. Proc. Natl. Acad. Sci. USA 96:10770-10775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Werler, P. J., E. Hartsuiker, and A. M. Carr. 2003. A simple Cre-loxP method for chromosomal N-terminal tagging of essential and non-essential Schizosaccharomyces pombe genes. Gene 304:133-141. [DOI] [PubMed] [Google Scholar]
- 65.Wilson, S., M. Tavassoli, and F. Z. Watts. 1998. Schizosaccharomyces pombe rad32 protein: a phosphoprotein with an essential phosphoesterase motif required for repair of DNA double strand breaks. Nucleic Acids Res. 26:5261-5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wilson, S., N. Warr, D. L. Taylor, and F. Z. Watts. 1999. The role of Schizosaccharomyces pombe Rad32, the Mre11 homologue, and other DNA damage response proteins in non-homologous end joining and telomere length maintenance. Nucleic Acids Res. 27:2655-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]