Abstract
Using a coimmunoprecipitation strategy, we showed that the Cdc34 ubiquitin (Ub)-conjugating enzyme from Saccharomyces cerevisiae self-associates in cell lysates, thereby indicating an in vivo interaction. The ability of Cdc34 to interact with itself is not dependent on its association with the ubiquitin ligase Skp1-Cdc53/Cul1-Hrt1-F-box complex. Rather, this interaction depends upon the integrity of the Cdc34∼Ub thiolester. Furthermore, several principal determinants within the Cdc34 catalytic domain, including the active-site cysteine, amino acid residues S73 and S97, and its catalytic domain insertion, also play a role in self-association. Mutational studies have shown that these determinants are functionally important in vivo and operate at the levels of both Cdc34∼Ub thiolester formation and Cdc34-mediated multi-Ub chain assembly. These determinants are spatially situated in a region that is close to the active site, corresponding closely to the previously identified E2-Ub interface. These observations indicate that the formation of the Cdc34∼Ub thiolester is important for Cdc34 self-association and that the interaction of Cdc34∼Ub thiolesters is in turn a prerequisite for both multi-Ub chain assembly and Cdc34's essential function(s). A conclusion from these findings is that the placement of ubiquitin on the Cdc34 surface is a structurally important feature of Cdc34's function.
Many important processes in eukaryotic cells are regulated by the covalent modification of protein targets by the highly conserved protein ubiquitin (Ub). This modification consists of a cascade of events involving three enzymes or enzyme complexes that activate Ub and transfer it to appropriately selected targets (reviewed in reference 14). In an ATP-dependent first step, the Ub-activating enzyme (E1) activates Ub by forming a high-energy thiolester bond (indicated by ∼) between its active-site cysteine and the carboxy terminus of Ub. Subsequently, Ub is transferred to the active-site cysteine of a Ub-conjugating enzyme (E2) via a transthiolation reaction. E2s, in conjunction with a Ub-ligase enzyme (E3), bind a protein target and transfer Ub to a lysine residue of the target. E3s comprise a diverse group of proteins frequently consisting of multisubunit complexes that function to provide the target recognition component of E2-E3 complexes. Once the initial Ub has been linked to the target, a lysine residue of that Ub may function as the site for further ubiquitination. Repetition of this process is thought to lead to the formation of multi-Ub chains linked to a target. These Ub chains can initiate a variety of actions, most often targeting the modified protein for degradation by the 26S proteasome (29).
The specific mechanism by which multi-Ub chains are assembled is poorly understood. The proper coordination of the E2∼Ub thiolester, the E3, and the target protein is required for target recognition, target ubiquitination, and subsequent multi-Ub chain assembly onto the target. One model has proposed that E2s form multimeric complexes with themselves to facilitate multi-Ub chain assembly (28). Evidence supporting self-association and heteroassociation of E2s has been reported. Several groups have observed that purified E2s can self-associate into dimers or higher-order complexes in vitro (9, 25). Furthermore, purified Caenorhabditis elegans Ubc1 (18), as well as Saccharomyces cerevisiae Ubc4 (12) and Cdc34 (26) proteins, can be cross-linked to themselves in vitro to form higher-order complexes. In vivo evidence for such interactions also exists with the S. cerevisiae Ubc7 protein, based on a two-hybrid interaction with itself (5). The in vivo observation made by Chen et al. (5) that the turnover of the S. cerevisiae MATα2 transcriptional regulator strongly correlates with an interaction between Ubc6 and Ubc7 suggests that these interactions may be relevant.
The relationship between E2-E2 interactions and E2 function is not entirely clear. Silver et al. have presented a model based on genetic evidence suggesting that the function of the S. cerevisiae Cdc34 protein may be dependent on its interaction with itself (28). A variety of essential biological processes are regulated by Cdc34 (11), including the degradation of many key regulators of cell cycle progression (7, 8, 27, 30). The function of Cdc34 is dependent on its ability to build multi-Ub chains onto its targets. Several determinants within Cdc34 are required for this function, including a number of key elements within the Cdc34 catalytic domain and a 125-amino-acid polyacidic carboxy-terminal extension (17, 28). In particular, the first 38 residues of this carboxy-terminal extension have been implicated in Cdc34's ability to interact with its E3 partners (21) and its ability to regulate the cell cycle (26).
We report here evidence that Cdc34 self-associates in vivo. Key residues involved in this interaction that are important for Cdc34's in vivo function as well as its ability to build multi-Ub chains in vitro have also been identified. Moreover, these residues overlap with residues that are important in Ub thiolester formation. Furthermore, we show that under reducing conditions that disrupt Cdc34∼Ub thiolester, self-association is also disrupted. Together our results suggest that Ub thiolester formation facilitates Cdc34 self-association and that self-association is required for Cdc34's catalytic function.
MATERIALS AND METHODS
Plasmids and yeast strains.
The plasmids and yeast strains used in this study are listed in Table 1. The pJD325 plasmid, containing the S. cerevisiae UBA1-his6 gene, was provided to us by Daniel Finley (Harvard Medical School). Versions of the S97D, S73K/S97D (SS), S73K/S97D/Δ12 (SS/Δ12), and Δ12 derivatives (19) were provided to us by Mark Goebl (Indiana University). Cdc34 carboxy-terminal truncations were generated as previously described (26). For Myc-Cdc34, the DNA sequences of CDC34 or cdc34 derivatives were amplified by PCR and inserted 3′ of the Myc epitope tag in the pESC(Trp) galactose-inducible expression plasmid (Stratagene) by using the BglII and PacI restriction enzyme sites. For Flag-Cdc34, the DNA sequences of CDC34 or cdc34 derivatives were amplified by PCR and inserted 3′ of the Flag epitope tag in the pESC(Trp) plasmid by using the XhoI and KpnI restriction enzyme sites. For Cdc53-3xHA, the DNA sequence of CDC53 was amplified by PCR and inserted into the pESC(URA) galactose-inducible expression plasmid (Stratagene) by using the BamHI and XhoI restriction enzyme sites. Three hemagglutinin (HA) epitope tags were cloned into a NotI restriction enzyme site that was engineered at the 3′ end of CDC53 before the stop codon. The yeast HA-Ub expression plasmid was generated by inserting a DNA segment encoding the HA epitope into the high-copy-number YEp352 Ub plasmid described previously (1), using the EcoRI and BglII restriction enzyme sites.
TABLE 1.
Yeast strains and plasmids used in this study
| Yeast strain or plasmid | Genotype or description | Source or reference |
|---|---|---|
| Strains | ||
| MHY501 | MATα his3-Δ200 leu2-3, 112 trp1-1 ura3-52 lys2-801 gal2 | M. Hochstrasser |
| YPH499 | MATaade2-101 his3-Δ200 leu2-Δ1 trp1-Δ63 ura3-52 lys2-801 | Stratagene |
| 15DaubΔ | MATabar1Δ ade1 his2 leu2-3, 112 trp1-1 ura3Δns | D. Stuart |
| DSY1105 | MATabar1Δ ade1 his2 leu2-3, 112 trp1-1 ura3Δns cdc53-1 | D. Stuart |
| Plasmids | ||
| Wild-type and control plasmids | ||
| pJD325 | 2μm PCUP1-UBA1-6xHis [LEU2] | D. Findley |
| pES5 | 2μm PCUP1-HA-Ub [URA3] | This study |
| pESC200 | 2μm PGAL1-CDC53-3xHA [URA3] | This study |
| pESC(Trp) | 2μm PGAL1-Myc, PGAL10-Flag [TRP1] | Stratagene |
| pESC(Ura) | 2μm PGAL1-Myc, PGAL10-Flag [URA3] | Stratagene |
| pESC1 | 2μm PGAL10-Flag-CDC34 [TRP1] | This study |
| pESC2 | 2μm PGAL1-Myc-CDC34 [TRP1] | This study |
| pESCA | 2μm PGAL1-Myc-CDC34, PGAL10-Flag-CDC34 [TRP1] | This study |
| Cdc34 catalytic domain derivative plasmids | ||
| pESC15 | 2μm PGAL10-Flag-cdc34 C95A [TRP1] | This study |
| pESC16 | 2μm PGAL1-Myc-cdc34 C95A [TRP1] | This study |
| pESCH | 2μm PGAL1-Myc-cdc34 C95A, PGAL10-Flag-cdc34 C95A [TRP1] | This study |
| pESC19 | 2μm PGAL10-Flag-cdc34 S97D [TRP1] | This study |
| pESC20 | 2μm PGAL1-Myc-cdc34 S97D [TRP1] | This study |
| pESCJ | 2μm PGAL1-Myc-cdc34 S97D, PGAL10-Flag-cdc34 S97D [TRP1] | This study |
| pESC3 | 2μm PGAL10-Flag-cdc34 S73K/S97D [TRP1] | This study |
| pESC4 | 2μm PGAL1-Myc-cdc34-S73K/S97D [TRP1] | This study |
| pESCB | 2μm PGAL1-Myc-cdc34 S73K/S97D, PGAL10-Flag-cdc34 S73K/S97D [TRP1] | This study |
| pESC5 | 2μm PGAL10-Flag-cdc34Δ12 [TRP1] | This study |
| pESC6 | 2μm PGAL1-Myc-cdc34Δ12 [TRP1] | This study |
| pESCC | 2μm PGAL1-Myc-cdc34Δ12, PGAL10-Flag-cdc34 Δ12 [TRP1] | This study |
| pESC7 | 2μm PGAL10-Flag-cdc34 S73K/S97D/Δ 12 [TRP1] | This study |
| pESC8 | 2μm PGAL1-Myc-cdc34 S73K/S97D/Δ 12 [TRP1] | This study |
| pESCD | 2μm PGAL1-Myc-cdc34 S73K/S97D/Δ 12, PGAL10-Flag-cdc34 S73K/S97D/Δ 12 [TRP1] | This study |
| Cdc34 carboxy-terminal truncation plasmids | ||
| pESC9 | 2μm PGAL10-Flag-cdc34Δ 170[TRP1] | This study |
| pESC10 | 2μm PGAL1-Myc-cdc34Δ 170[TRP1] | This study |
| pESCE | 2μm PGAL1-Myc-cdc34Δ 170, PGAL10-Flag-cdc34Δ 170 [TRP1] | This study |
| pESC11 | 2μm PGAL10-Flag-cdc34Δ 185[TRP1] | This study |
| pESC12 | 2μm PGAL1-Myc-cdc34Δ 185[TRP1] | This study |
| pESCF | 2μm PGAL1-Myc-cdc34Δ 185, PGAL10-Flag-cdc34 Δ185 [TRP1] | This study |
| pESC13 | 2μm PGAL10-Flag-cdc34Δ 209[TRP1] | This study |
| pESC14 | 2μm PGAL1-Myc-cdc34Δ 209[TRP1] | This study |
| pESCG | 2μm PGAL1-Myc-cdc34Δ 209, PGAL10-Flag-cdc34 Δ209 [TRP1] | This study |
| Bacterial expression plasmids | ||
| pREGB1 | pET-3a derivative + Ub | 15 |
| pREGB6 | pET-3a derivative + CDC34 | 26 |
| pCdc34Δ185 | pET-3a derivative + cdc34Δ185 | 26 |
| pCdc34Δ209 | pET-3a derivative + cdc34Δ209 | 26 |
| pBV4 | pET-3a derivative + cdc34C95A | This study |
| pBV8 | pET-3a derivative + cdc34S97D | This study |
| pBV9 | pET-3a derivative + cdc34S73K/S97D | This study |
A modified derivative of the pET3a plasmid was used to construct the Escherichia coli overexpression plasmids as previously described (26). The DNA sequences of CDC34 or the cdc34 derivatives were amplified by PCR and then inserted this into the pET3a derivative plasmid by using the SacI and KpnI sites.
All oligonucleotides used were synthesized by the Department of Biochemistry DNA Core Facility at the University of Alberta (Edmonton, Canada), and their sequences are available upon request. The restriction enzymes used were purchased from Promega and New England Biolabs. E. coli strain MC1061 was used as the host for all plasmid expression and was grown in Luria broth medium in the presence of required antibiotics (2). All plasmids were sequenced with a Beckman Coulter CEQ 2000 XL DNA analysis system.
Yeast strains 15DaubΔ and DSY1105 were provided by David Stuart (University of Alberta). Yeast strain MHY501 (23), used for Uba1 expression, was provided by Mark Hochstrasser (Yale University). All yeast strains were cultured in either a rich medium (1% yeast extract, 2% Bacto Peptone, and 2% glucose) or an SD medium (0.67% yeast nitrogen base without amino acids, 2% glucose, and the appropriate amino acids) as previously described (16).
Chemical cross-linking.
Yeast cells expressing Flag-Cdc34 were grown at 30°C to an optical density at 600 nm (OD600) of 0.8. The cells were then harvested, washed with water, and lysed in ice-cold cross-linking buffer (50 mM HEPES [pH 7.0], 100 mM NaCl, 1 mM EDTA, 0.1% NP-40, protease inhibitor cocktail [Sigma]) with the use of glass beads. The lysate was then centrifuged at 10,000 × g for 15 min at 4°C to remove debris. The cross-linker disuccinimidyl suberate (DSS) (Pierce) was then added to the lysate (5 mM final) and incubated for 30 min at 30°C. Free reactive groups of DSS were quenched by treatment of the sample with 50 mM Tris-Cl (pH 7.5) for 30 min at 30°C.
Immunoprecipitation and immunoblotting.
Coimmunoprecipitation experiments were carried out with the yeast strains YPH499, 15DaubΔ, and DSY1105 as described in Results. For the galactose-inducible expression of proteins in yeast, the recommended protocol of the manufacturer (Stratagene) was followed, using SSG medium (0.67% yeast nitrogen base without amino acids, 2% galactose, and the appropriate amino acids). Yeast cells expressing Myc-Cdc34, Flag-Cdc34, or Myc- and Flag-Cdc34 were grown to an OD600 of 0.8. Cells were harvested, washed with water, and lysed in ice-cold lysis buffer (50 mM HEPES [pH 7.5], 100 mM NaCl, 1 mM EDTA, 0.1% NP-40, protease inhibitor cocktail [Sigma]) by using glass beads. Lysates were then centrifuged at 10,000 × g for 15 min at 4°C to remove debris. The supernatants were precleared with 25 μl of protein G-agarose (Roche) for 1 h with gentle rotation at 4°C. This was followed by incubation of the lysates with 5 μl of anti-Myc antibody (0.4 mg/ml) (9E10; Roche) or 5 μl of anti-Flag antibody (0.4 mg/ml) (M2; Sigma) for 2 h at 4°C and then by incubation with 20 μl of protein G-agarose for 2 h with gentle rotation at 4°C. The immunoprecipitate-bead complexes were washed four times with ice-cold lysis buffer. The complexes were then boiled for 5 min in sodium dodecyl sulfate (SDS) loading buffer plus 10 mM dithiothreitol (DTT). The samples were subjected to SDS-12% polyacrylamide gel electrophoresis (12% SDS-PAGE), transferred to polyvinylidene fluoride membranes, and analyzed by immunoblotting with anti-Flag horseradish peroxidase (HRP)-conjugated antibody (M2; Sigma) or anti-Myc HRP-conjugated antibody (9E10; Roche).
The same procedure as described above was used for the experiment expressing Cdc53-3xHA in combination with Flag-Cdc34 or one of the Flag-Cdc34 derivatives, except that 5 μl of anti-HA antibody (0.4 mg/ml) (12CA5; Roche) was used for the immunoprecipitation and anti-HA HRP-conjugated antibody (3F10; Roche) was used for immunoblotting.
For the experiments involving HA-Ub, the cells were induced at an OD600 of 0.2 for HA-Ub expression by adding CuSO4 to the medium (0.1 mM final concentration). Immunoprecipitation was done as described above, as was the immunoblotting for Flag- and Myc-Cdc34. HA-Ub was analyzed by immunoblotting with an anti-HA HRP-conjugated antibody (3F10; Roche). For DTT treatment of samples prior to the immunoprecipitation, total cell extracts expressing HA-Ub, Flag-Cdc34, and Myc-Cdc34 were treated with or without DTT (100 mM final concentration) for 2 h at 30°C. The lysates were subsequently dialyzed into dialysis buffer (50 mM HEPES [pH 7.5], 100 mM NaCl) for 2 h at 4°C. Immunoprecipitations and immunoblotting of the treated lysates were carried out as described above.
Protein expression and purification.
Cdc34 and its derivatives were expressed from a modified derivative of pET3a (26). Cell harvesting and lysis were performed as previously described for Cdc34 (26). 35S-Ub was produced by using labeling procedures previously described (26). Purification of 35S-Ub was carried out by using a variation of a previously described technique (15). Briefly, E. coli extracts containing 35S-Ub were dialyzed at 4°C for 4 h against buffer A (50 mM Tris [pH 7.5], 1 mM DTT, 1 mM EDTA). The dialysates were then loaded onto a 1-ml HiTrap Q-Sepharose HP anion-exchange column (Pharmacia) equilibrated with buffer A and eluted with the same buffer at a flow rate of 1 ml/min. Flowthrough fractions containing 35S-Ub were then passed over a 1-ml HiTrap Q-Sepharose HP cation-exchange column (Pharmacia) equilibrated with buffer B (50 mM HEPES [pH 7.5], 1 mM DTT, 1 mM EDTA) and chromatographed with the same buffer at a flow rate of 1 ml/min. Flowthrough fractions containing 35S-Ub were then concentrated to 200 μl and passed over an HR 10/30 Superdex 75 gel filtration column (Pharmacia) equilibrated with buffer C (50 mM HEPES [pH 7.5], 150 mM NaCl, 1 mM EDTA, 0.1 mM DTT). 35S-Ub that was eluted off the column was concentrated and stored frozen in 10% glycerol at −80°C.
For the purification of S. cerevisiae Uba1, MHY501 yeast cells carrying the pJD325 plasmid were grown in SD medium lacking leucine to an OD600 of 0.2 at 30°C. CuSO4 was then added to the medium (0.1 mM final concentration), and the cells were grown to an OD600 of 1.0 at 30°C. Cells were then harvested, washed with water, and lysed in buffer D (50 mM Tris [pH 7.5], 1 mM EDTA, 1 mM beta-mercaptoethanol) with a protease inhibitor cocktail (Sigma). The lysate was then passed over a 5-ml HiTrap Q-Sepharose HP anion-exchange column (Pharmacia) equilibrated with buffer D, and the protein was eluted with an NaCl gradient of 0 to 1 M with buffer E (50 mM Tris [pH 7.5], 1 mM EDTA, 2 M NaCl, 1 mM beta-mercaptoethanol). Uba1-6His eluted at approximately 250 mM NaCl. The anion-exchange column fractions were then passed over a 5-ml HiTrap chelating column (Pharmacia) charged with CuSO4. The column was washed with four times its volume of 10 mM imidazole, and Uba1-6His was eluted with 500 mM imidazole. The eluted Uba1-6His was then concentrated and run over a High Load 16/60 Superdex 200 gel filtration column (Pharmacia) equilibrated with buffer C. Peak fractions of Uba1-6His were collected, pooled, and concentrated. Glycerol was added to a concentration of 10%, and the protein was frozen at −80°C. Uba1 concentrations were based on Ub-activating activity. A reaction mixture containing a sample of Uba1 and a known amount of 35S-Ub in 0.5 ml of ATP cocktail (30 mM HEPES [pH 7.5], 5 mM MgCl2, 5 mM ATP, 0.6 U of inorganic phosphatase per ml) was incubated at 30°C for 1 h. The sample was then run over an HR 10/30 Superdex 200 gel filtration column (Pharmacia) equilibrated with buffer C lacking DTT. The counts per minute of the 35S-Ub were monitored, and the incorporation of 35S-Ub into the Uba1 peak to form Uba1∼(Ub)2 was used to determine the concentration of Uba1.
In vitro Cdc34-Ub thiolester assays.
Thiolester reaction mixtures contained 100 nM Cdc34 or Cdc34 derivatives, 200 nM 35S-Ub (with a specific activity of 1.5 × 105 cpm/μg), and 10 nM Uba1-6xHis and were incubated at 30°C for 5 min in 2.5 ml of ATP cocktail. The 5-min reaction time ensures that little if any Cdc34-Ub conjugates form as a result of Cdc34 autoubiquitination. Reactions were stopped by adding 50 mM EDTA to chelate any unused Mg2+, thereby inactivating Uba1. Following incubation, each reaction mixture was immediately passed over a 1-ml HiTrap Q-Sepharose HP anion-exchange column (Pharmacia) equilibrated with buffer F (50 mM Tris [pH 7.5], 1 mM EDTA). Proteins were eluted with an NaCl gradient of 0 to 800 mM NaCl gradient with buffer G (50 mM Tris [pH 7.5], 1 mM EDTA, 2 M NaCl), and 0.5-ml fractions were collected. The radioactivity (counts per minute) fell into three well-resolved peaks corresponding either to 35S-Ub or to 35S-Ub that had been incorporated into Cdc34∼Ub thiolester or Uba1∼Ub thiolester.
All Cdc34 derivatives were analyzed for Ub thiolester formation by the above method, except Cdc34 Δ209 and Δ185, which could not be resolved in this way due to their reduced anionic character. For these two derivatives, reactions were carried out in the same manner as described above, but the mixtures were instead were loaded onto a High Load Superdex 75 16/60 gel filtration column (Pharmacia) equilibrated with buffer C lacking DTT. Proteins were eluted in 0.5-ml fractions, and again the radioactivity fell into three well-resolved peaks. Thiolester yields for all reactions were calculated by determining the specific radioactivity of the incorporated 35S-Ub and were expressed as percentages of the total Ub in the reaction.
Cdc34 autoubiquitination reactions.
All Cdc34-Ub conjugation reactions were performed by incubating 100 nM Cdc34 or Cdc34 derivatives with 200 nM 35S-Ub and 10 nM Uba1-6xHis at 30°C for 8 h in ATP cocktail. The reactions were terminated by the addition of 10 mM DTT, which was immediately followed by precipitation with 10% trichloroacetic acid. Samples were then resuspended and boiled for 5 min in SDS loading buffer, followed by separation by 12% SDS- PAGE and analysis by autoradiography as described previously (15).
RESULTS
Cdc34 self-associates.
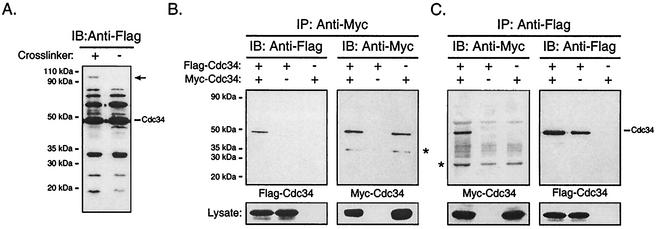
We have previously shown by using cross-linking studies that purified Cdc34 is capable of self-association in vitro (26). Our genetic observations have also suggested that Cdc34 may self-associate in vivo (26, 28). To investigate these observations further, we carried out cross-linking reactions with prepared whole-cell lysates from cells expressing Cdc34 amino-terminally tagged with the Flag epitope, Flag-Cdc34. Flag-Cdc34 was expressed in a wild-type yeast strain from a high-copy-number plasmid under control of a galactose-inducible promoter (Table 1). Cells were grown in galactose medium, harvested, and lysed. The lysate was subsequently treated with or without the cross-linker DSS and then subjected to SDS-PAGE. Flag-Cdc34 present within the lysates was visualized by immunoblotting with an anti-Flag antibody (Fig. 1A). In both the DSS-treated and untreated samples, multiple bands containing Flag-Cdc34 were detected. Bands having a molecular mass lower than that of Cdc34 likely correspond to partial degradation products. Bands having a molecular mass greater than that of Cdc34 and present in both the DSS-treated and untreated samples likely correspond to ubiquitinated and/or phosphorylated Cdc34, as previously described (10). Interestingly, the DSS-treated sample also exhibited a band that was absent in the untreated sample. This species corresponds to a specific cross-linked product containing Flag-Cdc34. The molecular mass of the cross-linked product was approximately double that of Cdc34, suggesting that it is a cross-linked dimer of Cdc34.
FIG. 1.
Cdc34 self-associates in vivo. (A) Cross-linking. Cell lysates from YPH499 yeast cells expressing Flag-Cdc34 were treated with or without the chemical cross-linker DSS. Cell lysates were subsequently analyzed by immunoblotting (IB) with an anti-Flag antibody. The position of Flag-Cdc34 is indicated, as is the position of a unique cross-linked product containing Flag-Cdc34 (arrow). (B and C) Coimmunoprecipitation. Total cell extracts of YPH499 cells expressing Flag-Cdc34, Myc-Cdc34, or both Flag-Cdc34 and Myc-Cdc34 were prepared. (B) Myc-Cdc34 was immunoprecipitated (IP) with an anti-Myc antibody, followed by immunoblotting with an anti-Myc antibody (right panel) to detect the amount of Myc-Cdc34 that had immunoprecipitated and with an anti-Flag antibody (left panel) to detect the amount of Flag-Cdc34 that had coimmunoprecipitated. (C) The reciprocal coimmunoprecipitation experiment to that for panel B was performed. Flag-Cdc34 was immunoprecipitated with an anti-Flag antibody, followed by immunoblotting with an anti-Flag antibody (right panel) and with an anti-Myc antibody (left panel). The position of Cdc34 is indicated, as is the position of a proteolytic product of Cdc34 (*). Protein expression levels within the lysates used for immunoprecipitations are shown at the bottoms of panels B and C.
Although this cross-linked product may have resulted from Cdc34 self-association, it remained a possibility that the unique band within the DSS-treated sample represented Flag-Cdc34 cross-linked to some other protein. To eliminate this possible ambiguity, we attempted to detect Cdc34 self-association by coimmunoprecipitation. Amino-terminally Flag- and Myc-tagged derivatives of Cdc34 were employed for coimmunoprecipitation. Each tagged version of Cdc34 was expressed in a wild-type yeast strain from a double-expression yeast plasmid from which both derivatives were under the control of galactose-inducible promoters, i.e., Gal10 for Flag-Cdc34, and Gal1 for Myc-Cdc34. The cells were induced by growth in galactose-containing medium and were subsequently lysed and subjected to immunoprecipitation with an anti-Myc antibody to isolate Myc-Cdc34 from the lysates. Immunoprecipitates were then analyzed by immunoblotting with an anti-Flag antibody to determine whether Flag-Cdc34 coimmunoprecipitated with Myc-Cdc34. As a control, cells expressing either Myc-Cdc34 or Flag-Cdc34 alone were also used. Flag-Cdc34 was detected by immunoblotting only when coexpressed with Myc-Cdc34, thereby demonstrating an interaction between the two (Fig. 1B). Similar results were also generated from the reciprocal experiment, in which an anti-Flag antibody was used for immunoprecipitation followed by immunoblotting with an anti-Myc antibody (Fig. 1C). Taken together, these observations indicate that Cdc34 self-associates in vivo.
The carboxy-terminal extension and the catalytic domain insertion of Cdc34 are not required for Cdc34 self-association.
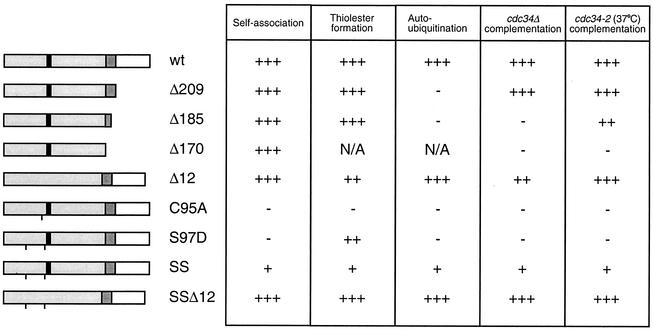
We wished to determine which sequence elements within Cdc34 were required for its ability to self-associate in cell lysates. The most obvious features of Cdc34 that distinguish it from other E2s include a carboxy-terminal extension and a catalytic domain insertion (see Fig. 8). Previous work showed that deletion of the catalytic domain 12-amino-acid insert (Δ12) does not eliminate the ability of Cdc34 to carry out its cell cycle function (19), whereas the carboxy-terminal extension, specifically residues 171 to 209, was required for its cell cycle function. Furthermore, these residues also appeared to stabilize the cross-linking of Cdc34 with itself in vitro, although residues 186 to 209 were dispensable at high concentrations (26).
FIG. 8.
Functional comparison of the Cdc34 derivatives. The primary structures of various Cdc34 derivatives are shown, with the catalytic domain shown in light gray and the 12-amino-acid insert that is present within the catalytic domain shown in black (amino acids 103 to 114). The carboxy-terminal extension is shown in white, and residues of this domain previously shown to be necessary and sufficient for Cdc34 function are shown in dark gray (amino acids 170 to 209) (21). A line at the appropriate position along the block diagram indicates the position of the point substitution(s) present in each derivative. The various derivatives are scored based on their abilities to self-associate, form Cdc34∼Ub thiolester, build multi-Ub chains in an autoubiquitination reaction, and complement either a cdc34 disruption strain (cdc34Δ) or a cdc34(Ts) strain (cdc34-2) (3, 19, 26). Each derivative was scored relative to wild-type (wt) Cdc34 (+++) such that partial function was scored as ++ or + and the absence of function was scored as −. N/A, not applicable.
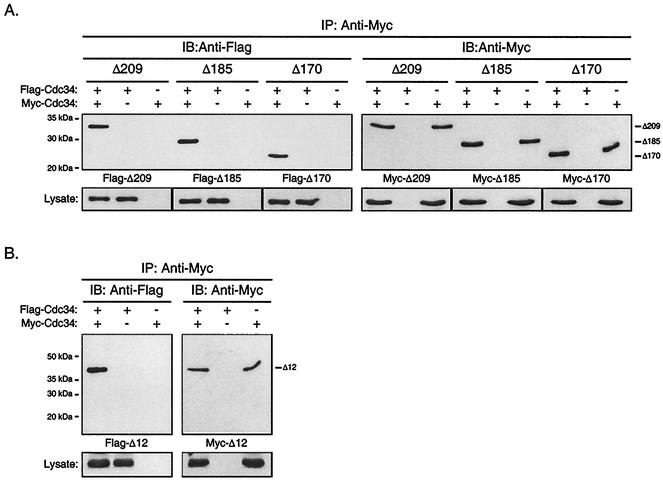
A Cdc34 derivative lacking the catalytic domain insert as well as several carboxy-terminal truncation derivatives were employed to assess whether these residues were required for Cdc34 self-association (Fig. 2). Flag- and Myc-tagged versions of each derivative were generated, and each was expressed from the double-expression yeast plasmid from either the Gal10 (Flag derivatives) or the Gal1 (Myc derivatives) promoter as described above for full-length Cdc34 (Table 1).
FIG. 2.
The Cdc34 carboxy-terminal extension and catalytic domain insertion are dispensable with respect to Cdc34 self-association. The same coimmunoprecipitation strategy as employed for Cdc34 (Fig. 1B) was employed for the Δ209 (residues 1 to 209), Δ185 (residues 1 to 185), and Δ170 (residues 1 to 170) carboxy-terminal truncation derivatives (A) as well as for the Δ12 (residues 103 to 114 deleted) catalytic domain deletion derivative (B). Flag- and Myc-tagged versions of these truncation derivatives were expressed in YPH499 cells. Cell lysates were extracted and subjected to immunoprecipitation (IP) with an anti-Myc antibody followed by immunoblotting (IB) with an anti-Myc antibody (right panels) and an anti-Flag antibody (left panels). Protein expression levels within the lysates used for immunoprecipitations are shown at the bottoms of the respective panels.
If either the catalytic domain insert or carboxy-terminal extension (in particular residues 171 to 209) were required for Cdc34 self-association, then a significant reduction or loss of signal corresponding to Flag-Cdc34 would be expected upon coimmunoprecipitation when these derivatives were employed. Deletion of neither the catalytic domain insertion (Fig. 2B) nor portions of the carboxy-terminal extension (Fig. 2A) had a significant affect on the ability of Cdc34 to self-associate. In fact, truncation of the entire carboxy-terminal extension (Δ170) had no observable effect on the interaction (Fig. 2A; also see Fig. 4). Thus, under the conditions employed here, both the catalytic domain insert and the carboxy-terminal extension of Cdc34 are dispensable with respect to Cdc34 self-association.
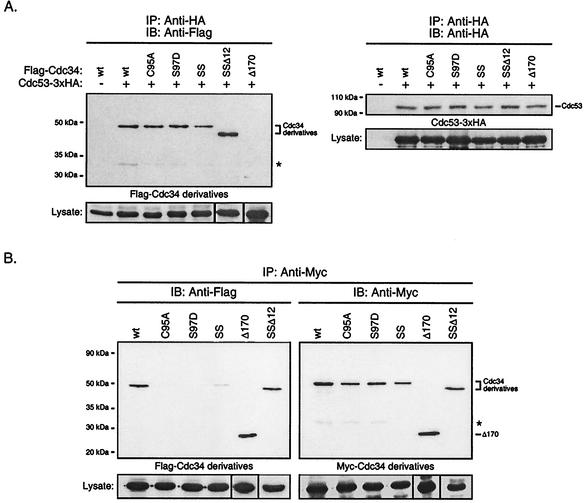
FIG. 4.
(A) Catalytic domain derivatives of Cdc34 interact with Cdc53 of the SCF complex. Total cell extracts were prepared from YPH499 cells coexpressing Cdc53-3xHA and Flag-Cdc34 or one of the Flag-tagged Cdc34 derivatives: C95A, S97D, S73K/S97D (SS), S73K/S97D/Δ12 (SSΔ12), or Δ170. Cdc53-3xHA was immunoprecipitated (IP) with an anti-HA antibody and was detected by immunoblotting (IB) with an anti-HA antibody (right panel). The amount of Flag-Cdc34 or Flag-Cdc34 derivative that coimmunoprecipitated was detected by immunoblotting with an anti-Flag antibody (left panel). The positions of Cdc53, Cdc34, and the Cdc34 derivatives are indicated. The asterisk indicates a proteolytic product of Cdc34. Protein expression levels within the lysates used for immunoprecipitations are shown at the bottoms of the respective panels. (B) Residues within the Cdc34 catalytic domain critical for self-association. The same coimmunoprecipitation strategy as employed for Cdc34 (Fig. 1B) was used for the C95A, S97D, S73K/S97D (SS), and S73K/S97D/Δ12 (SSΔ12) catalytic domain derivatives. Flag- and Myc-tagged versions of these derivatives were expressed in YPH499 cells. Cell lysates were extracted and subjected to immunoprecipitation with anti-Myc antibody followed by immunoblotting with anti-Myc antibody (right panel) and anti-Flag antibody (left panel). The asterisk indicates a proteolytic product of Cdc34. Protein expression levels within the lysates used for immunoprecipitations are shown at the bottoms of the respective panels. wt, wild type.
Cdc34 self-association is not dependent upon a functional SCF complex.
In previous work Cdc34 self-association had been demonstrated in vitro only by cross-linking experiments. Other physical means, however, such as gel filtration or analytical ultracentrifugation, indicated that under the conditions used, Cdc34 exhibited the hydrodynamic properties of an asymmetric monomer (26). Therefore, it appeared that Cdc34 self-association comprised a highly transient interaction, and we postulated that an auxiliary factor(s) is required to stabilize Cdc34's interaction with itself. The multisubunit Skp1-Cdc53/Cul1-Hrt1-F-box (SCF) complex, an E3 enzyme required for Cdc34's cell cycle function, has been shown to interact with Cdc34 (21) and thus was a potential candidate for such a factor. However, the interaction of Cdc34 with the SCF complex is dependent upon residues 170 to 209 of Cdc34's carboxy-terminal extension. As shown in Fig. 2A, under these conditions this region is dispensable with respect to Cdc34 self-association, suggesting that a Cdc34-SCF interaction is not required for self-association.
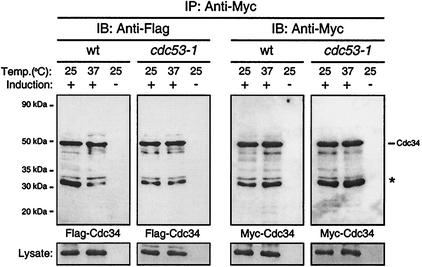
We tested the dependence of Cdc34 self-association on the SCF complex by performing the coimmunoprecipitation experiment described above with a yeast strain carrying a temperature-sensitive mutation in Cdc53, a core component of the SCF complex that directly interacts with Cdc34 (20). We used the cdc53-1 mutant strain, which has a single point mutation (R488C) in a region at the carboxy terminus of Cdc53 shown to be essential for Cdc34 binding (24). Expression of Myc-Cdc34 and Flag-Cdc34 was induced within the wild-type and cdc53-1 strains by growing each in galactose medium. Cells grown at either their permissive (25°C) or nonpermissive (37°C) temperature were subsequently lysed and subjected to immunoprecipitation followed by immunoblotting as described above. At the nonpermissive temperature, the cdc53-1 mutation disrupts critical SCF functions. If Cdc34 self-association required interaction with a functional SCF complex, it was expected that growth at the nonpermissive temperature would result in a significant reduction or loss of signal corresponding to Flag-Cdc34 for the cdc53-1 strain relative to the signal observed at the permissive temperature. Furthermore, this signal would also be reduced relative to that observed for the wild-type strain. As seen in Fig. 3 this comparison indicated that there is no significant loss of the ability of Cdc34 to self-associate within a strain defective in SCF function. This result coupled with the observation that the carboxy-terminal extension is not required for this interaction suggests that a Cdc34-SCF interaction is not mediating the ability of Cdc34 to interact with itself.
FIG. 3.
Cdc34 self-association is not dependent upon a functional SCF complex. Flag-Cdc34 and Myc-Cdc34 were expressed in 15DaubΔ (wild type [wt]) or DSY1105 (cdc53-1) cells at either the permissive (25°C) or nonpermissive (37°C) temperature. Cell extracts were prepared, and Myc-Cdc34 was immunoprecipitated (IP) with an anti-Myc antibody, followed by immunoblotting (IB) with either an anti-Myc antibody (right panel) or an anti-Flag antibody (left panel). The asterisk indicates a proteolytic product of Cdc34. Protein expression levels within the lysates used for immunoprecipitations are shown at the bottoms of the respective panels.
Residues within the Cdc34 catalytic domain that are required for self-association.
The inability to disrupt Cdc34 self-association by deleting either the carboxy-terminal extension or the catalytic domain insertion meant that other residues within the catalytic domain had to play a role in stabilizing the interaction. Furthermore, if Cdc34 self-association is necessary for its cell cycle function, then it was possible that residues previously identified as important to Cdc34 function could also be important in self-association. Of particular relevance to this study were several Cdc34 derivatives described by Liu et al. (19). Their genetic observations revealed an essential relationship between S73, S97, and the catalytic domain insertion (residues 103 to 114). They reasoned that because these residues differ from their counterparts in other E2s, they contribute to Cdc34's unique functionality. In particular, they found that replacement of S97 with aspartic acid, the residue found in the analogous position of other E2s, resulted in loss of function. Changing S73 to a lysine, the corresponding residue in other E2s, in combination with S97D slightly improved Cdc34 function but again could not fully complement the defect. However, when the S73K and S97D substitutions were made in combination with the deletion of the catalytic insertion (Δ12), full Cdc34 function was restored.
The functional importance of these residues was not clear, but they are distinct from the residues found at analogous positions in most other E2s, suggesting that they are required for the functional specificity of Cdc34 (19). The uniqueness of these residues to Cdc34 suggested that they might play a role in the interaction between Cdc34 and the E3 SCF complex. The effect of these changes on the ability of Cdc34 to interact with the SCF complex was tested by coimmunoprecipitation of Cdc34 with Cdc53, its interacting partner within the SCF complex. Cdc53 triply tagged with the HA epitope at its carboxy-terminus (Cdc53-3xHA) was coexpressed with each catalytic domain derivative of Cdc34 amino-terminally tagged with the Flag epitope. Cell lysates were immunoprecipitated with an anti-HA antibody to pull down Cdc53. Immunoblotting was subsequently carried out to determine whether each Cdc34 derivative as well as Cdc53 was present in the immunoprecipitates. As expected, the interaction between Cdc34 and Cdc53 was shown to be lost when a carboxy-terminal truncation of Cdc34 was employed (Fig. 4A, lanes Δ170). On the other hand, the catalytic domain derivatives tested (in particular the S97D and SS derivatives) (Fig. 4A) coimmunoprecipitated with Cdc53 to levels similar to those observed for wild-type Cdc34. These observations indicate that the in vivo defect associated with changes to residues 73 and 97 of the catalytic domain of Cdc34 do not affect its ability to interact with a core component of the SCF complex.
Based on this result, we tested the alternative possibility that these residues are required for the ability of Cdc34 to interact with itself. We therefore tested these derivatives for their ability to self-associate by coimmunoprecipitation, as described above, using amino-terminally tagged versions of each. Both Cdc34 and the Δ170 carboxy-terminal truncation derivative were employed as positive controls.
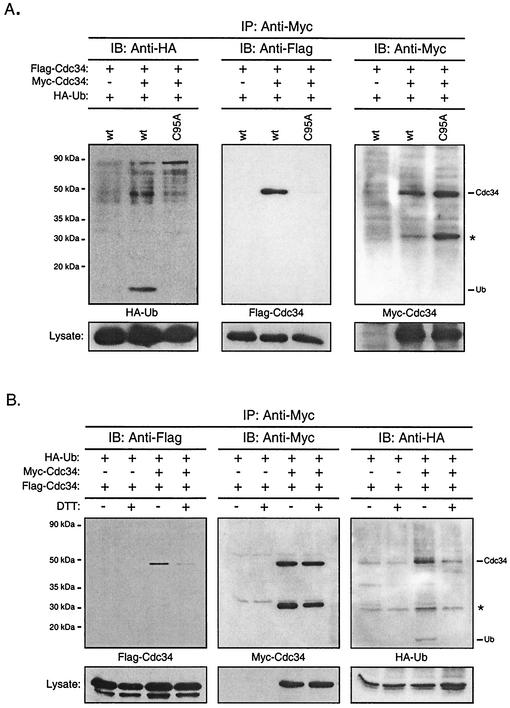
The modification of these catalytic domain residues proved to have a significant effect on the ability of Cdc34 to self-associate (Fig. 4B). In particular, the amino acid substitution S97D resulted in little, if any, detectable Flag-S97D in the immunoprecipitates, whereas Myc-S97D was detected at similar levels to those of both Cdc34 and Δ170. The complete loss of function for these derivatives in vivo provides further evidence that Cdc34 self-association is an important mechanistic feature of its function. Consistent with in vivo function, the effect of the S97D substitution was eliminated in part or fully by the introduction of additional changes to the catalytic domain. When the double substitution S73K/S97D was made, partial self-association was restored as evidenced by the weak Flag-S73K/S97D signal in the immunoprecipitate (Fig. 4B, lanes SS). When this derivative was further modified such that the catalytic domain insertion was deleted in combination with the S73K/S97D substitutions, this derivative fully restored the ability of Cdc34 to self-associate (Fig. 4B, lanes SSΔ12), thereby overcoming the self-association defect observed for the S97D derivative. Together these data indicate a supportive and interdependent role for these residues in both Cdc34 self-association and the cell cycle function of Cdc34. Furthermore, the ability of position 97 to mediate Cdc34 self-association is context dependent.
In vitro studies have previously suggested that an additional factor(s) is required to stabilize Cdc34 self-association (26). We had previously proposed that Cdc34∼Ub thiolester formation could be a prerequisite for self-association in which the covalently linked Ub of one Cdc34 monomer makes favorable contact with the other (26, 28). This model, together with the close proximity of residues defective in self-association to the catalytic cysteine residue (see Fig. 9), prompted us to examine the importance of the active site in self-association. If this model was correct, it would be expected that a cysteine-to-alanine amino acid substitution at the active site would eliminate Cdc34's ability to self-associate as a consequence of its inability to accept Ub. Significantly, the C95A active-site derivative exhibited considerable loss of its ability to self-associate. Moreover, this loss could not be attributed to an inability to immunoprecipitate Myc-C95A (Fig. 4B, lanes C95A). This result therefore is consistent with the idea that Cdc34 self-association is facilitated by the Cdc34∼Ub thiolester.
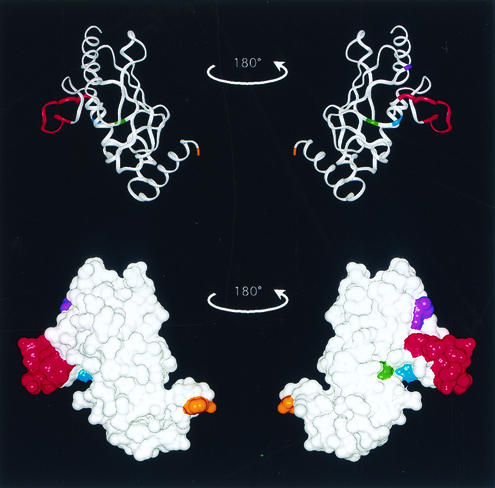
FIG. 9.
Determinants of Cdc34 self-association. The crystal structure of the S. cerevisiae Ubc7 catalytic domain (6) is used here as a template for highlighting key residues within Cdc34 and is represented as both a ribbon diagram (top) and a surface model (bottom). The amino terminus is found at the top of the structures and the carboxy terminus is found at the bottom. The residue that corresponds to residue 170, where the carboxy-terminal extension of Cdc34 would start, is highlighted (orange). Amino acid residues that play a role in Cdc34 self-association are also highlighted and include the active-site residue C95 (yellow), S73 (purple), S97 (light blue), and the catalytic domain insert (amino acids 103 to 114) (red).
Relationship between self-association, thiolester formation, and autoubiquitination.
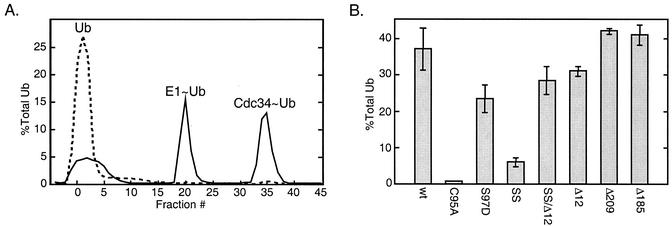
The apparent dependence of Cdc34 self-association on thiolester formation prompted us to address the converse question, i.e., does thiolester formation depend on self-association? We tested each of the Cdc34 derivatives described above with an in vitro thiolester formation assay that used purified E1, E2, and radiolabeled Ub as a probe (see Materials and Methods). Restricting reactions to 5 min eliminated the complication of Cdc34 autoubiquitination that results from much longer incubations. With two exceptions, the level of each Cdc34 thiolester derivative was measured by exploiting the observation that they elute from an anion-exchange column at an NaCl concentration different from those for Ub and Uba1∼Ub (Fig. 5A). The thiolester forms of the Δ209 and Δ185 tail deletion derivatives were observed to elute at lower NaCl concentrations as a consequence of the reduced anionic character that results from tail deletion. Consequently, these species were separated from the other reaction components by gel filtration. Each of the Cdc34 thiolester derivatives exhibited sensitivity to DTT, a characteristic feature of the thiolester covalent bond.
FIG. 5.
Ub thiolester formation. In vitro Ub thiolester formation was compared for a number of Cdc34 derivatives. Reaction mixtures containing Cdc34 or one of its derivatives (100 nM), Uba1 (10 nM), 35S-Ub (200 nM), and an ATP cocktail were incubated for 5 min at 30°C. Reactions were stopped by the addition of 50 mM EDTA, and the mixtures were then immediately loaded onto an anion-exchange column to separate reaction products. The Δ209 and Δ185 derivatives were separated by using a Superdex 75 HR10/30 size exclusion column (see Materials and Methods). (A) Example of the elution profile generated for a reaction mixture containing Cdc34 (solid line). Peaks containing 35S-Ub correspond to free Ub, Ub incorporated into Uba1 (E1∼Ub), and Cdc34 (Cdc34∼Ub) thiolester. In a separate reaction mixture, 10 mM DTT was added to disrupt thiolester (dashed line). (B) Incorporation of 35S-Ub into thiolester was determined for each Cdc34 derivative as a percentage of the total 35S-Ub added to each reaction mixture. The mean and standard deviation observed for three separate reactions for each derivative is shown. SS, S73K/S97D derivative; SSΔ12, S73K/S97D/Δ12 derivative; Δ12, catalytic domain insert deletion (residues 103 to 114 deleted); Δ209 and Δ185, carboxy-terminal truncation derivatives (residues 1 to 209 and 1 to 185, respectively).
From Fig. 5B it is evident that only the C95A and S73K/S97D derivatives exhibit a severe defect in thiolester formation, the former for obvious reasons and the latter for reasons that are unapparent (see Discussion). Notably, the S97D self-association defect is more severe than the S73K/S97D defect, yet exhibits a minimal defect in thiolester formation. Therefore, from these results alone, there is no clear dependence of thiolester formation on self-association. Greater insight into this issue, however, was gained from the experiment described below.
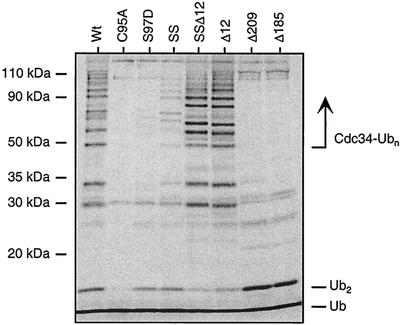
We subsequently examined the ability of each Cdc34 derivative to synthesize multi-Ub chains by extending the incubation time of the thiolester assay already described, followed by separation using SDS-PAGE (Fig. 6). The radiolabeled products that are detected in a reaction with Cdc34 include unincorporated Ub, free di-Ub (Ub2), and Ub chains that were covalently linked to Cdc34 as a consequence of autoubiquitination (Cdc34-Ubn).
FIG. 6.
Autoubiquitination. Cdc34 autoubiquitination was assayed for Cdc34 and its various derivatives by using an in vitro ubiquitination reaction. Reaction mixtures contained Cdc34 or one of its derivatives (100 nM), Uba1 (10 nM), 35S-Ub (200 nM), and an ATP cocktail and were incubated for 8 h at 30°C, representing an end point assay for autoubiquitination. DTT (100 mM) was added to stop the reactions, and the reaction products were analyzed by SDS-PAGE followed by autoradiography. The positions of free Ub (Ub), di-Ub (Ub2), and multi-Ub chains covalently linked to Cdc34 (Cdc34-Ubn) are indicated. SS, S73K/S97D derivative; SSΔ12, S73K/S97D/Δ12 derivative; Δ12, catalytic domain insert deletion (residues 103 to 114 deleted); Δ209 and Δ185, carboxy-terminal truncation derivatives (residues 1 to 209 and 1 to 185, respectively). A downward shift in molecular mass in the Ub chains on the Δ12 derivatives is observed, consistent with the catalytic domain insert deletion. Bands not indicated are degradation products of Cdc34-Ub.
The derivatives tested synthesized products that can be predicted either from previous observations or from their altered peptide sequence. Not surprisingly the C95A active-site derivative was completely inactive (3). The Δ209 and Δ185 carboxy-terminal truncation derivatives were capable of synthesizing free Ub2 but were incapable of catalyzing the formation of multi-Ub chains linked to Cdc34. The lack of Cdc34 conjugates in these derivatives can be explained by the absence of the key target lysines that have been previously shown to reside in the carboxy-terminal portion of the tail (4). Finally, the Δ12 derivative exhibited the capacity to autoubiquitinate itself (Fig. 6).
Analysis of the remaining derivatives provided some interesting observations. In addition to the self-association defect, the S97D derivative exhibited little, if any, autoubiquitination in spite of its ability to synthesize thiolester with Ub. The weakly associating S73/S97D derivative, however, exhibited relatively significant autoubiquitination even though it is severely compromised in its ability to synthesize thiolester. Furthermore, deletion of the catalytic insert in combination with the S73K and S97D substitutions (S73/S97D/Δ12) was found to alleviate the defects associated with autoubiquitination and self-association, as well to allow for full complementation of Cdc34 function in cdc34 mutant strains.
Cdc34 self-association depends on thiolester-linked Ub.
The C95A Cdc34 active-site derivative cannot form Ub thiolester and is defective in its ability to self-associate. These observations suggest that that the Ub thiolester form of Cdc34 facilitates self-association. However, Cdc34∼Ub thiolester is labile, and there is no guarantee that Cdc34 present in either the cell lysates or Cdc34 isolated via coimmunoprecipitation would be in the Cdc34∼Ub thiolester state. To confirm that Cdc34∼Ub thiolester was present during the coimmunoprecipitations used in this study, amino-terminally Flag- and Myc-tagged derivatives of Cdc34 were coexpressed with an amino-terminally HA-tagged derivative of Ub. Cell lysates were immunoprecipitated, followed by incubation in the presence of DTT to release and detect free Ub. Figure 7A verifies Cdc34 self-association, as Flag-Cdc34 (Fig. 7A, middle panel) coimmunoprecipitates with Myc-Cdc34 (Fig. 7A, right panel). Furthermore, free Ub released after DTT treatment corresponds to the presence of Cdc34∼(HA-Ub) thiolester in the immunoprecipitate (Fig. 7A, left panel). To ensure that the free Ub observed is specific to Cdc34, two negative controls were included. First, immunoprecipitation using an anti-Myc antibody was carried in the absence of expressed Myc-Cdc34 to ensure that free HA-Ub was not binding to the beads. Second, the C95A active-site derivative (incapable of forming the Cdc34∼Ub thiolester), which was defective in self-association (Fig. 7A, middle panel), showed no free Ub in the immunoprecipitate (Fig. 7A, left panel). These observations demonstrate the presence of Cdc34∼Ub thiolester in the immunoprecipitates.
FIG. 7.
Cdc34 self-association is facilitated by Ub thiolester formation. (A) Presence of Ub in Cdc34 coimmunoprecipitates. Myc- and Flag-tagged Cdc34 were coexpressed together with HA-tagged Ub in YPH499 cells. Myc-Cdc34 was immunoprecipitated (IP) from cell extracts by using an anti-Myc antibody. The Myc-Cdc34, Flag-Cdc34, and HA-Ub contained in the immunoprecipitate were separated by SDS-PAGE and detected by immunoblotting (IB) with anti-Myc (left panel), anti-Flag (middle panel), and anti-HA (left panel) antibodies. The replacement of Cdc34 by Cdc34 C95A served as the negative control. The asterisk indicates a proteolytic product of Cdc34. Protein expression levels within the lysates used for immunoprecipitations are shown at the bottoms of the respective panels. wt, wild type. (B) Dependence of Cdc34 self-association on thiolester formation. The procedure was the same as for panel A except that extracts were treated with or without DTT followed by dialysis to remove DTT prior to immunoprecipitation.
To demonstrate the dependence of Cdc34 self-association on the thiolester, the coimmunoprecipitation experiment described (Fig. 7A) was repeated with three important modifications: (i) each lysate was pretreated with or without DTT prior to immunoprecipitation, resulting in samples with and without thiolester; (ii) DTT-treated and untreated lysates were dialyzed to remove the DTT (which was previously found to be detrimental to antibody integrity) and then immunoprecipitated with the anti-Myc antibody; and (iii) at the final stage, all immunoprecipitates were treated with DTT to convert thiolester-linked Ub into the free form. Negative controls using cell lysates that lacked Myc-Cdc34 were included. Cdc34 self-association in the absence of DTT is demonstrated by the coimmunoprecipitation of Flag-Cdc34 with Myc-Cdc34 (Fig. 7B, left panel). Furthermore, the presence of HA-Ub in the same sample indicates the presence of the Cdc34∼Ub thiolester (Fig. 7B, right panel). In contrast, the same lysate pretreated with DTT prior to immunoprecipitation revealed little Flag-Cdc34 in the coimmunoprecipitation and therefore little of the self-associated form of CDC34 (Fig. 7B, left panel). Furthermore, the absence of free Ub indicates that the Cdc34∼Ub thiolester is also absent (Fig. 7B, right panel). Together these two observations indicate that Cdc34∼Ub is present under conditions where self-association is favored and, conversely, that the hydrolysis of the thiolester disfavors Cdc34's ability to self-associate.
Taken together, all of the results reported here can be mechanistically rationalized in terms of the previously reported model (28) with key modifications.
DISCUSSION
Using a coimmunoprecipitation strategy, we have shown directly that Cdc34 self-associates in cell lysates, thereby implying an in vivo interaction. We have also identified structural determinants within the catalytic domain that are not only important in self-association but also important for growth and either of two stages related to multi-Ub chain assembly, including thiolester formation and the covalent transfer of Ub to Ub. These results are entirely consistent with, and provide further insight into, a previously proposed mechanism for multi-Ub chain assembly (26, 28). In this model two interacting Cdc34∼Ub thiolester moieties interact in a manner that positions their Ub molecules for linkage. These and past findings make a credible circumstantial argument that (i) the interaction of one Cdc34 molecule with another is required for multi-Ub chain synthesis, (ii) the interaction is dependent on the formation of Cdc34∼Ub thiolester, and (iii) multi-Ub chain assembly is an obligatory step in Cdc34's function.
The interaction of Cdc34 with itself does not appear to be mediated by the E3. This conclusion is drawn from observations showing that Cdc34 self-associates in the absence of a functional SCF complex (Fig. 3) and that the carboxy-terminal extension of Cdc34, which is required for its interaction with the SCF complex (21), is dispensable for Cdc34 self-association (Fig. 2A). Rather, thiolester-linked Ub appears to stabilize the interaction between Cdc34 molecules, given that conditions that hydrolyze the thiolester bond also disrupt Cdc34 self-association (Fig. 7B). Similarly, residues in the catalytic domain of Cdc34 that play a role in Cdc34∼Ub thiolester formation also play a role in self-association, further strengthening the link between thiolester formation and self-association (see below). These observations indicate that Cdc34∼Ub self-association likely precedes its interaction with the SCF complex, resulting in a complex that is poised to build multi-Ub chains onto a target.
The dispensable nature of the carboxy-terminal extension in Cdc34 self-association observed here presents an apparent contradiction with previously described in vitro cross-linking reactions (26). These in vitro reactions employed purified truncation derivatives of Cdc34, and cross-linking was carried out in the absence of thiolester-linked Ub. Unlike the results of the cross-linking experiments, we find that in cell lysates Cdc34 immunoprecipitates as Cdc34∼Ub thiolester and that thiolester-linked Ub stabilizes Cdc34 self-association (Fig. 7). The presence of thiolester-linked Ub likely provides sufficient stability to self-association that additional stabilization provided by the tail is superfluous and the carboxy-terminal extension may be dispensed with in regard to this interaction. Rather, this study suggests that the principal role of the carboxy-terminal extension is to provide a site of interaction with the SCF complex through Cdc53, as previously described (21).
A causal relationship linking the essential function(s) of Cdc34 to its self-association is suggested by the growth behavior of the C95A, S97D, and S73K/S97D association-defective derivatives (3, 19). These growth defects do not appear to be a result of their inability to interact with the SCF complex, as each retains the ability to interact with the core SCF component Cdc53 (Fig. 4A). Rather, in each case the degree of the self-association defect directly corresponds to the degree of the growth defect (Fig. 8). Furthermore, there is an important mechanistic relationship between these residues and the catalytic insert, which is evident from a reversal in both the self-association defect and the growth defect in an S73K/S97D/Δ12 derivative (Fig. 8).
Based on these and the following observations, we believe that these residues play an important role in positioning the Ub thiolester correctly on the Cdc34 surface and that this requirement for orientation is necessary for the ability of Cdc34 to interact with itself and for multi-Ub chain assembly. First, these residues are found to cluster around the active-site cysteine when mapped onto the crystal structure of Ubc7, another E2 that contains a catalytic domain insert that is similar to that of Cdc34 (Fig. 9). Second, these residues coincide with analogous amino acid positions in other E2s that define key positions in the E2∼Ub thiolester interaction (13, 22). Third, amino acid substitutions of these positions affect their ability to form Ub thiolester and synthesize multi-Ub chains.
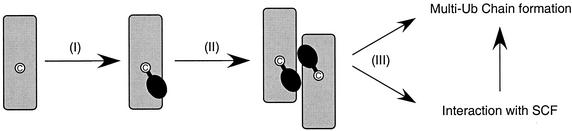
With respect to the third point, the C95A, S73K/S97D, and S73K/S97D/Δ12 derivatives exhibit a direct correlation between their ability to synthesize Ub thiolester and their ability to synthesize multi-Ub chains (Fig. 8). For example, the S73K/S97D derivative shows a strong defect in Ub thiolester formation that results in weak multi-Ub chain assembly. On the other hand, deletion of the catalytic domain insertion (S73K/S97D/Δ12) restores both Ub thiolester formation and multi-Ub chain assembly to wild-type levels. Furthermore, these activities parallel the ability of these derivatives both to self-associate and to carry out Cdc34's in vivo function, as mentioned above (Fig. 8). These analyses indicate that the in vivo function of Cdc34 is dependent upon three sequentially important events, i.e., (i) the formation of the Cdc34∼Ub thiolester, (ii) self-association, and (iii) assembly of multi-Ub chains, and that each of these mechanistic steps is dependent on the correct positioning of Ub on the Cdc34 surface (Fig. 10).
FIG. 10.
Hypothetical scheme for Cdc34 self-association and multi-Ub chain formation. Cdc34 is represented in gray, the catalytic cysteine residue is shown in white, and Ub is shown in black. (I) Ub is transferred to Cdc34 from an activated Uba1∼Ub thiolester to form Cdc34∼Ub thiolester. (II) Cdc34∼Ub thiolester formation mediates the self-association of two or more Cdc34s. (III) Self-associated Cdc34∼Ub thiolester is charged and positioned in such a way as to interact with a target either alone or through an Ub ligase, such as the SCF complex, and will proceed to build multi-Ub chains.
Of the substitutions discussed thus far, S97D would appear to be the one exception to the rule. Although the S97D derivative is capable of thiolester formation to near-wild-type levels, it also exhibits a complete loss of both self-association and multi-Ub chain assembly (Fig. 8), a result that is not immediately accommodated into the conclusions drawn above. The behavior of S97D can be understood in light of the following argument. First, the observation that S97D forms thiolester indicates that its defect occurs downstream of Ub thiolester formation. Second, this downstream defect includes the complete loss of both multi-Ub chain assembly and self-association. Thus, the defect is not in the synthesis of the Cdc34∼Ub thiolester but is within the thiolester complex itself. As mentioned above, it appears likely that amino acid positions 73 and 97 along with the catalytic domain insertion play a role in positioning Ub correctly on the Cdc34 surface following thiolester formation. Based on these results, we propose that the S97D substitution perturbs the orientation of the thiolester Ub in a manner that is unfavorable to self-association and multi-Ub chain assembly.
A common mechanistic relationship between Ub binding with positions 73 and 97 and the catalytic domain insertion is also supported by the following observations. The behavior of the S97D derivative changes with respect to its utilization of Ub when S73K is introduced (Fig. 8). The S73K/S97D derivative is less effective at thiolester formation than the S97D derivative but more effective than the S97D derivative in chain assembly. Therefore, S73K nullifies the multi-Ub chain assembly defect of S97D but results in reduced thiolester formation. Furthermore, the thiolester defect of the S73K/S97D derivative is dependent on the catalytic domain insertion, as its deletion restores both Ub thiolester formation and multi-Ub chain assembly to wild-type levels (Fig. 8). Thus, all of the derivatives examined here are at least consistent with the idea that the position Ub occupies on the Cdc34 surface is critical to Cdc34 activity.
The sequence of events that describe Cdc34 function in light of our present findings are summarized schematically in Fig. 10. (i) A monomer of Cdc34 is first activated with Ub by E1. (ii) Thiolester formation triggers the formation of a Cdc34∼Ub thiolester complex that results from favorable interactions that each Ub molecule makes across the boundary of the complex. (iii) The complex then either alone or in combination with the SCF complex directs the assembly of a multi-Ub chain on the targeted substrate. The catalytic heart of this pathway is a multimer of the Cdc34∼Ub thiolester that owes both its existence and its activity to the spatial relationship between Cdc34 and Ub.
Acknowledgments
This work was made possible by grants from the National Cancer Institute of Canada.
We thank the members of the Ellison lab for helpful discussion and D. Stuart for supplies and valuable advice.
REFERENCES
- 1.Arnason, T., and M. J. Ellison. 1994. Stress resistance in Saccharomyces cerevisiae is strongly correlated with assembly of a novel type of multiubiquitin chain. Mol. Cell. Biol. 14:7876-7883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1995. Short protocols in molecular biology, 3rd ed. John Wiley & Sons, Inc., New York, N.Y.
- 3.Banerjee, A., R. J. Deshaies, and V. Chau. 1995. Characterization of a dominant negative mutant of the cell cycle ubiquitin-conjugating enzyme Cdc34. J. Biol. Chem. 270:26209-26215. [DOI] [PubMed] [Google Scholar]
- 4.Banerjee, A., L. Gregori, Y. Xu, and V. Chau. 1993. The bacterially expressed yeast CDC34 gene product can undergo autoubiquitination to form a multiubiquitin chain-linked protein. J. Biol. Chem. 268:5668-5675. [PubMed] [Google Scholar]
- 5.Chen, P., P. Johnson, T. Sommer, S. Jentsch, and M. Hochstrasser. 1993. Multiple ubiquitin-conjugating enzymes participate in the in vivo degradation of the yeast MAT alpha 2 repressor. Cell 74:357-369. [DOI] [PubMed] [Google Scholar]
- 6.Cook, W. J., P. D. Martin, B. F. Edwards, R. K. Yamazaki, and V. Chau. 1997. Crystal structure of a class I ubiquitin conjugating enzyme (Ubc7) from Saccharomyces cerevisiae at 2.9 angstroms resolution. Biochemistry 36:1621-1627. [DOI] [PubMed] [Google Scholar]
- 7.Deshaies, R. J., V. Chau, and M. Kirschner. 1995. Ubiquitination of the G1 cyclin Cln2p by a Cdc34p-dependent pathway. EMBO J. 14:303-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drury, L. S., G. Perkins, and J. F. Diffley. 1997. The Cdc4/34/53 pathway targets Cdc6p for proteolysis in budding yeast. EMBO J. 16:5966-5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Girod, P. A., and R. D. Vierstra. 1993. A major ubiquitin conjugation system in wheat germ extracts involves a 15-kDa ubiquitin-conjugating enzyme (E2) homologous to the yeast UBC4/UBC5 gene products. J. Biol. Chem. 268:955-960. [PubMed] [Google Scholar]
- 10.Goebl, M. G., L. Goetsch, and B. Byers. 1994. The Ubc3 (Cdc34) ubiquitin-conjugating enzyme is ubiquitinated and phosphorylated in vivo. Mol. Cell. Biol. 14:3022-3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goebl, M. G., J. Yochem, S. Jentsch, J. P. McGrath, A. Varshavsky, and B. Byers. 1988. The yeast cell cycle gene CDC34 encodes a ubiquitin-conjugating enzyme. Science 241:1331-1335. [DOI] [PubMed] [Google Scholar]
- 12.Gwozd, C. S., T. G. Arnason, W. J. Cook, V. Chau, and M. J. Ellison. 1995. The yeast UBC4 ubiquitin conjugating enzyme monoubiquitinates itself in vivo: evidence for an E2-E2 homointeraction. Biochemistry 34:6296-6302. [DOI] [PubMed] [Google Scholar]
- 13.Hamilton, K. S., M. J. Ellison, K. R. Barber, R. S. Williams, J. T. Huzil, S. McKenna, C. Ptak, M. Glover, and G. S. Shaw. 2001. Structure of a conjugating enzyme-ubiquitin thiolester intermediate reveals a novel role for the ubiquitin tail. Structure 9:897-904. [DOI] [PubMed] [Google Scholar]
- 14.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425-479. [DOI] [PubMed] [Google Scholar]
- 15.Hodgins, R., C. Gwozd, T. Arnason, M. Cummings, and M. J. Ellison. 1996. The tail of a ubiquitin-conjugating enzyme redirects multi-ubiquitin chain synthesis from the lysine 48-linked configuration to a novel nonlysine-linked form. J. Biol. Chem. 271:28766-28771. [DOI] [PubMed] [Google Scholar]
- 16.Kaiser, C., S. Michaelis, and A. Mitchell. 1994. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 17.Kolman, C. J., J. Toth, and D. K. Gonda. 1992. Identification of a portable determinant of cell cycle function within the carboxyl-terminal domain of the yeast CDC34 (UBC3) ubiquitin conjugating (E2) enzyme. EMBO J. 11:3081-3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leggett, D. S., and P. M. Candido. 1997. Biochemical characterization of Caenorhabditis elegans UBC-1: self-association and auto-ubiquitination of a RAD6-like ubiquitin-conjugating enzyme in vitro. Biochem. J. 327(Part 2):357-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu, Y., N. Mathias, C. N. Steussy, and M. G. Goebl. 1995. Intragenic suppression among CDC34 (UBC3) mutations defines a class of ubiquitin-conjugating catalytic domains. Mol. Cell. Biol. 15:5635-5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mathias, N., S. L. Johnson, M. Winey, A. E. Adams, L. Goetsch, J. R. Pringle, B. Byers, and M. G. Goebl. 1996. Cdc53p acts in concert with Cdc4p and Cdc34p to control the G1-to-S-phase transition and identifies a conserved family of proteins. Mol. Cell. Biol. 16:6634-6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mathias, N., C. N. Steussy, and M. G. Goebl. 1998. An essential domain within Cdc34p is required for binding to a complex containing Cdc4p and Cdc53p in Saccharomyces cerevisiae. J. Biol. Chem. 273:4040-4045. [DOI] [PubMed] [Google Scholar]
- 22.Miura, T., W. Klaus, B. Gsell, C. Miyamoto, and H. Senn. 1999. Characterization of the binding interface between ubiquitin and class I human ubiquitin-conjugating enzyme 2b by multidimensional heteronuclear NMR spectroscopy in solution. J. Mol. Biol. 290:213-228. [DOI] [PubMed] [Google Scholar]
- 23.Papa, F. R., and M. Hochstrasser. 1993. The yeast DOA4 gene encodes a deubiquitinating enzyme related to a product of the human tre-2 oncogene. Nature 366:313-319. [DOI] [PubMed] [Google Scholar]
- 24.Patton, E. E., A. R. Willems, D. Sa, L. Kuras, D. Thomas, K. L. Craig, and M. Tyers. 1998. Cdc53 is a scaffold protein for multiple Cdc34/Skp1/F-box protein complexes that regulate cell division and methionine biosynthesis in yeast. Genes Dev. 12:692-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pickart, C. M., and I. A. Rose. 1985. Functional heterogeneity of ubiquitin carrier proteins. J. Biol. Chem. 260:1573-1581. [PubMed] [Google Scholar]
- 26.Ptak, C., J. A. Prendergast, R. Hodgins, C. M. Kay, V. Chau, and M. J. Ellison. 1994. Functional and physical characterization of the cell cycle ubiquitin-conjugating enzyme CDC34 (UBC3). Identification of a functional determinant within the tail that facilitates CDC34 self-association. J. Biol. Chem. 269:26539-26545. [PubMed] [Google Scholar]
- 27.Schwob, E., T. Bohm, M. D. Mendenhall, and K. Nasmyth. 1994. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell 79:233-244. [DOI] [PubMed] [Google Scholar]
- 28.Silver, E. T., T. J. Gwozd, C. Ptak, M. Goebl, and M. J. Ellison. 1992. A chimeric ubiquitin conjugating enzyme that combines the cell cycle properties of CDC34 (UBC3) and the DNA repair properties of RAD6 (UBC2): implications for the structure, function and evolution of the E2s. EMBO J. 11:3091-3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Voges, D., P. Zwickl, and W. Baumeister. 1999. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. 68:1015-1068. [DOI] [PubMed] [Google Scholar]
- 30.Yaglom, J., M. H. Linskens, S. Sadis, D. M. Rubin, B. Futcher, and D. Finley. 1995. p34Cdc28-mediated control of Cln3 cyclin degradation. Mol. Cell. Biol. 15:731-741. [DOI] [PMC free article] [PubMed] [Google Scholar]