Abstract
Mitogen-activated protein kinase (MAPK)-activated protein kinase 2 (MAPKAPK2) mediates multiple p38 MAPK-dependent inflammatory responses. To define the signal transduction pathways activated by MAPKAPK2, we identified potential MAPKAPK2 substrates by using a functional proteomic approach consisting of in vitro phosphorylation of neutrophil lysate by active recombinant MAPKAPK2, protein separation by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and phosphoprotein identification by peptide mass fingerprinting with matrix-assisted laser desorption ionization mass spectrometry (MALDI-MS) and protein database analysis. One of the eight candidate MAPKAPK2 substrates identified was the adaptor protein, 14-3-3ζ. We confirmed that MAPKAPK2 interacted with and phosphorylated 14-3-3ζ in vitro and in HEK293 cells. The chemoattractant formyl-methionyl-leucyl-phenylalanine (fMLP) stimulated p38-MAPK-dependent phosphorylation of 14-3-3 proteins in human neutrophils. Mutation analysis showed that MAPKAPK2 phosphorylated 14-3-3ζ at Ser-58. Computational modeling and calculation of theoretical binding energies predicted that both phosphorylation at Ser-58 and mutation of Ser-58 to Asp (S58D) compromised the ability of 14-3-3ζ to dimerize. Experimentally, S58D mutation significantly impaired both 14-3-3ζ dimerization and binding to Raf-1. These data suggest that MAPKAPK2-mediated phosphorylation regulates 14-3-3ζ functions, and this MAPKAPK2 activity may represent a novel pathway mediating p38 MAPK-dependent inflammation.
A diverse group of cellular responses are elicited by activation of a highly conserved family of mitogen-activated protein kinase (MAPK) signaling pathways, which includes extracellular signal-regulated kinases (ERKs), c-jun N-terminal kinases (JNKs), ERK5, and p38 MAPKs. A large body of evidence indicates that p38 MAPK activity is critical to immune and inflammatory responses. p38 MAPK is activated in macrophages, neutrophils, and T cells by numerous extracellular mediators of inflammation, including chemoattractants, cytokines, chemokines, and bacterial lipopolysaccharide (LPS) (reviewed in reference 31). p38 MAPK participates in LPS-induced proinflammatory cytokine production in macrophages and regulates multiple neutrophil functional responses, including respiratory burst activity, chemotaxis, granular exocytosis, adherence, interleukin-8 (IL-8) synthesis, priming, and apoptosis (8, 25, 29, 30, 37, 39). p38 MAPK also mediates T-cell differentiation and apoptosis by regulating gamma interferon production (27, 34). Inhibition of p38 MAPK in mice prevented the progression of collagen-induced arthritis (13) and resulted in a significant decrease in LPS-induced tumor necrosis factor (TNF-α) release and neutrophil infiltration into the lungs (37).
Multiple p38 MAPK-dependent inflammatory responses are mediated by a serine-threonine kinase, MAPK-activated protein kinase 2 (MAPKAPK2). Zu et al. reported that introduction of a MAPKAPK2 inhibitory peptide into neutrophils blocked formyl-methionyl-leucyl-phenylalanine (fMLP), but not phorbol ester, stimulation of respiratory burst activity (42). We used the same peptide to confirm that MAPKAPK2 regulates fMLP-stimulated respiratory burst activity and to show that MAPKAPK2 plays a role in fMLP-stimulated chemotaxis and TNF-α-stimulated exocytosis in human neutrophils (4). MAPKAPK2-deficient mice displayed a significant reduction in LPS-induced TNF-α production and hypotension, and neutrophils from these mice demonstrated impaired chemotaxis (9, 20).
The substrates of MAPKAPK2 that mediate these responses have not been clearly defined. One mechanism by which MAPKAPK2 induces TNF-α production is by stabilizing TNF-α mRNA via phosphorylation of the zinc finger protein tristetraprolin (24). Heat shock protein 27 (Hsp27), leukocyte-specific protein 1 (LSP1), and 5-lipoxygenase (5-LO) were identified previously as MAPKAPK2 substrates in neutrophils (12, 28, 40). Hsp27 binds actin filaments, and Hsp27 expression and phosphorylation are implicated in regulation of cytoskeletal organization (18). The functional role of Hsp27 in neutrophils, however, remains to be determined. Neutrophils from LSP1-deficient mice demonstrate reduced chemotaxis, impaired cytoskeletal organization, and enhanced respiratory burst activity (10, 15). 5-LO catalyzes the initial steps in the production of leukotrienes, inflammatory mediators derived from arachidonic acid (39). We showed recently that MAPKAPK2 phosphorylates and activates PKB/Akt in human neutrophils, providing an antiapoptotic activity (19, 33). The large number of inflammatory responses regulated by MAPKAPK2 suggests that multiple substrates remain to be identified.
The present study was designed to identify substrates of MAPKAPK2 in human neutrophils. We developed a functional proteomic approach using a combination of in vitro MAPKAPK2 phosphorylation of neutrophil lysate, separation of phosphorylated proteins by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and phosphoprotein identification by peptide mass fingerprinting using matrix-assisted laser desorption ionization mass spectrometry (MALDI-MS) and protein database analysis. One of the eight MAPKAPK2 substrates identified by this approach was 14-3-3ζ. The 14-3-3 proteins function as adaptor or scaffolding proteins by spontaneously forming homo- and heterodimers that interact with phosphoserine- and phosphothreonine-containing sequences in protein ligands (23, 41). 14-3-3 proteins interact with over 100 proteins and thereby participate in many cellular functions, including cell signaling, the cell cycle, and apoptosis (reviewed in reference 38). We have reported that activation of Akt in neutrophils is p38 MAPK dependent and that MAPKAPK2, but not p38 MAPK, phosphorylates and activates Akt in vitro (33). We recently reported that Akt interacts with and phosphorylates 14-3-3ζ (32). Based on the multiple functions of 14-3-3 proteins and association with MAPKAPK2 signaling, we examined the interaction of MAPKAPK2 with 14-3-3ζ in greater detail. Our results show that MAPKAPK2 interacts with and phosphorylates 14-3-3ζ at Ser-58, and analysis of 14-3-3 mutants suggests this phosphorylation regulates 14-3-3ζ dimerization and ligand binding.
MATERIALS AND METHODS
Expression vectors.
Expression of glutathione S-transferase (GST)-MAPKAPK2 and GST-p38α MAPK fusion proteins was accomplished by constructing a pGEX-5X-2 (Amersham Pharmacia Biotech, Piscataway, N.J.) expression plasmid with the human cDNA for MAPKAPK2 or p38α MAPK downstream of GST. pcDNA3.1-MK2-EE, a constitutively active mutant, was obtained from Matthias Gaestel, Martin-Luther-Universität, Germany (6). The construct for GST-14-3-3ζ was obtained from Thierry Dubois, University of Edinburgh, Edinburgh, United Kingdom. Recombinant His-14-3-3ζ was expressed by cloning a BamHI-EcoRI insert from the pGEX-2T-14-3-3ζ construct into pRSET A (Invitrogen, Carlsbad, Calif.). MBP fusion proteins of 14-3-3ζ were prepared by using the cDNA for human 14-3-3ζ cloned into pMalc-2 (New England Biolabs), as described previously (41). Mutation of Ser-58 to Asp was performed with the Quikchange Mutagenesis kit (Stratagene). [35S]methionine-labeled MAPKAPK2 and p38α MAPK were expressed with the TNT Quick Master expression kit (Promega, Madison, Wis.) using a yeast expression plasmid, pGBKT7. Ser-58 on pRSET-14-3-3ζ was mutated to alanine or aspartic acid with a Clontech site-directed mutagenesis kit using 5′-CCGTAGGTCAGCGTGGAGGGTCG-3′ as the mutation primer for alanine, 5′-CCGTAGGTCAGATTGGAGGGTCG-3′ as the mutation primer for aspartic acid, and 5′-CAGCAGGTGGCTCGGAATACAGAGA-3′ as the selection primer for both mutations. The appropriate mutations were verified by DNA sequencing. Recombinant proteins were expressed and isolated from the BL21 strain of Escherichia coli.
Neutrophil isolation.
Neutrophils were isolated from healthy donors using plasma-Percoll gradients as described by Haslett et al. (11). Trypan blue staining revealed that at least 97% of cells were neutrophils with >95% viability. After isolation, neutrophils were suspended in Krebs-Ringer phosphate buffer (pH 7.2) at the desired concentration. The Human Studies Committee of the University of Louisville approved the use of human donors.
Neutrophil lysate preparation for MAPKAPK2 substrate identification.
Neutrophils (108) were lysed in 500 μl of lysis buffer containing 2 M thiourea, 7 M urea, 65 mM CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, 58 mM dithiothreitol (DTT), and 4.5% ampholytes (pH 3 to 10). Lysates were centrifuged at 12,000 × g for 20 min at 15°C. Prior to addition of exogenous MAPKAPK2, lysate urea was reduced to 1 M by size exclusion chromatography. One-milliliter spin-out columns (Chemicon International, Inc., Temecula, Calif.) were equilibrated in 3 ml of kinase buffer (25 mM HEPES, 25 mM β-glycerophosphate, 25 mM MgCl2, 2 mM DTT, 0.1 mM NaVO3, 65 mM CHAPS) containing 4 or 1 M urea. Neutrophil lysate (500 μl) was loaded on the column, equilibrated with buffer containing 4 M urea, and centrifuged at 500 × g for 1 min, and then the procedure was repeated with buffer containing 1 M urea.
MAPKAPK2 substrate identification.
Neutrophil lysates (400 μg of total protein) were incubated with 10 μCi of [γ-32P]ATP (ICN Biomedicals, Inc.) in the presence and absence of 400 ng of recombinant active MAPKAPK2 (Upstate Biotechnology, Lake Placid, N.Y.) at 30°C for 3 h. Proteins were then separated by SDS-PAGE (10% polyacrylamide), and phosphorylation was visualized by autoradiography. Autoradiographs were compared to Coomassie blue-stained gels to identify potential MAPKAPK2 substrates. Bands representing potential substrates were cut from gels and prepared for mass spectrometric analysis.
Trypsin digestion and MS analysis.
To obtain peptides for MS analysis, Coomassie blue-stained bands were cut from gels and digested with trypsin by a modification of the method of Jensen et al. (14). The excised gel pieces were incubated for 15 min in 100 mM NH4HCO3 and 50% acetonitrile and dried by vacuum centrifugation. Proteins were then reduced by incubation with 20 mM DTT at 56°C for 45 min, followed by alkylation with 65 mM iodoacetamide in the dark at room temperature for 30 min. After alkylation, gel pieces were incubated for 15 min in 50 mM NH4HCO3 and 50% acetonitrile and dried by vacuum centrifugation, and then proteins were hydrolyzed by incubation in 20 ng of modified trypsin per ml (Promega) at 37°C overnight.
Trypsin-generated peptides were applied by a thin-film spotting procedure for MALDI-MS analysis using α-cyanohydroxycinnamic acid as a matrix on stainless steel targets, as described by Jensen et al. (14). Mass spectral data were obtained with a Tof-Spec 2E instrument (Micromass) and a 337-nm N2 laser at 20 to 35% power in the reflector mode. Spectral data were obtained by averaging 10 spectra, each of which was the composite of 10 laser firings. Mass axis calibrations were accomplished by using peaks from tryptic auto-hydrolysis. Peptide masses obtained by MALDI-MS analysis were used to search the National Center for Biotechnology Information (NCBI) database to identify the intact proteins.
In vitro kinase assays.
Phosphorylation of recombinant His-14-3-3ζ by MAPKAPK2 and p38α MAPK was examined by incubation of active recombinant MAPKAPK2 (40 ng) or active recombinant p38α MAPK (25 ng) (Upstate Biotechnology) with 10 μCi of [γ-32P]ATP and 0.5 μg of recombinant His-14-3-3ζ in 30 μl of kinase buffer (25 mM HEPES, 25 mM β-glycerophosphate, 25 mM MgCl2, 2 mM DTT, 0.1 mM NaVO3). The MAPKAPK2 assay was performed in the presence or absence of 200 μM MAPKAPK2 inhibitory peptide (AFHRAFNRQLANGVAEIR) or 200 μM scrambled peptide (REVNCRFRFAHANQAGAI) (Commonwealth Technologies, Richmond, Va.). The MAPKAPK2 inhibitory peptide was characterized previously by Zu et al. (42). Reaction mixtures were incubated at 30°C for 30 min. Following the 30-min incubation, reactions were terminated with Laemmli SDS sample dilution buffer, proteins were separated by SDS-PAGE (10% polyacrylamide), and phosphorylation was visualized by autoradiography.
To determine the stoichiometry of phosphorylation, the optimal time of His-14-3-3ζ was first determined by incubation of 1 μg of His-14-3-3ζ with 40 ng of recombinant active MAPKAPK2 and 10 μCi of [γ-32P]ATP at 30°C for times ranging from 10 min to 4 h. Optimal phosphorylation was seen by 2 h. Stoichiometry was determined by incubation of 1 μg of His-14-3-3ζ with 40 ng of recombinant active MAPKAPK2, 2 pmol of [γ-32P]ATP, and 200 pmol of ATP at 30°C for 2 h. Separate reaction mixtures containing 200 pmol of ATP and 2 pmol of [γ-32P]ATP were used to determine the specific activity of ATP in each reaction. Samples were subjected to SDS-PAGE (10% polyacrylamide) and stained with Coomassie blue, bands corresponding to 14-3-3ζ were excised and counted by scintillation spectrometry, and the molar ratio of phosphorylation was calculated.
14-3-3ζ phosphorylation in HEK293 cells.
HEK293 cells (60% confluent) were incubated with a 6:1 ratio of Lipofectamine to plasmid in OPTI medium (Gibco, Grand Island, N.Y.) at 37°C for 6 h. Medium was then replaced with Dulbecco's modified Eagle's medium (DMEM) (Gibco) containing 10% fetal bovine serum, and cells were incubated at 37°C overnight. HEK293 cells were transfected with pcDNA3.1-MK2-EE (constitutively active MAPKAPK2) or vector. Twenty-four hours after transfection, the medium was replaced with 3 ml of phosphate-free minimal essential medium (MEM) (Gibco) and incubated at 37°C for 1 h. The medium was then replaced with 1 ml of MEM containing 1 mCi of [32P]orthophosphate (ICN Biomedicals, Inc.) and incubated at 37°C for 4 h. Cells were then lysed with 500 μl of lysis buffer containing 20 mM Tris (pH 7.4), 1% Triton X-100, 0.5% NP-40, 150 mM NaCl, 25 mM MgCl2, 20 mM NaF, 0.2 mM NaVO3, 1 mM EDTA, 1 mM EGTA, and 5 mM phenylmethylsulfonyl fluoride (PMSF). Cell lysates were incubated with 4 μg of polyclonal anti-14-3-3ζ (Santa Cruz Biotechnology, Santa Cruz, Calif.) overnight at 4°C, followed by a 2-h incubation with 30 μl (1:1 slurry in phosphate-buffered saline [PBS]) of protein A-Sepharose beads. Beads were washed twice with lysis buffer, and proteins were eluted with Laemmli SDS sample dilution buffer, separated by SDS-PAGE (10% polyacrylamide), and transferred to nitrocellulose membrane. Incorporation of 32P was visualized by autoradiography, and expression of 14-3-3 proteins was determined by immunoblot analysis (monoclonal anti-14-3-3; Stressgen).
14-3-3ζ phosphorylation in neutrophils.
Human neutrophils (2 × 107) were incubated with or without 300 nM fMLP for 1, 3, or 10 min at 37°C. For some experiments, neutrophils were pretreated with 3 μM SB203580 (Calbiochem) for 60 min prior to addition of fMLP. Following incubations, cells were lysed in 100 μl of buffer containing 2 M thiourea, 7 M urea, 65 mM CHAPS, 58 mM DTT, and 4.5% ampholytes (pH 4 to 6.5). Some samples were treated with PP2A phosphatase. Size exclusion chromatography was used to exchange lysate (900 μg of protein) into a buffer containing 1 M urea, 25 mM MOPS (morpholinepropanesulfonic acid), 1 mM MgCl2, 1 mM DTT, 0.1 mM MnCl2, 1 mM EGTA, and 60 mM β-mercaptoethanol prior to in vitro phosphatase reaction. Following buffer exchange, lysate was incubated with 0.5 U of PP2A (Upstate Biotechnology) for 1.5 h at 30°C. Proteins from each sample were separated by isoelectric focusing with immobilized-pH-gradient (IPG) strips (7 cm, pH 3.9 to 5.1; Bio-Rad) and by SDS-PAGE (10% polyacrylamide). IPG strips were hydrated with neutrophil lysate (100 μg of protein) or phosphatase reaction mixture and rehydration buffer (7 M urea, 2 M thiourea, 2% CHAPS, 0.01 M DTT, 2% ampholytes [pH 3 to 10], 0.01% bromophenol blue) to give a total volume of 135 μl. Following overnight hydration, IPG gels were isoelectrically focused (IEF) at a maximum voltage of 8,000 and 50 μA/gel for 20,000 V-h. Following IEF, IPG gels were sequentially incubated for 10 min in equilibration buffer I (6 M urea, 2% DTT, 30% glycerol) and equilibration buffer II (6 M urea, 2.5% iodoacetamide, 30% glycerol). IPG gels were then used for SDS-PAGE (10% polyacrylamide), followed by immunoblot analysis for 14-3-3.
GST pull-down.
GST or GST-14-3-3ζ glutathione-coupled Sepharose (10 μl) was incubated with recombinant [35S]MAPKAPK2 or [35S]p38α MAPK in 50 μl of kinase buffer containing 25 mM HEPES, 25 mM β-glycerophosphate, 25 mM MgCl2, 2 mM DTT, 0.1 mM NaVO3, and 10% glycerol overnight at 4°C. Beads were then washed twice with kinase buffer, proteins were eluted with Laemmli SDS sample dilution buffer and separated by SDS-PAGE (10% polyacrylamide), and 35S-labeled proteins were detected by autoradiography.
GST, GST-MAPKAPK2, or GST-p38α MAPK glutathione-coupled Sepharose was incubated with 250 ng of recombinant His-14-3-3ζ in 50 μl of kinase buffer or with neutrophil lysate (400 μg of protein) overnight at 4°C. Neutrophil lysates were prepared by suspending 2 × 107 cells in 200 μl of lysis buffer containing 20 mM Tris (pH 7.4), 1% Triton X-100, 0.5% NP-40, 150 mM NaCl, 25 mM MgCl2, 20 mM NaF, 0.2 mM NaVO3, 1 mM EDTA, 1 mM EGTA, 5 mM PMSF, and 10% glycerol. Following incubation, beads were washed twice with lysis buffer. Proteins were eluted with Laemmli SDS sample dilution buffer and separated by SDS-PAGE (10% polyacrylamide), and 14-3-3 was detected by immunoblotting.
Coimmunoprecipitation.
Neutrophil lysates (400 μg of protein) were incubated with 40 μl (1:1 slurry) of protein A-Sepharose, or protein A-Sepharose and 4 μg of polyclonal anti-MAPKAPK2 (H-66; Santa Cruz Biotechnology) overnight at 4°C. Following incubation, beads were washed twice with lysis buffer. Proteins were eluted with Laemmli SDS sample dilution buffer and separated by SDS-PAGE (10% polyacrylamide), and 14-3-3 was detected by immunoblotting.
14-3-3ζ dimerization.
Gel migration and cross-linking experiments were performed with recombinant 14-3-3ζ-WT (wild type), 14-3-3ζ-(S58A), and 14-3-3ζ-(S58D). For gel migration experiments, 2 μg of recombinant 14-3-3ζ-WT, 14-3-3ζ-(S58A), or 14-3-3ζ-(S58D) was separated by SDS-PAGE (10% polyacrylamide) or nondenaturing PAGE, and proteins were visualized by Coomassie blue staining. For cross-linking experiments, 100 ng of recombinant 14-3-3ζ-WT, 14-3-3ζ-(S58A), or 14-3-3ζ-(S58D) was incubated in the presence and absence of 5 mM chemical cross-linker disuccinimidyl suberate (DSS; Pierce Biotechnology) in 20 μl of PBS at room temperature for 2 h before or after addition of 40 ng of recombinant active MAPKAPK2. Reactions were stopped by incubation with 50 mM Tris-HCl (pH 7.4) at room temperature for 15 min. Following incubations, proteins were denatured with Laemmli SDS dilution buffer, separated by SDS-PAGE (10% polyacrylamide), and immunoblotted for 14-3-3 (polyclonal anti-14-3-3ζ antibody; Santa Cruz Biotechnology).
14-3-3ζ-Raf binding assays.
U2OS cells were cultured in DMEM supplemented with 10% fetal calf serum and antibiotics (50 U of penicillin per ml and 50 μg of streptomycin per ml). Cells from ∼70% confluent cultures were lysed by addition of Tris-buffered saline (pH 7.4) containing 1 mM MgCl2; 1 mM CaCl2; 4 μg each of AEBSF, leupeptin, and pepstatin A per ml; 8 μg of aprotinin per ml; 5 mM sodium orthovanadate; 25 mM NaF; 10 mM β-glycerophosphate; 1 mM DTT; and 1% NP-40 for 15 min at 4°C, followed by centrifugation at 12,000 × g for 15 min at 4°C. Lysates were incubated with amylose beads containing equal amounts of immobilized MBP-14-3-3ζ-wild-type, MBP-14-3-3ζ-S58D, or MBP alone overnight at 4°C. Following overnight incubations, beads were washed three times with PBS-0.5% NP-40 and then three times with PBS. Proteins were eluted with Laemmli SDS sample dilution buffer and separated by SDS-PAGE (10% polyacrylamide), and c-Raf-1 was detected by immunoblot analysis.
Calculation of 14-3-3ζ dimerization energetics and residual surface potential.
The structure of the 14-3-3 dimer bound to two copies of its mode 1 binding peptide (41) (PDB accession no. 1QJB) was used as a model. All waters were removed from the structure, as were the bound peptides. Histidines were assigned neutral charge with protonation of the epsilon nitrogen chosen as the most favorable state based on hydrogen bonding pattern. To build Ser-58-phosphorylated 14-3-3 and S58D mutant 14-3-3 homodimers, as well as heterodimers with an unmodified wild type, Ser-58 on one or both monomers was replaced with phosphoserine or aspartic acid, respectively. The HBUILD facility of CHARMM was then used with the PARAM22 parameter set to place all hydrogen atoms (17). All atom positions not determined crystallographically were minimized freely. Parameters for phosphoserine were created from existing PARAM22 parameters for dianionic methyl phosphate. Seven model structures were generated: one for dimeric wild-type 14-3-3, one for dimeric phosphorylated 14-3-3, one for dimeric S58D 14-3-3, and two each for heterodimers of S58D/wild-type 14-3-3 and pSer58/wild-type 14-3-3. Unminimized structures contained strong van der Waals clashes at the S58 position, and were rectified using the SHAPES facility of CHARMM to perform minimizations on each structure, followed by HBUILD to place hydrogens correctly. Binding free energies were calculated for both the unminimized and minimized structures as the sum of electrostatic, van der Waals, and surface area contributions. The van der Waals contributions were calculated by using the PARAM22 parameter set, surface area contributions were calculated as −5 cal/Å2 of molecular surface area buried (35; data not shown [nearly constant for all dimers]), and electrostatic binding free energy was calculated using DELPHI (35). Solvent was treated as a continuum with a dielectric constant of 80 and a salt concentration of 0.145 M, and protein was treated as a continuum with dielectric constant of 4 and embedded charges at atom centers. Numeric solutions were obtained at a final grid spacing of 0.21 Å, and our results are the average of 10 translations of the molecule on the grid. Residual surface potential, calculated as the sum of the desolvation potential of one 14-3-3 monomer and the interaction potential of its binding partner, was calculated using DELPHI (35), with a grid spacing of 0.16 Å and with no translational averaging. Desolvation potential is the difference in electrostatic potential at the binding surface of a single 14-3-3 protein in its dimeric and monomeric states. Interaction potential is the potential generated by the second 14-3-3 monomer on the first monomer in the dimer. Similar results were obtained for residual potentials calculated by using either the unminimized or minimized model structures.
RESULTS
Identification of MAPKAPK2 substrates.
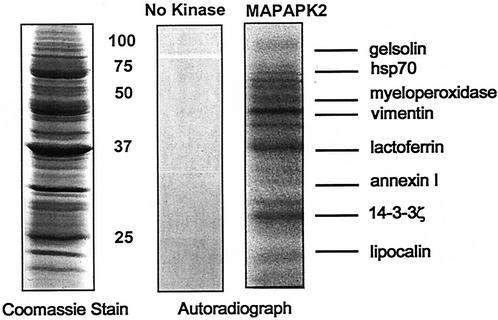
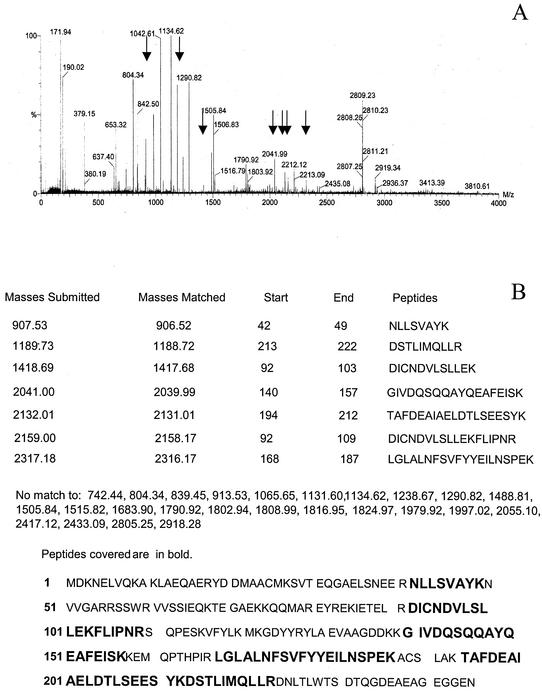
To screen for MAPKAPK2 substrates, human neutrophil lysates prepared using a urea-based buffer were incubated with [γ-32P]ATP in the presence and absence of active recombinant MAPKAPK2, proteins were separated by SDS-PAGE, and phosphorylated proteins were visualized by autoradiography. Autoradiographs were compared to Coomassie blue-stained gels to identify potential MAPKAPK2 substrates (Fig. 1). Bands corresponding to phosphorylated proteins were cut from gels and subjected to in-gel trypsin digestion, and proteins were identified by peptide mass fingerprinting using MALDI-MS and protein database analysis (NCBI; www.matrixscience.com). A significant match was indicated by an assigned MOWSE score of >71, representing the probability that a match was not a random event. No phosphorylation was observed following incubation of neutrophil lysates with [γ-32P]ATP in the absence of recombinant active MAPKAPK2 (Fig. 1), indicating the absence of endogenous kinase activity under the conditions of neutrophil lysate preparation. Eight proteins identified as candidate MAPKAPK2 substrates by this method are shown in Fig. 1, including a previously identified MAPKAPK2 substrate, vimentin. The 28-kDa phosphorylated band was identified as 14-3-3ζ, with a MOWSE score of 75 and 37% protein coverage. Figure 2A shows the mass spectra of the peptides obtained by MALDI-MS that were queried to the theoretical masses in the NCBI protein database using the Mascot search engine. Figure 2B shows the masses of seven peptides, after excluding autolytic trypsin masses, which identified 14-3-3ζ. The large number of peptides in the mass spectra is consistent with the presence of several proteins within the band that identified 14-3-3ζ. After excluding the masses of these seven peptides, the masses of the remaining 27 peptides were analyzed. Although no significant matches were found, the highest MOWSE scores identified phosphoglycerate mutase 1 (MOWSE score, 56; accession no. gi16757984) and an unknown mouse protein (MOWSE score, 65; accession no. gi12805565). Based on the role of 14-3-3 proteins in cell signaling, cell cycle control, and apoptosis, further studies were performed to confirm the interaction of MAPKAPK2 with 14-3-3ζ.
FIG. 1.
Recombinant MAPKAPK2 phosphorylation of neutrophil lysate. Candidate MAPKAPK2 substrates were identified by phosphorylation of neutrophil lysates with [32P]ATP in the presence and absence of recombinant active MAPKAPK2. Proteins were separated by SDS-PAGE, and phosphoproteins were detected by comparing autoradiographs to Coomassie blue-stained gels. No endogenous kinase activity resulted from incubation of neutrophil lysate without recombinant active MAPKAPK2. Peptide mass fingerprinting of trypsin-digested phosphoprotein bands using MALDI-MS and protein database analysis identified eight candidate MAPKAPK2 substrates.
FIG. 2.
Identification of 14-3-3ζ as a MAPKAPK2 substrate. (A) The mass spectra showing peptide masses (in mass per charge [m/z] units) obtained by MALDI-time of flight MS analysis of the phosphorylated protein band at 28 kDa. Thirty-four mass spectrum peaks were present and were queried to the theoretical masses in the entire NCBI protein database by using the Mascot search engine. A maximal 150-ppm error window and one missed tryptic cleavage were allowed. The seven peptide masses that identified 14-3-3ζ are marked by arrows. (B) The peptide masses used to identify 14-3-3ζ are compared to theoretical masses of peptides from 14-3-3ζ. The peptide sequence for each mass and the peptide coverage of 14-3-3ζ are shown. There was no significant match by peptide mass fingerprinting to the 27 peptide masses listed.
MAPKAPK2 interacts with and phosphorylates 14-3-3ζ in vitro.
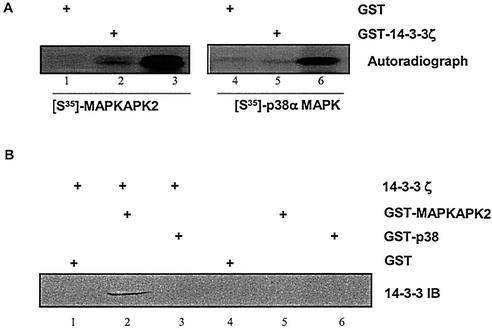
The ability of 14-3-3ζ to directly interact with MAPKAPK2 was examined by a GST pull-down approach. Recombinant GST or GST-14-3-3ζ coupled to glutathione-Sepharose was incubated with recombinant [35S]methionine-labeled MAPKAPK2 or p38α MAPK. Precipitated proteins were separated by SDS-PAGE, and 35S-labeled proteins were visualized by autoradiography. Figure 3A shows that MAPKAPK2, but not p38α MAPK, precipitated with GST-14-3-3ζ. Next, recombinant GST, GST-MAPKAPK2, or GST-p38α MAPK coupled to glutathione-Sepharose was incubated with recombinant His-14-3-3ζ. Precipitated proteins were separated by SDS-PAGE, followed by immunoblot analysis for 14-3-3. Figure 3B shows that 14-3-3ζ precipitated with MAPKAPK2 but not p38α MAPK. Neither 14-3-3ζ nor MAPKAPK2 precipitated with GST-Sepharose. These data demonstrate that 14-3-3ζ directly interacts with MAPKAPK2 and that this interaction is specific for MAPKAPK2.
FIG. 3.
14-3-3ζ interacts with MAPKAPK2. (A) Recombinant [35S]methionine-labeled MAPKAPK2 (lanes 1 and 2) and p38α MAPK (lanes 4 and 5) were incubated with GST or GST-14-3-3ζ glutathione-coupled Sepharose. Lanes 3 and 6 show the migration of [35S]MAPKAPK2 and [35S]p38α MAPK. Proteins were separated by SDS-PAGE and subjected to autoradiography. The autoradiograph shows that MAPKAPK2, but not p38α MAPK, precipitated with 14-3-3ζ. (B) Recombinant His-14-3-3ζ was incubated with GST (lane 1), GST-MAPKAPK2 (lane 2), or GST-p38α MAPK glutathione-coupled Sepharose (lane 3). Controls were GST (lane 4), GST-MAPKAPK2 (lane 5), and GST-p38α MAPK (lane 6) without recombinant 14-3-3ζ. Precipitated proteins were separated by SDS-PAGE, followed by immunoblot (IB) analysis for 14-3-3. The immunoblot shows that 14-3-3ζ precipitated with MAPKAPK2, but not p38α MAPK.
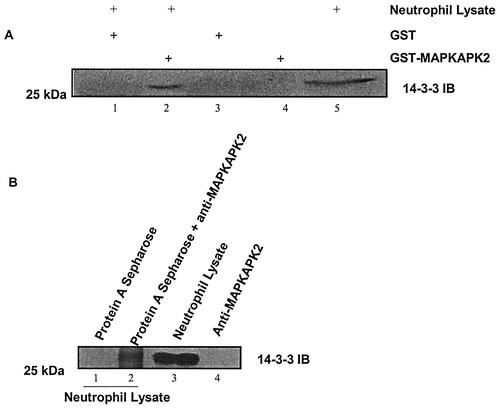
To determine if MAPKAPK2 and 14-3-3 interact in neutrophil lysates, GST pull-down and coimmunoprecipitation assays were performed. Neutrophil lysates were incubated with recombinant GST-MAPKAPK2 or a MAPKAPK2 antibody coupled to protein A-Sepharose. Precipitated proteins were separated by SDS-PAGE, followed by immunoblot analysis for 14-3-3. Figure 4A shows that endogenous 14-3-3 precipitated with recombinant GST-MAPKAPK2, but not GST-glutathione-Sepharose. Figure 4B shows that 14-3-3 coimmunoprecipitated with MAPKAPK2 from neutrophil lysate, while protein A-Sepharose did not precipitate 14-3-3. These data suggest that MAPKAPK2 and 14-3-3 are physically associated in human neutrophils. In neither assay was the amount of 14-3-3 precipitated equal to the total amount in the lysate. Saturating concentrations of GST-MAPKAPK2 or anti-MAPKAPK2, however, were not used in these experiments. Additionally, the antibody employed recognizes three isoforms of 14-3-3, 14-3-3ɛ, 14-3-3β, and 14-3-3ζ, not all of which may interact with MAPKAPK2.
FIG. 4.
MAPKAPK2 and 14-3-3ζ interact in neutrophil lysate. (A) Neutrophil lysates (800 μg of protein) were incubated with recombinant GST (lane 1) or GST-MAPKAPK2 (lane 2) glutathione-coupled Sepharose. Controls were GST (lane 3), GST-MAPKAPK2 (lane 4), or neutrophil lysate (200 μg of protein) (lane 5). Proteins were separated by SDS-PAGE, followed by immunoblot (IB) analysis for 14-3-3. The immunoblot shows that endogenous 14-3-3 precipitated with recombinant MAPKAPK2. (B) Neutrophil lysates (800 μg of protein) were incubated with protein A-coupled Sepharose (lane 1) or protein A-coupled Sepharose and MAPKAPK2 antibody (lane 2). Controls were neutrophil lysate (200 μg of protein) (lane 3) or MAPKAPK2 antibody alone (lane 4). Proteins were separated by SDS-PAGE, followed by immunoblot analysis for 14-3-3. The immunoblot shows that 14-3-3 immunoprecipitated with MAPKAPK2 from neutrophil lysate.
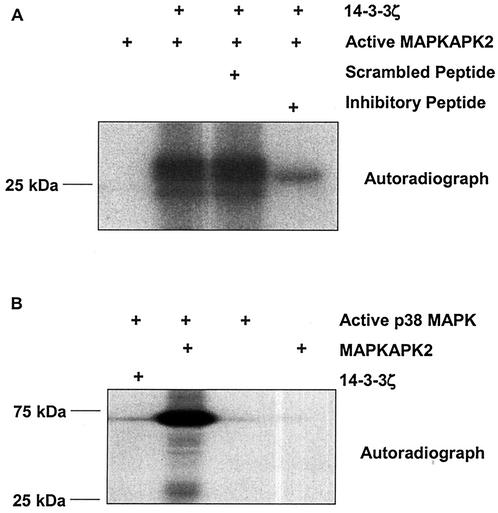
To determine if MAPKAPK2 is capable of phosphorylating 14-3-3ζ, we employed an in vitro kinase assay. Active recombinant MAPKAPK2 was incubated with recombinant 14-3-3ζ and [γ-32P]ATP in the presence or absence of a MAPKAPK2 inhibitory peptide or a scrambled peptide. Following kinase reactions, proteins were separated by SDS-PAGE, and phosphorylated proteins were visualized by autoradiography. Figure 5A shows that MAPKAPK2 phosphorylates 14-3-3ζ, and this phosphorylation is specifically inhibited by the MAPKAPK2 inhibitory peptide. The ability of p38α MAPK, an upstream kinase for MAPKAPK2, to phosphorylate 14-3-3ζ in vitro was also examined. Figure 5B shows that active recombinant p38α MAPK phosphorylates recombinant MAPKAPK2, but not recombinant 14-3-3ζ. Thus, both the physical interaction and phosphorylation of 14-3-3ζ are specific for MAPKAPK2.
FIG. 5.
In vitro MAPKAPK2 phosphorylation of 14-3-3ζ. (A) Active recombinant MAPKAPK2 was incubated with [32P]ATP and recombinant His-14-3-3ζ in the presence of a scrambled peptide, MAPKAPK2 inhibitory peptide, or without peptide. Proteins were separated by SDS-PAGE, and phosphorylation was visualized by autoradiography. The autoradiograph shows that MAPKAPK2 phosphorylates 14-3-3ζ, and 14-3-3ζ phosphorylation is inhibited by the MAPKAPK2 inhibitory peptide. (B) Active recombinant p38α MAPK was incubated with [32P]ATP and recombinant 14-3-3ζ or recombinant MAPKAPK2. Proteins were separated by SDS-PAGE, and phosphorylation was visualized by autoradiography. The autoradiograph shows that p38α MAPK phosphorylated MAPKAPK2, but not 14-3-3ζ.
MAPKAPK2 phosphorylates 14-3-3ζ in intact cells.
The ability of MAPKAPK2 to phosphorylate 14-3-3ζ in intact cells was examined by using HEK293 cells. Cells were transfected with constitutively active MAPKAPK2 (MK2-EE) or vector. Twenty-four hours after transfection, cells were loaded with [32P]orthophosphate and lysed, and 14-3-3ζ was immunoprecipitated. Precipitated proteins were separated by SDS-PAGE and transferred to nitrocellulose membrane, and phosphorylated proteins were visualized by autoradiography. Immunoblot analysis (Fig. 6A) shows equal amounts of 14-3-3ζ were precipitated from MAPKAPK2- or vector-transfected cells. The corresponding autoradiograph shows that 14-3-3ζ was phosphorylated in cells transfected with constitutively active MAPKAPK2- but not vector-transfected cells. These results indicate that MAPKAPK2 phosphorylates 14-3-3 in intact cells.
FIG. 6.
14-3-3 phosphorylation in HEK293 cells and human neutrophils. (A) HEK293 cells were transfected with vector or constitutively active MAPKAPK2 (MK2-EE). After 24 h of transfection, cells were loaded with [32P]orthophosphate, and endogenous 14-3-3ζ was immunoprecipitated. Precipitated proteins were separated by SDS-PAGE and transferred to nitrocellulose membrane, and phosphorylation of 14-3-3ζ was detected by autoradiography. The autoradiograph shows that transfection of active MAPKAPK2, but not vector, resulted in phosphorylation of 14-3-3ζ. Immunoblot (IB) analysis of the same nitrocellulose membrane shows that an equal amount of 14-3-3ζ was precipitated from MK2-EE- or vector-transfected cells. (B) Human neutrophils were incubated with or without fMLP for 1, 3, or 10 min. An aliquot of the 10-min fMLP lysate was treated with 0.5 U of PP2A phosphatase prior to two-dimensional separation. Neutrophils were pretreated with 3 μM SB203580 for 60 min prior to incubation with fMLP for 10 min. Proteins were separated by two-dimensional gel electrophoresis and immunoblotted (IB) for 14-3-3. The immunoblot shows that 3- and 10-min fMLP treatments, but not the 1-min fMLP treatment, resulted in a negative isoelectric shift of a portion of 14-3-3. This pI shift was reversed by both phosphatase and SB203580 treatment. These results indicate that fMLP stimulates p38 MAPK-dependent phosphorylation of 14-3-3 in neutrophils.
The ability of the chemoattractant fMLP to stimulate 14-3-3 phosphorylation in human neutrophils was examined. Neutrophils were incubated with or without fMLP for 1, 3, or 10 min. Following fMLP stimulation, cells were lysed, proteins were separated by two-dimensional gel electrophoresis, and 14-3-3 was detected by immunoblotting. Figure 6B shows three 14-3-3 spots of similar molecular mass, but different pIs in unstimulated cells. The monoclonal 14-3-3 antibody used for these studies detects three different isoforms of similar molecular weights, but different pIs: 14-3-3β (pI 4.78), 14-3-3ɛ (pI 4.72), and 14-3-3ζ (pI 4.70). Thus, the three 14-3-3 spots may represent these different isoforms. Stimulation of neutrophils with fMLP for 10 min resulted in appearance of an additional spot at a more negative pI, suggestive of a phosphorylation event. A similar shift of a small amount of 14-3-3 was also seen at 3 min. To confirm that the shift in protein location resulted from phosphorylation, an aliquot of the 10-min fMLP lysate was treated with PP2A phosphatase prior to two-dimensional separation. Figure 6B shows that PP2A treatment partially reversed the pI shift, indicating that fMLP induced 14-3-3 phosphorylation in neutrophils. Failure of PP2A treatment to return the shifted spot back to that observed for the control might be due to incomplete dephosphorylation or fMLP stimulation of other negatively charged posttranslational modifications.
To determine if fMLP-stimulated 14-3-3 phosphorylation is regulated by the p38 MAPK cascade, neutrophils were treated with 3 μM SB203580, a pharmacological p38α MAPK inhibitor, prior to 10 min of fMLP stimulation. Figure 6B shows that SB203580 treatment prevented the shift in 14-3-3, confirming that the p38α MAPK cascade participates in fMLP-induced 14-3-3 phosphorylation in neutrophils.
MAPKAPK2 phosphorylates 14-3-3ζ at Ser-58.
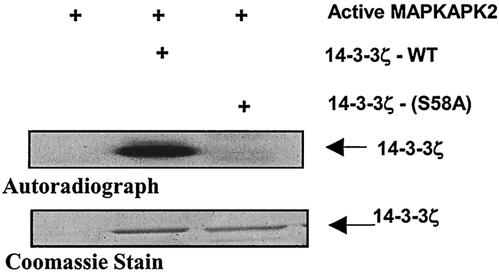
A search of the 14-3-3ζ amino acid sequence revealed a sequence (GARRSS58) similar to the consensus MAPKAPK2 phosphorylation motif (HXRXXS). To determine if MAPKAPK2 phosphorylates 14-3-3ζ on Ser-58, this amino acid residue was mutated to alanine. The ability of recombinant active MAPKAPK2 to phosphorylate recombinant 14-3-3ζ-WT and recombinant 14-3-3ζ-(S58A) was determined by an in vitro kinase assay. Figure 7 shows that MAPKAPK2 phosphorylates 14-3-3ζ-WT but not 14-3-3ζ-(S58A), suggesting that the primary MAPKAPK2 phosphorylation site on 14-3-3ζ is Ser-58.
FIG. 7.
MAPKAPK2 phosphorylation of 14-3-3ζ at Ser-58. Recombinant active MAPKAPK2 was incubated with [32P]ATP and recombinant 14-3-3ζ-WT or 14-3-3ζ-(S58A). Proteins were separated by SDS-PAGE, and phosphorylation was visualized by autoradiography. To demonstrate equal loading of 14-3-3 proteins, gels were also stained with Coomassie blue. The autoradiograph shows that MAPKAPK2 phosphorylated 14-3-3ζ-WT, but not 14-3-3ζ-(S58A).
Dimerization and Raf binding by 14-3-3ζ.
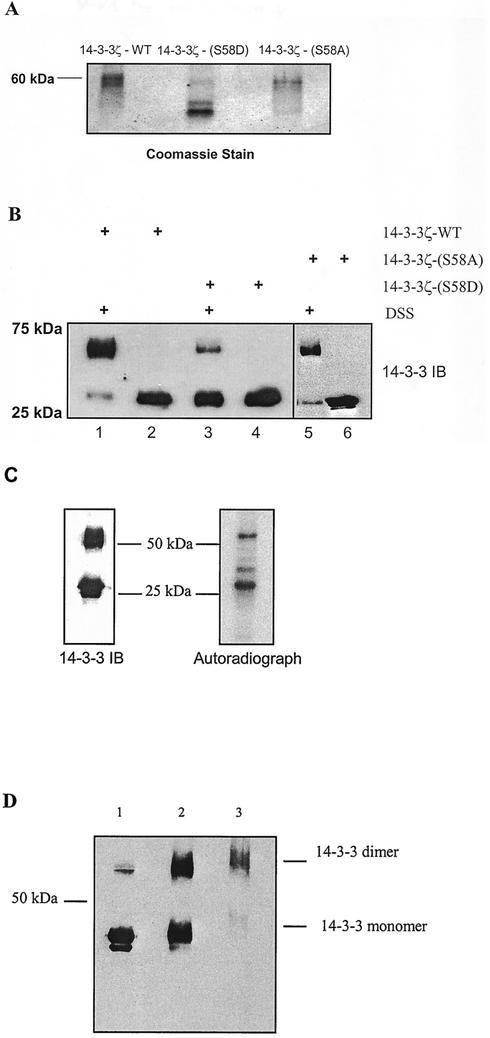
The X-ray crystal structure of 14-3-3ζ shows Ser-58 is located at the dimer interface (22). To determine if phosphorylation at Ser-58 might regulate dimerization, a phosphorylation-mimicking mutant, 14-3-3ζ-(S58D), was compared to 14-3-3ζ-(S58A) and 14-3-3ζ-WT in nondenaturing PAGE migration and intermolecular cross-linking experiments. Figure 8A shows that in nondenaturing PAGE, 14-3-3ζ-WT and 14-3-3ζ-(S58A) migrated at a larger molecular size than 14-3-3ζ-(S58D), suggesting that 14-3-3ζ-WT and 14-3-3ζ-(S58A) form dimers, while 14-3-3ζ-(S58D) does not dimerize. To confirm the reduction of 14-3-3ζ-(S58D) dimerization, chemical cross-linking with DSS was performed. Figure 8B shows a significant reduction in 60-kDa dimer formation of 14-3-3ζ-(S58D), compared to 14-3-3ζ-WT and 14-3-3ζ-(S58A). Only 30-kDa monomers were seen in the absence of DSS. These data indicate that mimicking phosphorylation at Ser-58 by mutation to Asp inhibits 14-3-3ζ dimerization.
FIG. 8.
Role of 14-3-3ζ phosphorylation in dimerization. (A) Recombinant 14-3-3ζ-WT, recombinant 14-3-3ζ-(S58D), and recombinant 14-3-3ζ-(S58A) were separated by nondenaturing PAGE, and proteins were detected by Coomassie blue staining. 14-3-3ζ-WT and 14-3-3ζ-(S58A) migrated at a larger molecular size than 14-3-3ζ-(S58D). (B) 14-3-3ζ-WT, 14-3-3ζ-(S58D), and 14-3-3ζ-(S58A) were incubated in the presence (lanes 1, 3, and 5) and absence (lanes 2, 4, and 6) of a chemical cross-linker, DSS. Proteins were separated by SDS-PAGE and immunoblotted (IB) for 14-3-3. The immunoblot shows a substantial reduction in 60-kDa dimer formation of 14-3-3ζ-(S58D), compared to 14-3-3ζ-WT and 14-3-3ζ-(S58A). (C) To determine the distribution of phosphorylated 14-3-3ζ between monomer and dimer, His-14-3-3ζ-WT was incubated with active recombinant MAPKAPK2 and [32P]ATP for 2 h at 30°C and then cross-linked with DSS. Under these conditions, about 20% of 14-3-3ζ was phosphorylated (data not shown). Following cross-linking, the reaction mixture was denatured with Laemmli buffer, and proteins were separated bySDS-PAGE (10% polyacrylamide). Proteins were then electrophoretically transferred to nitrocellulose. Autoradiography (right panel) and immunoblot analysis for 14-3-3 (left panel) are shown. Under these conditions, approximately equal amounts of 14-3-3 appear as monomer and dimer. The autoradiograph demonstrates that there is significantly more phosphorylation of monomer than dimer. (D) To determine the effect of MAPKAPK2 phosphorylation on dimer formation, His-14-3-3ζ-WT was incubated with 40 ng of MAPKAPK2 under three conditions prior to separation by SDS-PAGE (10% polyacrylamide), transfer to nitrocellulose, and immunoblot analysis for 14-3-3. Lane 1 contains 14-3-3ζ that was incubated with MAPKAPK2 for 2 h, but was not cross-linked with DSS, prior to SDS-PAGE. Lane 2 contains 14-3-3ζ that was phosphorylated by MAPKAPK2 for 2 h and then cross-linked with DSS prior to SDS-PAGE. Lane 3 contains 14-3-3ζ that was cross-linked with DSS prior to incubation with MAPKAPK2 for 2 h. The immunoblot shows that in the absence of cross-linking, the majority of 14-3-3ζ exists as monomer. Cross-linking after phosphorylation by MAPKAPK2 (lane 2) resulted in an equal distribution of monomer and dimer. Cross-linking before phosphorylation by MAPKAPK2 (lane 3) resulted in the majority of 14-3-3ζ migrating as dimer. Thus, phosphorylation by MAPKAPK2 resulted in a shift in 14-3-3ζ from dimer to monomer.
The effect of phosphorylation on dimerization was examined by two types of cross-linking experiments. To determine the distribution of phosphorylated 14-3-3ζ between monomer and dimer, recombinant 14-3-3ζ was incubated with recombinant active MAPKAPK2 and [32P]ATP prior to cross-linking with DSS. Cross-linked 14-3-3ζ was subjected to SDS-PAGE and transferred to nitrocellulose membrane, followed by autoradiography and immunoblotting for 14-3-3. Figure 8C shows that the amounts of monomeric and dimeric 14-3-3ζ were approximately equal. On the other hand, autoradiography showed that 14-3-3 monomer was phosphorylated to a greater degree than dimer. To compare the amounts of dimerization in the presence or absence of phosphorylation, 14-3-3ζ was cross-linked with DSS either prior to or after incubation with recombinant active MAPKAPK2 or was incubated with MAPKAPK2 without cross-linking. The reaction mixtures were subjected to SDS-PAGE (10% polyacrylamide), transferred to nitrocellulose, and immunoblotted for 14-3-3. Figure 8D shows that in the absence of cross-linking (lane 1), a very small amount of 14-3-3 migrated as a dimer. Phosphorylation by MAPKAPK2 prior to cross-linking resulted in an equal distribution of 14-3-3 between monomer and dimer (lane 2), as seen in Fig. 8C. On the other hand, cross-linking prior to incubation with MAPKAPK2 resulted in the majority of 14-3-3 migrating as dimer. Thus, MAPKAPK2 phosphorylation of 14-3-3ζ resulted in a shift from primarily dimer to an equal amount of dimer and monomer. The failure of MAPKAPK2 phosphorylation of 14-3-3ζ to completely prevent cross-linking may be due to the relatively low stoichiometry of in vitro phosphorylation, between 20 and 25%. These data support the findings obtained with 14-3-3ζ-(S58D) suggesting that phosphorylation regulates dimerization.
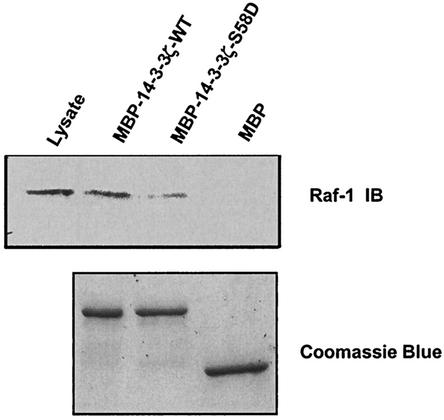
To investigate the possible role of Ser-58 phosphorylation by MAPKAPK2 in 14-3-3ζ ligand binding, an MBP fusion protein of 14-3-3ζ-WT and 14-3-3ζ-(S58D) was constructed. Incubation of U2OS cell lysates with MBP-14-3-3ζ-WT and MBP-14-3-3ζ-(S58D) proteins demonstrated a significant reduction in c-Raf-1 binding by the S58D mutant (Fig. 9). These results using a phosphorylation-mimicking mutant suggest that phosphorylation at Ser-58 may also regulate 14-3-3ζ interactions with ligand.
FIG. 9.
The S58D mutation in 14-3-3ζ impairs Raf binding. U2OS cell lysates were incubated with bead-immobilized MBP fusion proteins of wild-type 14-3-3ζ, the S58D mutant, or MBP alone. Bound proteins were analyzed by SDS-PAGE followed by immunoblotting (IB) for Raf (upper panel). An aliquot of the initial lysate is shown in the first lane. Coomassie staining (lower panel) verified that the beads contained equivalent amounts of protein.
Computer modeling indicates that phosphorylation of 14-3-3ζ at Ser-58 inhibits dimerization.
The energetics of dimerization of wild-type 14-3-3, 14-3-3 phosphorylated at Ser-58, and 14-3-3-(S58D) were evaluated computationally. The modeling and calculations indicate that phosphorylation strongly destabilizes dimer formation. Table 1 shows that, relative to the unphosphorylated wild-type 14-3-3, there is a significant increase in dimerization free energies for either a mixed heterodimer of the phosphorylated and unphosphorylated forms (24 kcal/mol) or a homodimer of phosphoproteins (90 kcal/mol). These large destabilizing contributions result from both electrostatics and packing. The magnitude of destabilization indicates that Ser-58 phosphorylation strongly disfavors dimerization. We similarly computed the binding free energy for the 14-3-3-(S58D) mutant used as a mimic for Ser-58 phosphorylation in the dimerization assays. While the electrostatic contribution of binding a wild-type 14-3-3 monomer to an S58D mutant monomer is more favorable than binding to the pSer-58 phosphorylated form, there is a reduced van der Waals interaction that leads to a net penalty relative to dimerization of either the unphosphorylated wild-type form or heterodimerization of the phosphorylated and unphosphorylated forms. Based on these results, homodimerization of phosphorylated 14-3-3 is expected to be even less favorable than homodimerization of 14-3-3-(S58D).
TABLE 1.
Calculated differences in binding free energy between wild-type and modified 14-3-3ζ dimers
| Dimer type (A/B) | ΔΔG (kcal/mol)a
|
|||
|---|---|---|---|---|
| Electrostatic
|
Total
|
|||
| No minimization | Rigid body minimization | No minimization | Rigid body minimization | |
| WT/WT | 0.0 | 0.0 | 0.0 | 0.0 |
| S58D/WT | 3.3 | 8.1 | 28.1 | 35.4 |
| WT/S58D | 3.6 | 7.9 | 31.4 | 34.1 |
| pSer58/WT | 4.1 | 12.4 | 92.9 | 21.3 |
| WT/pSer58 | 12.6 | 12.6 | 28.0 | 27.4 |
| S58D/S58D | 7.0 | 14.3 | 58.8 | 52.7 |
| pSer58/pSer58 | 25.2 | 23.4 | 384.8 | 90.3 |
Each column indicates the difference in total or electrostatic component binding free energy between a particular 14-3-3ζ dimer and the wild-type (WT) homodimer using the same minimization scheme.
It is fairly easy to understand the basis for worsened van der Waals interactions in the dimerization of phosphorylated and mutant 14-3-3 relative to the wild type, because both modifications lead to the packing of extra atoms into a small area. Electrostatic effects are somewhat less intuitive, due to the unpredictable contribution of solvent to dimerization. One way to visualize the electrostatics of a binding event is through calculation of the residual potential (16, 21). Residual potential is a measure of the electrostatic complementarity of a macromolecule for its binding partner and is defined as the sum, at the surface of one macromolecule, of the desolvation potential of that molecule and the interaction potential with its binding partner. For a molecule with optimal complementarity to its partner, the residual potential is zero at all points on the surface (16). To qualitatively visualize the relative electrostatic fitness of 14-3-3 variants for dimerization, we plotted deviation from ideal complementarity colorimetrically on the molecular surface of 14-3-3 (Fig. 10). While the interface of unphosphorylated wild-type 14-3-3 is reasonably complementary, as indicated by the expanse of white surface in Fig. 10A, phosphorylation of Ser-58 or mutation to Asp generates large regions of noncomplementarity at the site of modification (red regions in panels D and E). Similar noncomplementarity was also seen in heterodimers of either S58D or pSer-58 with wild-type 14-3-3ζ (Fig. 10B and C). This noncomplementarity graphically indicates why phosphorylation of Ser-58 on 14-3-3 or mutation of Ser-58 to aspartic acid, incurring a higher energetic desolvation penalty than is recovered by improved electrostatic interaction with nearby positively charged residues, causes the modified 14-3-3 to be less suitable for dimerization.
FIG. 10.
Computational modeling of 14-3-3 dimerization. Residual surface potential for the wild-type unmodified 14-3-3ζ homodimer (A), the heterodimer of S58D mutant and wild-type unmodified 14-3-3ζ (B), the heterodimers of pSer58 and wild-type 14-3-3ζ (C), the homodimer of 14-3-3ζ-S58D mutant (D), and the homodimer of Ser-58-phosphorylated 14-3-3ζ (E) plotted at the surface of a monomer. All plots show the chain treated as ligand, with the unsatisfied residual potential color scale ranging from deep red (−40 kT/e) to deep blue (+40 kT/e).
DISCUSSION
The proteomic approach described in this study possesses the advantage that lysate preparation in a urea-based buffer results in the absence of endogenous kinase activity, allowing kinase-specific substrate identification. Eight candidate substrates of MAPKAPK2 were identified, including one previously known substrate, vimentin (3). The artificial conditions of the in vitro kinase reaction, however, may permit MAPKAPK2 phosphorylation of proteins that are not substrates in intact cells. Additionally, a band from SDS-PAGE may contain more than one protein. Thus, proteins identified by MALDI-MS analysis require confirmation that these candidates are true substrates.
14-3-3ζ was confirmed as a novel substrate of MAPKAPK2. GST pull-down assays with recombinant proteins demonstrated a physical interaction between MAPKAPK2 and 14-3-3ζ, and coimmunoprecipitation showed that endogenous MAPKAPK2 and 14-3-3ζ interact in neutrophils. Active recombinant MAPKAPK2 phosphorylated recombinant 14-3-3ζ, and transfection of HEK293 cells with constitutively active MAPKAPK2 resulted in phosphorylation of 14-3-3ζ. These results establish that MAPKAPK2 interacts with and phosphorylates 14-3-3ζ both in vitro and in intact cells. As indicated by a shift in pI, fMLP stimulated 14-3-3 phosphorylation in human neutrophils. This isoelectric shift was inhibited by addition of a p38 MAPK inhibitor, SB203580, indicating that fMLP-stimulated 14-3-3 phosphorylation is p38 MAPK dependent. Since p38α MAPK did not phosphorylate or directly interact with 14-3-3ζ in vitro, these results suggest that fMLP-stimulated 14-3-3 phosphorylation in neutrophils resulted from p38α MAPK activation of MAPKAPK2. We recently reported evidence that activation of Akt in intact neutrophils was p38 MAPK dependent, and MAPKAPK2, but not p38α MAPK, phosphorylated and activated Akt in vitro (33). We also reported Akt interacts with and phosphorylates 14-3-3ζ (32). Thus, p38-MAPK-dependent phosphorylation of 14-3-3 in neutrophils may be mediated by direct MAPKAPK2 phosphorylation or may also be indirectly mediated by MAPKAPK2, through activation of Akt.
Phosphorylation of 14-3-3ζ is a regulatory mechanism for 14-3-3ζ binding and activity. Aitken et al. reported that 14-3-3δ is the Ser-184 phosphorylated form of 14-3-3ζ and that phosphorylation of Ser-184 increases interaction with and inhibition of PKC in vitro (1, 2). Sphingosine-dependent protein kinase-1 (SDK1) phosphorylates Ser-58 (26), but the effect of this phosphorylation on 14-3-3 function was not determined. Casein kinase I phosphorylates Thr-232, leading to inhibition of 14-3-3ζ interaction with Raf-1 in HEK293 cells (5).
The present study demonstrates that MAPKAPK2 phosphorylates 14-3-3ζ at Ser-58. 14-3-3 proteins spontaneously form homo- and heterodimers, and dimerization is reported to regulate ligand binding to 14-3-3 proteins (23, 36, 41). The location of Ser-58 at the 14-3-3ζ dimer interface (22) suggested the hypothesis that phosphorylation at Ser-58 regulated 14-3-3ζ dimerization. Several approaches were used to examine the effect of phosphorylation at Ser-58 on 14-3-3ζ dimerization. Nondenaturing PAGE and chemical cross-linking experiments demonstrated experimentally that the S58D mutation significantly impaired 14-3-3ζ dimerization. The distribution of MAPKAPK2-mediated radiolabeled 14-3-3 between monomer and dimer was determined. The majority of radiolabeled 14-3-3ζ was monomeric. The distribution of 14-3-3ζ between monomer and dimer was compared when cross-linking was performed before or after phosphorylation by MAPKAPK2. Cross-linking prior to phosphorylation resulted in primarily dimer formation, while cross-linking after phosphorylation resulted in an equal distribution between monomer and dimer. Finally, computer modeling was used to assess the effect of Ser-58 phosphorylation on 14-3-3 dimerization. Computational modeling and calculation of theoretical binding energies predicted that 14-3-3ζ phosphorylated at Ser-58 and mutation of Ser-58 to Asp would severely compromise the ability of 14-3-3ζ to dimerize. Attempts to directly demonstrate the effect of 14-3-3ζ phosphorylation on dimerization were unsuccessful due to low stoichiometry of phosphorylation under the experimental conditions. In addition to an effect on dimer formation, studies using a fusion protein pull-down assay indicated that Raf-1 binding to 14-3-3ζ-(S58D) was impaired. Taken together, our data suggest that phosphorylation at Ser-58 may behave like the 14-3-3ζ-(S58D) mutant in regulating 14-3-3ζ dimerization and interaction with ligands. Analysis of 14-3-3 isoforms indicates that three other isoforms, 14-3-3β, 14-3-3ɛ, and 14-3-3γ, contain a serine in a MAPKAPK2 consensus phosphorylation site at a location similar to that of 14-3-3ζ. Thus, MAPKAPK2 may regulate the function of several 14-3-3 isoforms.
14-3-3ζ is reported to interact with a number of components of cell signaling pathways, including G protein-coupled IL receptors (IL-3R and IL-9R), regulator of G protein signaling proteins (RGS7), protein kinases (Raf-1, protein kinase C [PKC], Akt, PI3K, ASK1, MEKK1, and PKUα), protein phosphatases (IP5P and PTPH1), and transcription factors (FKHRL1 and GR) (reviewed in reference 38). These interactions lead to a role for 14-3-3ζ in numerous cellular functions, including cell proliferation, apoptosis, vesicle transport, cytoskeletal organization, and gene expression. While the function of 14-3-3ζ in neutrophils is unknown, our data suggest the possibility that MAPKAPK2-mediated regulation of 14-3-3 protein function may play a role in the inflammatory process. Consistent with this hypothesis, Fagerholm et al. recently reported that 14-3-3ζ interacts with leukocyte-specific adhesion molecules, CD11/CD18, suggesting a possible role for 14-3-3ζ in neutrophil adhesion (7).
Acknowledgments
We thank Jian Cai for expert technical assistance in the MALDI-MS analysis. We also thank Michael Altman for the use of his residual potential plotting software, Barry Honig for making available the DelPhi software package, and Martin Karplus for CHARMM.
This study was supported by NIH R21 DK06289 (K.R.M.), NIH R01 HL66358 and NIH R21 DK62086 (J.B.K.), Department of Veterans Affairs Merit Reviews (J.B.K. and K.R.M.), American Heart Association Ohio Valley Affiliate grants (K.M.R. and M.J.R.), and NIH R01 GM60594 and R01 GM59281 and Burroughs-Wellcome Career Development Award (M.B.Y.).
REFERENCES
- 1.Aitken, A., S. Howell, D. Jones, J. Madrazo, and Y. Patel. 1995. 14-3-3 alpha and delta are the phosphorylated forms of Raf-activating 14-3-3 beta and zeta. J. Biol. Chem. 270:5706-5709. [DOI] [PubMed] [Google Scholar]
- 2.Aitken, A., S. Howell, D. Jones, J. Madrazo, H. Martin, Y. Patel, and K. Robinson. 1995. Post-translationally modified 14-3-3 isoforms and inhibition of protein kinase C. Mol. Cell. Biochem. 149-150:41-49. [DOI] [PubMed] [Google Scholar]
- 3.Cheng, T., and Y. Lai. 1998. Identification of mitogen-activated protein kinase-activated protein kinase as vimentin kinase activated by okadaic acid in 9L rat brain tumor cells. J. Cell Biochem. 71:169-181. [PubMed] [Google Scholar]
- 4.Coxon, P. Y., M. J. Rane, S. Uriarte, D. W. Powell, S. Singh, W. Butt, Q. Chen, and K. R. McLeish. MAPK-activated protein kinase-2 participates in p38-dependent and ERK-dependent functions in human neutrophils. Cell. Signal., in press. [DOI] [PubMed]
- 5.Dubois, T., C. Rommel, S. Howell, U. Steinhussen, Y. Soneji, N. Morrice, K. Moelling, and A. Aitken. 1997. 14-3-3 is phosphorylated by casein kinase I on residue 232. Phosphorylation at this site in vivo regulates Raf/14-3-3 interaction. J. Biol. Chem. 272:28882-28888. [DOI] [PubMed] [Google Scholar]
- 6.Engel, K., H. Schultz, F. Martin, A. Kotlyarov, K. Plath, M. Hahn, U. Heinemann, and M. Gaestel. 1995. Constitutive activation of mitogen-activated protein kinase-actived protein kinase 2 by mutation of phosphorylation sites and an A-helix motif. J. Biol. Chem. 270:27213-27221. [DOI] [PubMed] [Google Scholar]
- 7.Fagerholm, S., N. Morrice, C. Gahmberg, and P. Cohen. 2002. Phosphorylation of the cytoplasmic domain of the integrin CD18 chain by protein kinase C isoforms in leukocytes. J. Biol. Chem. 277:1728-1738. [DOI] [PubMed] [Google Scholar]
- 8.Frasch, C., J. Nick, V. Fadok, D. Bratton, S. Worthen, and P. Henson. 1998. p38 mitogen-activated protein kinase-dependent and -independent intracellular signal transduction pathways leading to apoptosis in human neutrophils. J. Biol. Chem. 273:8389-8397. [DOI] [PubMed] [Google Scholar]
- 9.Hannigan, M., L. Zhan, Y. Ai, and C. Huang. 2001. Leukocyte-specific gene 1 protein (LSP1) is involved in chemokine KC-activated cytoskeletal reorganization in murine neutrophils in vitro. J. Leukoc. Biol. 69:497-504. [PubMed] [Google Scholar]
- 10.Hannigan, M., L. Zhan, Y. Ai, A. Kotlyarov, M. Gaestel, and C. Huang. 2001. Abnormal migration phenotype of mitogen-activated protein kinase-activated protein kinase 2−/− neutrophils in Zigmond chambers containing formyl-methionyl-leucyl-phenylalanine gradients. J. Immunol. 167:3953-3961. [DOI] [PubMed] [Google Scholar]
- 11.Haslett, C., L. Guthrie, M. Kopaniak, J. Johnston, and P. Henson. 1985. Modulation of multiple neutrophil functions by preparative methods of trace concentrations of bacterial lipopolysaccharide. Am. J. Pathol. 119:101-110. [PMC free article] [PubMed] [Google Scholar]
- 12.Huang, C., L. Zhan, Y. Ai, and J. Jongstra. 1996. LSP1 is the major substrate for mitogen-activated protein kinase-activated protein kinase 2 in human neutrophils. J. Biol. Chem. 272:17-19. [DOI] [PubMed] [Google Scholar]
- 13.Jackson, J., B. Bolognese, L. Hillegass, S. Kassis, J. Adams, D. Griswold, and J. Winkler. 1998. Pharmacological effects of SB220025, a selective inhibitor of p38 mitogen-activated protein kinase, in angiogenesis and chronic inflammatory disease models. J. Pharmacol. Exp. Ther. 284:687-692. [PubMed] [Google Scholar]
- 14.Jensen, O., M. Wilm, A. Shevchenko, and M. Mann. 1999. Sample preparation methods for mass spectrometric peptide mapping directly from 2-DE gels. Methods Mol. Biol. 112:513-530. [DOI] [PubMed] [Google Scholar]
- 15.Jongstra-Bilen, J., V. Misener, C. Wang, H. Ginzberg, A. Auerbach, A. Joyner, G. Downey, and J. Jongstra. 2000. LSP1 modulates leukocyte populations in resting and inflamed peritoneum. Blood 96:1827-1835. [PubMed] [Google Scholar]
- 16.Kangas, E., and B. Tidor. 1999. Charge optimization leads to favorable electrostatic binding free energy. J. Chem. Phys. 109:7522-7545. [DOI] [PubMed] [Google Scholar]
- 17.Karplus, M. 2002. Molecular dynamics simulations of biomolecules. Acc. Chem. Res. 35:321-323. [DOI] [PubMed] [Google Scholar]
- 18.Kiemer, A., N. Weber, R. Furst, N. Bildner, S. Kulhanek-Heinze, and A. Vollmar. 2002. Inhibition of p38 MAPK activation via induction of MKP-1: atrial natriuretic peptide reduces TNF-alpha-induced actin polymerization and endothelial permeability. Circ. Res. 90:874-881. [DOI] [PubMed] [Google Scholar]
- 19.Klein, J., M. Rane, J. Scherzer, P. Coxon, R. Kettritz, J. Mathiesen, A. Buridi, and K. McLeish. 2000. Granulocyte-macrophage colony-stimulating factor delays neutrophil constitutive apoptosis through phosphoinositide 3-kinase and extracellular signal-regulated kinase pathways. J. Immunol. 164:4286-4291. [DOI] [PubMed] [Google Scholar]
- 20.Kotlyarov, A., A. Neininger, C. Schubert, R. Eckert, C. Birchmeirer, H. Volk, and M. Gaestel. 1999. MAPKAP kinase 2 is essential for LPS-induced TNF-alpha biosynthesis. Nat. Cell Biol. 1:94-97. [DOI] [PubMed] [Google Scholar]
- 21.Lee, L., and B. Tidor. 2001. Optimization of binding electrostatics: charge complementarity in the barnase-barstar protein complex. Protein Sci. 10:362-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu, D., J. Bienkowska, C. Petosa, J. Collier, H. Fu, and R. Liddington. 1995. Crystal structure of the zeta isoforms of the 14-3-3 protein. Nature 376:191-194. [DOI] [PubMed] [Google Scholar]
- 23.Luo, Z., X. Zhang, U. Rapp, and J. Avruch. 1995. Identification of the 14.3.3ζ domains important for self-association and Raf binding. J. Biol. Chem. 270:23681-23687. [DOI] [PubMed] [Google Scholar]
- 24.Mahtani, K. R., M. Brook, J. L. E. Dean, G. Sully, J. Saklatvala, and A. R. Clark. 2001. Mitogen-activated protein kinase p38 controls the expression and posttranslational modification of tristetraprolin, a regulator of tumor necrosis factor alpha mRNA stability. Mol. Cell. Biol. 21:6461-6469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McLeish, K., C. Knall, R. Ward, P. Gerwins, P. Coxon, J. Klein, and G. Johnson. 1998. Activation of mitogen-activated protein kinase cascades during priming of human neutrophils by TNF-alpha and GM-CSF. J. Leukoc. Biol. 64:537-545. [PubMed] [Google Scholar]
- 26.Megidish, T., J. Cooper, L. Zhang, H. Fu, and S. Hakomori. 1998. A novel sphingosine-dependent protein kinase (SDK1) specifically phosphorylates certain isoforms of 14-3-3 protein. J. Biol. Chem. 273:21834-21845. [DOI] [PubMed] [Google Scholar]
- 27.Merritt, C., H. Enslen, N. Diehl, D. Conze, R. J. Davis, and M. Rincon. 2000. Activation of p38 mitogen-activated protein kinase in vivo selectively induces apoptosis of CD8+ but not CD4+ T cells. Mol. Cell. Biol. 20:936-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nahas, N., T. Molski, G. Fernandez, and R. Sha'afi. 1996. Tyrosine phosphorylation and activation of a new mitogen-activated protein (MAP)-kinase cascade in human neutrophils stimulated with various agonists. Biochem. J. 318:247-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nick, J., N. Avdi, S. Young, C. Knall, P. Gerwins, G. Johnson, and S. Worthen. 1997. Common and distinct intracellular signaling pathways in human neutrophils utilized by platelet activating factor and FMLP. J. Clin. Investig. 99:975-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nick, J., N. Avdi, S. Young, L. Lehman, P. McDonald, S. Frasch, M. Billstrom, P. Henson, G. Johnson, and S. Worthen. 1999. Selective activation and functional significance of p38alpha mitogen-activated protein kinase in lipopolysaccharide-stimulated neutrophils. J. Clin. Investig. 103:851-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ono, K., and J. Han. 2000. The p38 signal transduction pathway activation and function. Cell. Signal. 12:1-13. [DOI] [PubMed] [Google Scholar]
- 32.Powell, D., M. Rane, Q. Chen, S. Singh, and R. McLeish. 2002. Identification of 14-3-3ζ as a protein kinase B/Akt substrate. J. Biol. Chem. 277:21639-21642. [DOI] [PubMed] [Google Scholar]
- 33.Rane, M., P. Coxon, D. Powell, R. Webster, J. Klein, W. Pierce, P. Ping, and K. McLeish. 2001. P38 kinase-dependent MAPKAPK-2 activation functions as 3-phosphoinositide-dependent kinase-2 for Akt in human neutrophils. J. Biol. Chem. 276:3517-3523. [DOI] [PubMed] [Google Scholar]
- 34.Rincon, M., H. Enslen, J. Raingeaud, M. Recht, T. Zapton, M. Su, L. Penix, R. Davis, and R. Flavell. 1998. Interferon-γ expression by Th1 effector T cells mediated by the p38 MAP kinase signaling pathway. EMBO J. 17:2817-2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sitkoff, D., K. Sharp, and B. Honig. 1994. Correlating solvation free energies and surface tensions of hydrocarbon solutes. Biophys. Chem. 51:397-403. [DOI] [PubMed] [Google Scholar]
- 36.Tzivion, G., L. Zhijun, and J. Avruch. 1998. A dimer 14-3-3 protein is an essential cofactor for Raf kinase activity. Nature 394:88-92. [DOI] [PubMed] [Google Scholar]
- 37.Underwood, D., R. Osborn, S. Bochnowicz, E. Webb, D. Rienman, D. Lee, A. Romanic, J. Adams, D. Hay, and D. Griswold. 2000. SB 239063, a p38 MAPK inhibitor, reduces neutrophilia, inflammatory cytokines, MMP-9, and fibrosis in lungs. Am. J. Physiol. Lung Cell. Mol. Physiol. 279:895-902. [DOI] [PubMed] [Google Scholar]
- 38.van Hermert, M., H. Steensma, and P. van Heusden. 2001. 14-3-3 proteins: key regulators of cell division, signaling, and apoptosis. BioEssays 23:936-946. [DOI] [PubMed] [Google Scholar]
- 39.Ward, R., M. Nakamura, and K. McLeish. 2000. Priming of the neutrophil respiratory burst involves p38 mitogen-activated protein kinase-dependent exocytosis of flavocytochrome b558-containing granules. J. Biol. Chem. 275:36713-36719. [DOI] [PubMed] [Google Scholar]
- 40.Werz, O., J. Klemm, B. Samuelsson, and O. Radmark. 2000. 5-Lipoxygenase is phosphorylated by p38 kinase-dependent MAPKAP kinases. Proc. Natl. Acad. Sci. USA 97:5262-5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yaffe, M., K. Rittinger, S. Volinia, P. Caron, A. Aitken, H. Leffers, S. Gamblin, S. Smerdon, and L. Cantley. 1997. The structural basis for 14-3-3:phosphopeptide binding specificity. Cell 91:961-971. [DOI] [PubMed] [Google Scholar]
- 42.Zu, Y., Y. Ai, A. Gilchrist, M. Labadia, R. Sha’afi, and C. Haung. 1996. Activation of MAP kinase-activated protein kinase 2 in human neutrophils after phorbol ester of fMLP peptide stimulation. Blood 87:5287-5296. [PubMed] [Google Scholar]