Abstract
The p53 tumor suppressor is regulated by MDM2-mediated ubiquitination and degradation. Mitogenic signals activate p53 by induction of ARF expression, which inhibits p53 ubiquitination by MDM2. Recent studies showed that the MDM2 homolog MDMX is also an important regulator of p53. We present evidence that MDM2 promotes MDMX ubiquitination and degradation by the proteasomes. This effect is stimulated by ARF and correlates with the ability of ARF to bind MDM2. Promotion of MDM2-mediated MDMX ubiquitination requires the N-terminal domain of ARF, which normally inhibits MDM2 ubiquitination of p53. An intact RING domain of MDM2 is also required, both to interact with MDMX and to provide E3 ligase function. Increase of MDM2 and ARF levels by DNA damage, recombinant ARF adenovirus infection, or inducible MDM2 expression leads to proteasome-mediated down-regulation of MDMX levels. Therefore, MDMX and MDM2 are coordinately regulated by stress signals. The ARF tumor suppressor differentially regulates the ability of MDM2 to promote p53 and MDMX ubiquitination and activates p53 by targeting both members of the MDM2 family.
The p53 protein is regulated by multiple mechanisms, which is critical for its ability to respond to stress and function as a tumor suppressor. p53 turnover is regulated by MDM2, which functions as a ubiquitin E3 ligase to promote p53 ubiquitination and degradation by proteasomes (33). Stress signals such as DNA damage induce p53 accumulation by phosphorylation (22). Mitogenic signals activate p53 by induction of the ARF tumor suppressor encoded by an alternative open reading frame in the p16INK4a locus, which inhibits the ability of MDM2 to ubiquitinate p53 (24, 30, 33).
MDMX is a recently identified homolog of MDM2 (28). MDMX shares strong homology to MDM2 at the amino acid sequence level and can bind to p53 and inhibit its transcription function in transient-transfection assays. However, unlike MDM2, MDMX does not promote p53 ubiquitination or degradation in vivo (9, 29). Furthermore, expression of MDMX is not induced by DNA damage (23, 28). MDM2 is well established as an important regulator of p53 activity during embryonic development. Knock out of MDM2 in mice results in embryonic lethality due to hyperactivation of p53 (20). However, recent studies showed that MDMX-null mice also die in utero in a p53-dependent fashion, which can be rescued by crossing into the p53-null background (5, 19, 21). Therefore, MDMX is also an important regulator of p53 during embryonic development, having a function that cannot be substituted by endogenous MDM2.
MDM2 is a ubiquitin E3 ligase that promotes ubiquitination of itself, p53, and several other cellular proteins, including androgen receptor, Tip60, glucocorticoid receptor, and beta 2-adrenergic receptor (12, 14, 25, 27). Ubiquitination of p53 and MDM2 itself requires the integrity of the C-terminal RING domain, which in other RING-containing E3 ligases is involved in direct interaction with the E2 ubiquitin-conjugating enzyme (11). Efficient ubiquitination of p53 also requires the central acidic domain of MDM2, which contains the binding site for ARF (17). The central region of MDM2 may be important for correct positioning of E2 after binding to p53 and facilitates the ubiquitination of p53. Deletion of the acidic domain or binding of ARF to this region inhibits the ability of MDM2 to ubiquitinate p53 (17).
The ability of MDM2 to promote self-ubiquitination results in its rapid degradation, with a half-life of approximately 30 min. In contrast, MDMX does not appear to have ubiquitin ligase function and has a relatively long half-life in cells. The RING domain of MDMX has been shown to bind to the RING domain of MDM2, which results in stabilization of MDM2 and p53 in cotransfection assays (26, 29). The regulation of MDMX level and function is still not well understood. Similar to MDM2, MDMX is a substrate for caspase 3 and may be degraded by caspases during apoptosis (3, 6). Here we present evidence that MDMX is regulated by MDM2-mediated ubiquitination. ARF promotes MDMX ubiquitination by MDM2, suggesting a novel mechanism by which ARF efficiently activates p53 function and connects MDMX to other signaling pathways through MDM2 and ARF.
MATERIALS AND METHODS
Cell lines, plasmids, and reagents.
H1299 (non-small cell lung carcinoma; p53-null), U2OS (osteosarcoma; p53 wild type; ARF deficient), and human primary foreskin fibroblast (HFF; passage 20) cells were maintained in Dulbecco's modified Eagle's medium with 10% fetal bovine serum. Human MDMX cDNAs (myc-MDMX and MDMX-myc-His6) were kindly provided by Donna George (26). MDM2/p53 double-null mouse embryo fibroblast 174.1 cells were a kind gift from Guillermina Lozano (16). ARF/p53 double-null mouse embryo fibroblasts were a kind gift from Gerry Zambetti. The myc epitope tag fused to the MDMX cDNA was removed by subcloning to generate a nontagged MDMX. A Flag epitope tag was added to the C terminus of myc-MDMX by PCR to create double-tagged myc-MDMX-Flag. A His6-ubiquitin expression plasmid was kindly provided by David Lane (31). ARF deletion mutants were kind gifts from Yue Xiong (32). MDM2 deletion mutants were described previously (1, 2). Point mutants of MDM2 were created using the QuikChange kit (Stratagene). All p53, MDM2, and ARF constructs used in this study were of human origin. Monoclonal antibody 8C6 against human MDMX was generated in our laboratory and reacts specifically with MDMX in a region between residues 101 and 393 (13). Adenovirus expressing human ARF was kindly provided by Yue Xiong and was used as described previously (15). A U2OS stable cell line expressing tetracycline-repressible human MDM2 was created by cloning MDM2 cDNA into the pUHD15.1 vector and cotransfecting with pUHG10.3 (7).
Western blotting.
Cells were lysed in lysis buffer (50 mM Tris-HCl [pH 8.0], 5 mM EDTA, 150 mM NaCl, 0.5% NP-40, 1 mM phenylmethylsulfonyl fluoride) and centrifuged for 5 min at 10,000 × g, and the insoluble debris were discarded. Cell lysate (10 to 50 μg of protein) was fractionated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and transferred to Immobilon P filters (Millipore). The filter was blocked for 1 h with phosphate-buffered saline containing 5% nonfat dry milk and 0.1% Tween 20. The following monoclonal antibodies were used: 3G9 for MDM2 (2), DO-1 for p53 (Pharmingen), 14PO2 for ARF (Neomarkers), and 8C6 for MDMX. The filter was developed using ECL-plus reagent (Amersham).
In vivo ubiquitination assay.
H1299 and U2OS cells in 9-cm plates were transfected with combinations of 1 μg of green fluorescent protein (GFP) expression plasmid, 5 μg of His6-ubiquitin expression plasmid, 1 to 5 μg of human MDMX, 5 μg of MDM2, and 5 μg of ARF expression plasmids using a conventional calcium phosphate precipitation method. Thirty-two hours after transfection, cells from each plate were collected into two aliquots. One aliquot (10%) was used for conventional Western blotting to confirm expression and degradation of transfected proteins. The remaining cells (90%) were used for purification of His6-tagged proteins by Ni2+-nitrilotriacetic acid (NTA) beads. The cell pellet was lysed in buffer A (6 M guanidinium-HCl, 0.1 M Na2HPO4/NaH2PO4, 0.01 M Tris-Cl [pH 8.0], 5 mM imidazole, 10 mM β-mercaptoethanol) and incubated with Ni2+-NTA beads (Qiagen) for 4 h at room temperature. The beads were washed with buffer A, buffer B (8 M urea, 0.1 M Na2PO4/NaH2PO4, 0.01 M Tris-Cl [pH 8.0], 10 mM β-mercaptoethanol), and buffer C (8 M urea, 0.1 M Na2PO4/NaH2PO4, 0.01 M Tris-Cl [pH 6.3], 10 mM β-mercaptoethanol), and bound proteins were eluted with buffer D (200 mM imidazole, 0.15 M Tris-Cl [pH 6.7], 30% glycerol, 0.72 M β-mercaptoethanol, 5% SDS). The eluted proteins were analyzed by Western blotting for the presence of conjugated MDMX by using 8C6 antibody.
In vitro ubiquitination assay.
Glutathione S-transferase (GST)-MDM2 full-length and deletion mutants were expressed in Escherichia coli and purified by binding to glutathione-agarose beads. The substrate MDMX was produced by in vitro translation in rabbit reticulocyte lysate, using the TNT system (Promega) in the presence of [35S]methionine. Fifteen microliters of packed beads loaded with ∼2 μg of GST-fusion proteins was incubated with 4 μl of MDMX in vitro translation product, 250 ng of GST-Ubc5Hb (Boston Biochem), 250 ng of purified rabbit E1 (AG Scientific), 2 μg of His6-ubiquitin (AG Scientific), and 20 μl of reaction buffer (50 mM Tris [pH 7.5], 2.5 mM MgCl2, 15 mM KCl, 1 mM dithiothreitol, 0.01% Triton X-100, 1% glycerol, 4 mM ATP). The mixture was incubated at 37°C for 1 h with shaking, boiled in SDS sample buffer, and fractionated by SDS-polyacrylamide gel electrophoresis. The gel was dried, and MDMX was detected by autoradiography.
RESULTS
MDM2 stimulates MDMX ubiquitination and degradation.
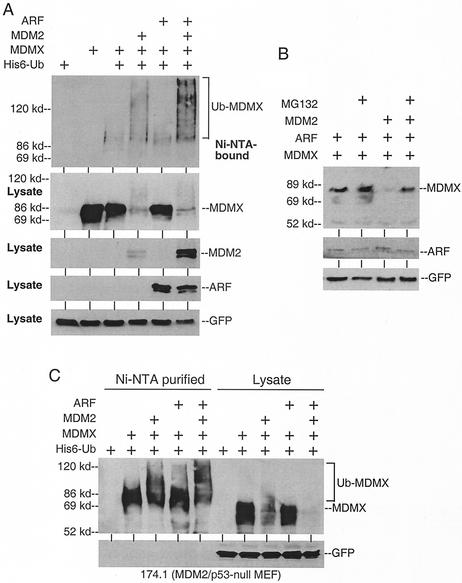
MDM2 is rapidly degraded by the proteasome due to self-ubiquitination (4). In contrast, little is known about the regulation of MDMX turnover. We tested whether it is subjected to ubiquitination in vivo. To detect MDMX ubiquitination, we coexpressed MDMX and His6-ubiquitin by transient transfection into H1299 cells. MDMX conjugated to His6-ubiquitin was purified by using Ni2+-NTA beads under denaturing conditions and detected by Western blotting with MDMX-specific monoclonal antibody 8C6 (13). The result showed that a small portion of MDMX was conjugated to ubiquitin in vivo (mainly monoubiquitination, based on a slightly slower mobility than unmodified MDMX). Transfection of MDM2 strongly stimulated MDMX polyubiquitination (Fig. 1A). Interestingly, cotransfection of MDM2 and ARF did not inhibit but rather stimulated MDMX polyubiquitination (Fig. 1A). The MDM2 expression level was also increased in the presence of ARF (likely due to stabilization of the protein) and may account for the increase in MDMX ubiquitination.
FIG. 1.
MDMX ubiquitination and degradation by MDM2. (A) MDMX was coexpressed with His6-ubiquitin in H1299 cells by transient transfection. His6-ubiquitinated proteins were purified, and MDMX was detected by Western blotting with 8C6 antibody (top panel). The lower panels are control Western blots of nonpurified cell lysate (10% of the amount used for Ni-NTA purification). (B) H1299 cells transfected with the indicated plasmids were treated with 30 μM MG132 for 4 h prior to Western blot analysis for MDMX. Transfection efficiency was verified by expression of cotransfected GFP plasmid. (C) MDM2/p53-null 174.1 cells were transfected with the indicated plasmids. Ni-NTA-purified ubiquitinated MDMX and cell lysate were run on the same gel to show the molecular weight shift due to MDM2-independent monoubiquitination.
Direct Western blotting also showed that the MDMX expression level was significantly reduced when coexpressed with MDM2 (Fig. 1A), suggesting that ubiquitination of MDMX led to subsequent degradation by the proteasomes. Consistent with this hypothesis, treatment with the proteasome inhibitor MG132 for 4 h prior to cell harvest partially prevented MDMX down-regulation by MDM2 cotransfection (Fig. 1B). In the absence of cotransfected MDM2, MDMX was relatively stable and did not accumulate in the presence of MG132.
To determine whether MDM2 and ARF are required for the basal ubiquitination of MDMX, the assay was repeated using MDM2/p53-null or ARF/p53-null mouse fibroblasts. The results showed that monoubiquitination of MDMX occurred in the absence of endogenous MDM2 (Fig. 1C) and ARF (data not shown). Therefore, MDMX can undergo ARF- and MDM2-independent monoubiquitination. Cotransfection of MDM2 or MDM2 and ARF also stimulated MDMX polyubiquitination in mouse cells.
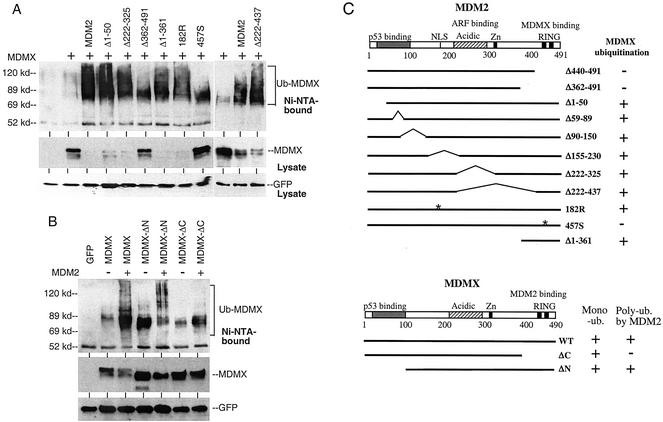
The MDM2 RING domain is sufficient for promoting MDMX ubiquitination.
Next, a panel of MDM2 mutants was tested for ubiquitination of MDMX. The results showed that the RING domain of MDM2 was important for promoting MDMX ubiquitination (representative mutants are shown in Fig. 2A). MDM2 deletion mutants without the p53-binding or ARF-binding domains were still capable of stimulating MDMX ubiquitination, indicating that these regions were not essential for this function. An MDM2 nuclear localization signal point mutant (182R) that prevents nuclear translocation was also able to ubiquitinate and degrade MDMX. Furthermore, the C-terminal 130 amino acids of MDM2 containing the RING domain were sufficient to ubiquitinate MDMX (Fig. 2A). Deletion of the MDMX C-terminal RING domain (Δ394-490) also prevented polyubiquitination by MDM2 (Fig. 2B). Therefore, these results showed that polyubiquitination of MDMX by MDM2 required formation of an MDMX-MDM2 complex through RING domain-mediated heterodimerization and possibly the E3 function of MDM2.
FIG. 2.
MDM2 domains required for promoting MDMX ubiquitination. (A) H1299 cells were transfected with His6-ubiquitin, MDMX, and representative MDM2 mutants. MDMX ubiquitination and degradation were detected by Ni-NTA purification and Western blotting. (B) H1299 cells were transfected with His6-ubiquitin, MDM2, and MDMX mutants. MDMX ubiquitination was detected by Ni-NTA purification and Western blotting. (C) Diagram of MDM2 and MDMX mutants and summary of results. MDMX-ΔN has a deletion of residues 1 to 100. MDMX-ΔC has a deletion of residues 394 to 490.
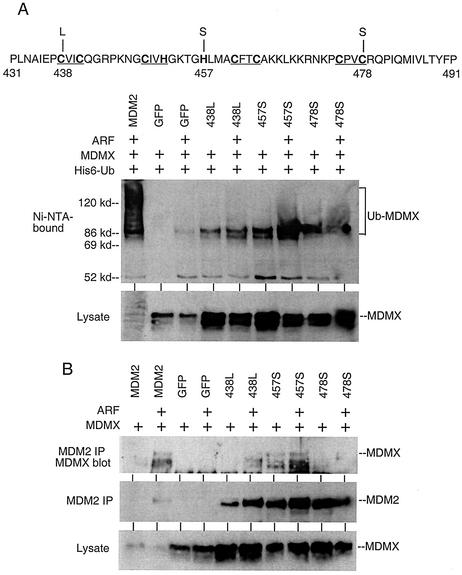
Ubiquitination of MDMX requires MDM2 ubiquitin ligase function.
To test whether the E3 function of MDM2 is required for polyubiquitination of MDMX, we mutated three key cysteine and histidine residues in the RING domain of MDM2 that a previous study had found to be important for E3 function (438C-L, 457H-S, and 478C-S) (Fig. 3A) (4). These mutations prevented polyubiquitination of MDMX in the presence of ARF (Fig. 3A). When analyzed by MDM2 immunoprecipitation and MDMX Western blotting, the 438L and 478S mutants lost most or all of the MDMX binding, indicating that these mutations affected both E3 and MDMX binding functions (Fig. 3B). The 457S mutant was still capable of binding to MDMX, but the efficiency was significantly lower than that of wild-type MDM2, considering the higher expression levels of both proteins in the transfected cells due to lack of degradation (Fig. 3B). Since the absolute amount of the 457S-MDMX complex was comparable to that of MDM2-MDMX, yet MDMX ubiquitination was much weaker in the 457S-MDMX combination, we concluded that the E3 function of MDM2 was critical for ubiquitination of MDMX. However, it was clear from these results that MDMX binding and E3 functions were both dependent on the integrity of the MDM2 RING domain and could not be completely separated by these mutations.
FIG. 3.
Ubiquitination of MDMX requires MDM2 E3 function. (A) Position of point mutations in the MDM2 RING domain. C and H residues important for zinc coordination are underlined. Ubiquitination of MDMX by MDM2 mutants was determined by transfection of H1299 cells as described in the legend for Fig. 1A. (B) Binding of MDMX to MDM2 point mutants. MDMX and MDM2 mutants were cotransfected into H1299 cells and analyzed by MDM2 immunoprecipitation followed by MDMX Western blotting.
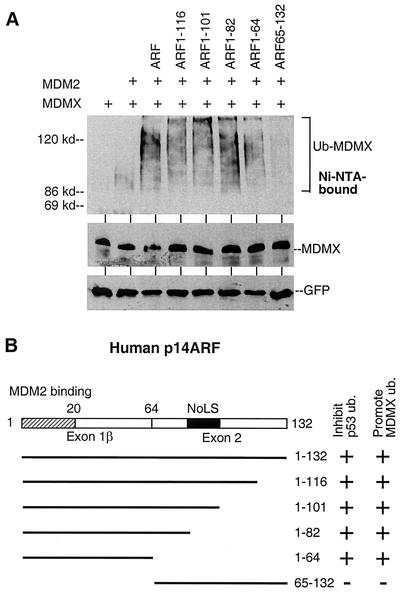
The ARF N-terminal domain is required to cooperate with MDM2.
It is well established that ARF is an inhibitor of MDM2 E3 ligase function on p53 (33). The results in Fig. 1 showed that its effect on MDM2-mediated MDMX ubiquitination was opposite to that of p53. Therefore, we analyzed a panel of ARF deletion mutants to identify the domain required for stimulating MDMX ubiquitination. The results showed that the N-terminal domain of ARF containing the MDM2-binding region was required for stimulating MDMX ubiquitination in the presence of MDM2 (Fig. 4A). Deleting the nucleolar targeting signal between residues 82 and 101 did not prevent stimulation of MDMX ubiquitination (32). Interestingly, the N-terminal region of ARF contains the MDM2 binding site and is sufficient to inhibit p53 ubiquitination by MDM2 (17). Therefore, ARF binding to MDM2 differentially regulates its ability to ubiquitinate p53 and MDMX.
FIG. 4.
The ARF N-terminal domain is sufficient to cooperate with MDM2. (A) H1299 cells were transfected with MDMX, a suboptimal amount of MDM2, and different ARF mutants. His6-ubiquitin-conjugated MDMX was purified and detected by Western blotting. (B) Diagram of ARF mutants and summary of results.
Although ARF stimulated MDMX ubiquitination, MDM2 alone was sufficient to induce significant degradation of MDMX even in ARF-deficient cells (data not shown), indicating that ARF is not essential for this process. To observe stimulation of MDMX degradation by ARF in H1299 cells requires careful titration to limit the amount of MDM2 plasmid (data not shown). The effect of ARF on MDMX degradation was more evident and reproducible in mouse cells (Fig. 1C), suggesting that cell type may affect the effect of ARF in this transfection assay.
Regulation of endogenous MDMX levels by ARF and MDM2.
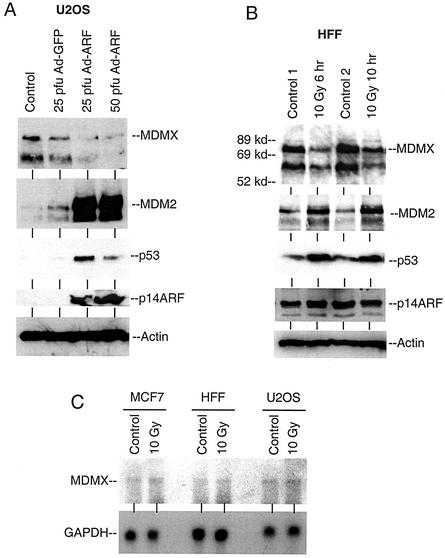
To determine whether the endogenous MDMX expression level was regulated by ARF, ARF-deficient U2OS cells were infected with ARF adenovirus to restore ARF expression. As expected, expression of ARF resulted in strong induction of p53 and MDM2 levels. This was accompanied by reduction of MDMX expression (Fig. 5A). In this experiment, infection with Ad-ARF at a high titer did not result in a higher p53 level, possibly due to complicating effects such as apoptosis. In the second experiment, primary HFFs were treated with ionizing radiation to induce MDM2 expression. DNA damage resulted in induction of p53 and MDM2 expression levels in HFFs and a corresponding reduction of MDMX expression (Fig. 5B). Reduction of MDMX levels was also observed after induction of MDM2 expression by irradiation of U2OS cells or induction of MDM2 by activating a temperature-sensitive p53 mutant in H1299 cells (data not shown).
FIG. 5.
DNA damage and ARF expression down-regulate MDMX level. (A) U2OS cells were infected with ARF adenovirus at the indicated multiplicity for 24 h and analyzed for MDMX, MDM2, and p53 expression levels by direct Western blotting. (B) Primary HFFs were treated with 10 Gy of gamma radiation and analyzed for MDMX, MDM2, p53, and ARF levels by Western blotting at the indicated times after treatment. (C) Gamma irradiation does not reduce the MDMX mRNA level. Cells were treated with gamma radiation, and 6 h later total RNA was isolated and hybridized with an MDMX cDNA probe in a Northern blotting analysis.
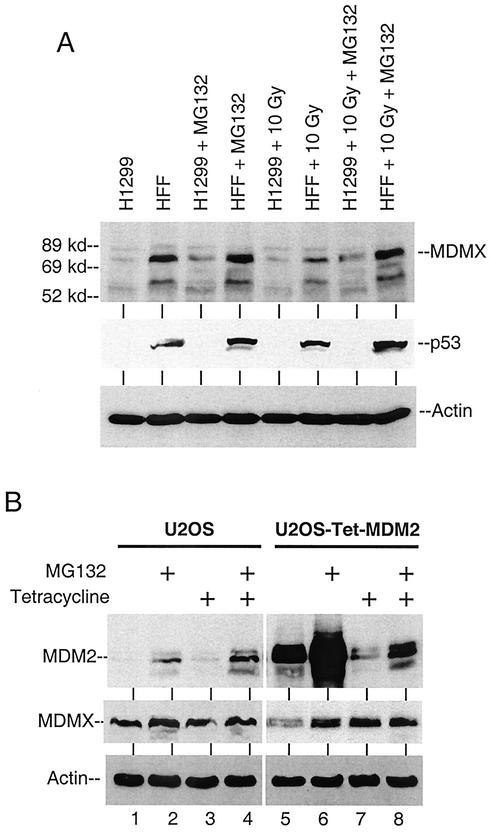
To determine whether the reduced MDMX level was due to degradation by the proteasomes, irradiated HFF cells were treated with MG132 for 4 h prior to analysis. The decrease in MDMX level after irradiation was blocked by MG132 (Fig. 6A), suggesting it was mediated by proteasomes. In contrast, the p53-null H1299 cells did not express elevated MDM2 after irradiation and the MDMX level was not reduced. Northern blot analysis using several cell lines confirmed that the MDMX mRNA level was not reduced by radiation (Fig. 5C). These results suggested that ionizing radiation induced proteasome-mediated degradation of MDMX, which may be due to p53 activation and induction of MDM2 expression.
FIG. 6.
Induction of MDM2 results in MDMX degradation by proteasomes. (A) HFFs and H1299 cells were treated with 10 Gy of gamma radiation for 6 h. MG132 (30 μM) was added immediately after radiation. The MDMX level was determined by Western blotting. (B) U2OS cells stably expressing tetracycline-repressible MDM2 were cultured in the presence or absence of 0.5 μg of tetracycline/ml for 20 h. MG132 was added for the last 4 h. MDMX and MDM2 levels were determined by Western blotting.
To test whether a change in the MDM2 level independent of p53 activation or irradiation was sufficient to regulate MDMX level, we created a U2OS cell line expressing tetracycline-repressible MDM2. Induction of MDM2 expression by removal of tetracycline correlated with a decrease of MDMX level (Fig. 6B, lanes 5 and 7), which was restored by a 4-h MG132 treatment. Therefore, increase of MDM2 expression alone was sufficient to promote MDMX degradation. However, the moderate decrease of MDMX was not proportional to the strong induction of MDM2, suggesting that other factors may limit the efficiency of MDMX degradation by MDM2.
Epitope tagging prevents MDMX degradation by MDM2.
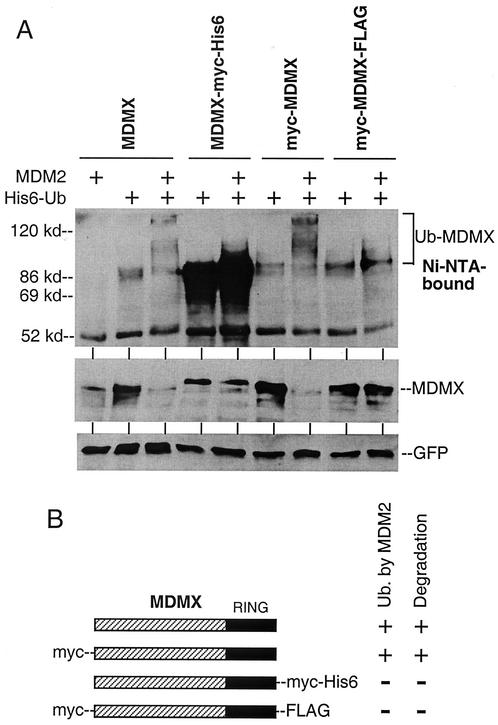
Previous studies using epitope-tagged MDMX did not indicate an effect of MDM2 on MDMX degradation (10, 26). We also had inconsistent results in early experiments when tagged and nontagged versions of MDMX were used interchangeably. A direct comparison of nontagged MDMX with several versions of epitope-tagged MDMX revealed that addition of c-myc or Flag epitopes to the C terminus of MDMX significantly inhibited polyubiquitination by MDM2 and completely prevented degradation (Fig. 7). Therefore, addition of epitopes to the RING domain of MDMX interferes with ubiquitination and degradation by MDM2. Because of this finding, all of the results described above were from repeat experiments using the nontagged version of MDMX.
FIG. 7.
Inhibition of MDMX degradation by C-terminal epitope tags. (A) H1299 cells were transfected with MDMX with different epitope tags at the N or C terminus and subjected to Ni-NTA purification and Western blotting. The MDMX-myc-His6 protein was completely recovered by Ni-NTA beads as expected. (B) Diagrams of epitope-tagged MDMX and summary of results. C-terminal epitope tags block polyubiquitination and degradation by MDM2.
MDM2 promotes MDMX polyubiquitination in vitro.
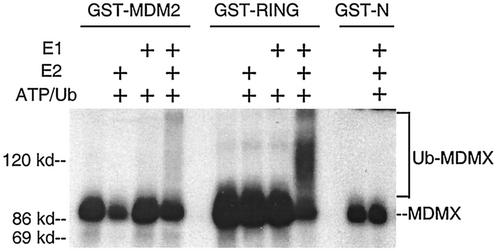
In order to confirm the role of MDM2 as an E3 for the polyubiquitination of MDMX, we established an in vitro ubiquitination assay for MDMX. In this assay, GST-MDM2 was able to stimulate the polyubiquitination of in vitro-translated MDMX in the presence of purified E1 and E2 (GST-UbcH5b) (Fig. 8). Furthermore, the C-terminal fragment of MDM2 containing the RING domain was sufficient to promote MDMX ubiquitination, whereas an MDM2 N-terminal fragment had no effect. These results corroborate the in vivo ubiquitination results shown in Fig. 2 and demonstrate that MDM2 functions as a ubiquitin E3 ligase for MDMX.
FIG. 8.
MDM2 promotes MDMX polyubiquitination in vitro. GST-MDM2, GST-RING (containing MDM2 residues 290 to 491), and GST-N (containing MDM2 residues 1 to 150) were purified using glutathione agarose beads. Loaded beads were incubated with in vitro-translated MDMX in the presence or absence of E1 and E2 in a ubiquitination reaction as described in Materials and Methods. Polyubiquitinated MDMX appears as a high-molecular-weight smear above the unmodified MDMX band.
DISCUSSION
The results described above show that MDMX is subjected to modification by ubiquitin conjugation. Interaction with MDM2 leads to strong stimulation of MDMX polyubiquitination and degradation by proteasomes. MDM2 appears to function as a ubiquitin E3 ligase in this process, since the MDM2457S RING domain mutant that prevents p53 or self-ubiquitination also inhibits ubiquitination of MDMX, while still partially retaining MDMX binding. Furthermore, purified recombinant GST-MDM2 is able to stimulate MDMX polyubiquitination under cell-free conditions. MDMX also undergoes basal monoubiquitination independent of MDM2, suggesting interaction with other ubiquitination factors. It is noteworthy that previous studies using epitope-tagged MDMX did not observe an effect of MDM2 on MDMX degradation (10, 26). We found that MDMX tagged with c-myc or Flag epitopes at the C terminus were resistant to ubiquitination and degradation by MDM2, possibly due to altered RING domain conformation. Therefore, use of nontagged or N-terminally tagged MDMX appears to be critical for detecting its efficient ubiquitination and degradation by MDM2.
Surprisingly, ARF stimulates MDMX ubiquitination by MDM2, which is opposite to its effect on p53 ubiquitination. The stimulating effect of ARF is carried out by the N-terminal domain that normally inhibits MDM2 ubiquitination of p53. Therefore, ARF interaction with MDM2 differentially affects its E3 function toward different substrates rather than inactivating its E3 function in general. This effect is plausible because ARF does not directly interact with the RING domain of MDM2, which may be involved in recruiting E2. Binding of ARF to the acidic domain of MDM2 may simply alter its ability to properly orient E2 for the transfer of ubiquitin to certain substrates. In the case of p53, ubiquitin conjugation is blocked. But in the case of MDMX, ubiquitination is not affected or even stimulated. ARF increases the MDM2 expression level by stabilization, which may account for increased MDMX ubiquitination. However, ARF may also qualitatively stimulate the E3 activity of MDM2 toward MDMX. Further experiments using the in vitro ubiquitination system will be required to determine whether this is the case.
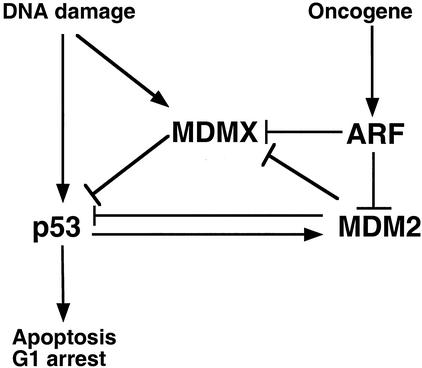
These results suggest that ARF is a more effective and multifunctional regulator of the p53 pathway than previously appreciated. By blocking the ability of MDM2 to ubiquitinate p53, ARF is sufficient to induce p53 stabilization and activation. Additionally, by increasing MDMX ubiquitination, ARF also simultaneously promotes the elimination of another important inhibitor of p53. The combination of these two functions should result in more potent activation of p53. Because p53 activation induces MDM2 expression, a higher MDM2 level should further cooperate with ARF to degrade MDMX, resulting in a positive feedback effect. Activation of p53 by other stress signals, such as DNA damage, also leads to strong induction of MDM2 expression. An increased MDM2 level may in fact facilitate p53 activation by degrading MDMX while its own interaction with p53 is temporarily blocked by phosphorylation (Fig. 9).
FIG. 9.
MDMX is regulated by stress signals. MDM2 and MDMX cooperate to inhibit p53 activity in the absence of stress. Stress signals activate p53 by targeting both MDM2 and MDMX. Oncogenes such as E2F1 and c-myc induce ARF expression, which activates p53 by inhibiting p53 ubiquitination and promoting MDMX ubiquitination by MDM2. DNA damage activates p53 and induces MDM2 expression, which in turn degrades MDMX and leads to further activation of p53. DNA damage also promotes MDMX nuclear translocation, which may expose it to degradation by MDM2 but also enables it to more efficiently inhibit p53 at certain stages of the damage response.
The MDM2 negative feedback loop is an important mechanism in p53 regulation. However, knockout experiments showed that MDMX is also critical for regulating p53 (5, 19, 21). Recent studies suggested that MDMX and MDM2 depend on each other for efficient inhibition of p53. MDM2 stimulates MDMX by targeting it to the nucleus, whereas MDMX enhances MDM2's ability to degrade p53 in a highly dose-dependent manner (8, 18). Therefore, MDMX may be part of a more complicated MDM2 feedback loop by regulating MDM2 function and acting as a receiver of other signals. The ability of MDM2 to degrade MDMX ensures that it also exists in a balance with MDM2 and p53. Although MDMX can inhibit p53 function, the dependence on a suicidal interaction with MDM2 enables MDMX to be regulated by other signals. The MDMX degradation mechanism also ensures that stress signals can efficiently activate p53 by coordinately regulating both p53 inhibitors (Fig. 9).
Recent studies showed that ectopically expressed MDMX is mainly localized in the cytoplasm and undergoes nuclear translocation by both MDM2-dependent and -independent mechanisms (8, 13, 18). Nuclear entry of MDMX is critical for its ability to inhibit p53 (8, 18), suggesting that MDMX plays an active role in limiting p53 activation during the DNA damage response. However, results presented in this report show that DNA damage can induce degradation of MDMX, which would facilitate p53 activation. Therefore, the net outcome of these regulatory events on the function of MDMX may depend on the level of damage and different stages of the stress response. Further work will be required to elucidate the role of MDMX in p53 regulation during DNA damage and other stress conditions.
In summary, our findings reveal that the MDMX expression level is directly regulated by MDM2-mediated ubiquitination. The results also identify ARF as a more dynamic regulator of MDM2 ubiquitin ligase function and suggest a potential mechanism of MDMX regulation by a variety of stress signals through MDM2 and ARF. Elucidating how MDMX ubiquitination and degradation are regulated should lead to a better understanding of its role in the p53 pathway and tumorigenesis.
Acknowledgments
We thank Donna George for providing the MDMX construct which made our work possible. We also thank Yue Xiong for ARF constructs and helpful discussions.
This work was supported by grants from the American Cancer Society and National Institutes of Health to J. Chen.
REFERENCES
- 1.Chen, J., J. Lin, and A. J. Levine. 1995. Regulation of transcription functions of the p53 tumor suppressor by the mdm-2 oncogene. Mol. Med. 1:142-152. [PMC free article] [PubMed] [Google Scholar]
- 2.Chen, J., V. Marechal, and A. J. Levine. 1993. Mapping of the p53 and mdm-2 interaction domains. Mol. Cell. Biol. 13:4107-4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, L., V. Marechal, J. Moreau, A. J. Levine, and J. Chen. 1997. Proteolytic cleavage of the mdm2 oncoprotein during apoptosis. J. Biol. Chem. 272:22966-22973. [DOI] [PubMed] [Google Scholar]
- 4.Fang, S., J. P. Jensen, R. L. Ludwig, K. H. Vousden, and A. M. Weissman. 2000. Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J. Biol. Chem. 275:8945-8951. [DOI] [PubMed] [Google Scholar]
- 5.Finch, R. A., D. B. Donoviel, D. Potter, M. Shi, A. Fan, D. D. Freed, C. Y. Wang, B. P. Zambrowicz, R. Ramirez-Solis, A. T. Sands, and N. Zhang. 2002. Mdmx is a negative regulator of p53 activity in vivo. Cancer Res. 62:3221-3225. [PubMed] [Google Scholar]
- 6.Gentiletti, F., F. Mancini, M. D'Angelo, A. Sacchi, A. Pontecorvi, A. G. Jochemsen, and F. Moretti. 2002. MDMX stability is regulated by p53-induced caspase cleavage in NIH3T3 mouse fibroblasts. Oncogene 21:867-877. [DOI] [PubMed] [Google Scholar]
- 7.Gossen, M., and H. Bujard. 1992. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. USA 89:5547-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gu, J., H. Kawai, L. Nie, H. Kitao, D. Wiederschain, A. G. Jochemsen, J. Parant, G. Lozano, and Z. M. Yuan. 2002. Mutual dependence of MDM2 and MDMX in their functional inactivation of p53. J. Biol. Chem. 277:19251-19254. [DOI] [PubMed] [Google Scholar]
- 9.Jackson, M. W., and S. J. Berberich. 2000. MdmX protects p53 from Mdm2-mediated degradation. Mol. Cell. Biol. 20:1001-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jackson, M. W., M. S. Lindstrom, and S. J. Berberich. 2002. MdmX binding to ARF affects Mdm2 protein stability and p53 transactivation. J. Biol. Chem. 276:25336-25341. [DOI] [PubMed] [Google Scholar]
- 11.Joazeiro, C. A., and A. M. Weissman. 2002. RING finger proteins: mediators of ubiquitin ligase activity. Cell 102:549-552. [DOI] [PubMed] [Google Scholar]
- 12.Legube, G., L. K. Linares, C. Lemercier, M. Scheffner, S. Khochbin, and D. Trouche. 2002. Tip60 is targeted to proteasome-mediated degradation by Mdm2 and accumulates after UV irradiation. EMBO J. 21:1704-1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li, C., C. Lihong, and C. Jiandong. 2002. DNA damage induces MDMX nuclear translocation by p53-dependent and -independent mechanisms. Mol. Cell. Biol. 22:7562-7571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin, H. K., L. Wang, Y. C. Hu, S. Altuwaijri, and C. Chang. 2002. Phosphorylation-dependent ubiquitylation and degradation of androgen receptor by Akt require Mdm2 E3 ligase. EMBO J. 21:4037-4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu, W., J. Lin, and J. Chen. 2002. Expression of p14ARF overcomes tumor resistance to p53. Cancer Res. 62:1305-1310. [PubMed] [Google Scholar]
- 16.McMasters, K. M., R. Montes de Oca Luna, J. R. Pena, and G. Lozano. 1996. mdm2 deletion does not alter growth characteristics of p53-deficient embryo fibroblasts. Oncogene 13:1731-1736. [PubMed] [Google Scholar]
- 17.Midgley, C. A., J. M. Desterro, M. K. Saville, S. Howard, A. Sparks, R. T. Hay, and D. P. Lane. 2001. An N-terminal p14ARF peptide blocks Mdm2-dependent ubiquitination in vitro and can activate p53 in vivo. Oncogene 19:2312-2323. [DOI] [PubMed] [Google Scholar]
- 18.Migliorini, D., D. Danovi, E. Colombo, R. Carbone, P. G. Pelicci, and J. C. Marine. 2002. Hdmx recruitment into the nucleus by Hdm2 is essential for its ability to regulate p53 stability and transactivation. J. Biol. Chem. 277:7318-7323. [DOI] [PubMed] [Google Scholar]
- 19.Migliorini, D., E. L. Denchi, D. Danovi, A. Jochemsen, M. Capillo, A. Gobbi, K. Helin, P. G. Pelicci, and J. C. Marine. 2002. Mdm4 (Mdmx) regulates p53-induced growth arrest and neuronal cell death during early embryonic mouse development. Mol. Cell. Biol. 22:5527-5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oca Luna, R. M., D. S. Wagner, and G. Lozano. 1995. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature 378:203-206. [DOI] [PubMed] [Google Scholar]
- 21.Parant, J., A. Chavez-Reyes, N. A. Little, W. Yan, V. Reinke, A. G. Jochemsen, and G. Lozano. 2002. Rescue of embryonic lethality in Mdm4-null mice by loss of Trp53 suggests a nonoverlapping pathway with MDM2 to regulate p53. Nat. Genet. 29:92-95. [DOI] [PubMed] [Google Scholar]
- 22.Prives, C., and P. A. Hall. 1999. The p53 pathway. J. Pathol. 187:112-126. [DOI] [PubMed] [Google Scholar]
- 23.Ramos, Y. F., R. Stad, J. Attema, L. T. Peltenburg, A. J. van der Eb, and A. G. Jochemsen. 2002. Aberrant expression of HDMX proteins in tumor cells correlates with wild-type p53. Cancer Res. 61:1839-1842. [PubMed] [Google Scholar]
- 24.Ries, S. J., C. H. Brandts, A. S. Chung, C. H. Biederer, B. C. Hann, E. M. Lipner, F. McCormick, and W. M. Korn. 2001. Loss of p14ARF in tumor cells facilitates replication of the adenovirus mutant dl1520 (ONYX-015). Nat. Med. 6:1128-1133. [DOI] [PubMed] [Google Scholar]
- 25.Sengupta, S., and B. Wasylyk. 2002. Ligand-dependent interaction of the glucocorticoid receptor with p53 enhances their degradation by Hdm2. Genes Dev. 15:2367-2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharp, D. A., S. A. Kratowicz, M. J. Sank, and D. L. George. 1999. Stabilization of the MDM2 oncoprotein by interaction with the structurally related MDMX protein. J. Biol. Chem. 274:38189-38196. [DOI] [PubMed] [Google Scholar]
- 27.Shenoy, S. K., P. H. McDonald, T. A. Kohout, and R. J. Lefkowitz. 2002. Regulation of receptor fate by ubiquitination of activated beta 2-adrenergic receptor and beta-arrestin. Science 294:1307-1313. [DOI] [PubMed] [Google Scholar]
- 28.Shvarts, A., W. T. Steegenga, N. Riteco, T. van Larr, P. Dekker, M. Bazuine, R. C. A. van Ham, W. V. D. H. van Oordt, G. Hateboer, A. J. van der Eb, and A. G. Jochemsen. 1996. MDMX: a novel p53-binding protein with some functional properties of MDM2. EMBO J. 15:5349-5357. [PMC free article] [PubMed] [Google Scholar]
- 29.Stad, R., N. A. Little, D. P. Xirodimas, R. Frenk, A. J. van der Eb, D. P. Lane, M. K. Saville, and A. G. Jochemsen. 2001. Mdmx stabilizes p53 and Mdm2 via two distinct mechanisms. EMBO Rep. 2:1029-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stott, F. J., S. Bates, M. C. James, B. B. McConnell, M. Starborg, S. Brookes, I. Palmero, K. Ryan, E. Hara, K. H. Vousden, and G. Peters. 1998. The alternative product from the human CDKN2A locus, p14(ARF), participates in a regulatory feedback loop with p53 and MDM2. EMBO J. 17:5001-5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xirodimas, D., M. K. Saville, C. Edling, D. P. Lane, and S. Lain. 2002. Different effects of p14ARF on the levels of ubiquitinated p53 and Mdm2 in vivo. Oncogene 20:4972-4983. [DOI] [PubMed] [Google Scholar]
- 32.Zhang, Y., and Y. Xiong. 1999. Mutations in human ARF exon 2 disrupt its nucleolar localization and impair its ability to block nuclear export of MDM2 and p53. Mol. Cell 3:579-591. [DOI] [PubMed] [Google Scholar]
- 33.Zhang, Y., and Y. Xiong. 2001. Control of p53 ubiquitination and nuclear export by MDM2 and ARF. Cell Growth Differ. 12:175-186. [PubMed] [Google Scholar]