Abstract
RNF5 is a RING finger protein found to be important in the growth and development of Caenorhabditis elegans. The search for RNF5-associated proteins via a yeast two-hybrid screen identified a LIM-containing protein in C. elegans which shows homology with human paxillin. Here we demonstrate that the human homologue of RNF5 associates with the amino-terminal domain of paxillin, resulting in its ubiquitination. RNF5 requires intact RING and C-terminal domains to mediate paxillin ubiquitination. Whereas RNF5 mediates efficient ubiquitination of paxillin in vivo, protein extracts were required for in vitro ubiquitination, suggesting that additional modifications and/or an associated E3 ligase assist RNF5 targeting of paxillin ubiquitination. Mutant Ubc13 efficiently inhibits RNF5 ubiquitination, suggesting that RNF5 generates polychain ubiquitin of the K63 topology. Expression of RNF5 increases the cytoplasmic distribution of paxillin while decreasing its localization within focal adhesions, where it is primarily seen under normal growth. Concomitantly, RNF5 expression results in inhibition of cell motility. Via targeting of paxillin ubiquitination, which alters its localization, RNF5 emerges as a novel regulator of cell motility.
A growing number of RING (really interesting new gene) finger proteins have been implicated as key regulators of cell growth and in organ and tissue development (1). RINGs are cysteine-rich, zinc-binding domains defined by a pattern of conserved cysteine and histidine residues (reviewed in reference 1) that exert their regulatory function through association with and effects on signal transduction and cell cycle control proteins. In many cases RINGs elicit E3 ligase activity, which serves to limit their availability and to alter the stability and/or localization of their associated proteins (1, 4, 18, 20).
RINGs that directly target ubiquitination of their associated substrates via their E3 ligase activity include the mammalian homologues of seven in absentia (Siah), AO7, BRCA1, and Mdm2 (1, 12, 18, 20); Parkin, which is associated with autosomal juvenile parkinsonism (36); and the Cbl protein family, which regulates cell signaling (18, 37). In the process of searching for RING finger proteins resembling mammalian RINGs known to exhibit E3 ligase activities in Caenorhabditis elegans, we identified and characterized RNF5 (C16C10.7), the C. elegans homologue of the human RING-finger protein 5 (RNF5) (17) and Arabidopsis thaliana RMA1 (17, 22). RNF5 shows structural homology within the RING domain with BRCA1, Cbl, and Mdm2 and is important to development in C. elegans (Broday et al., unpublished data). RNF5 expression is reduced in human tumors (our unpublished observations), partly due to transcriptional suppression mediated by the polycomb group protein EZH2, which is involved in prostate cancer progression (44). The search for RNF5-associated proteins identified a LIM domain-containing protein in C. elegans which shows homology with human paxillin. Here we demonstrate that through its association with paxillin, RNF5 affects paxillin ubiquitination and localization.
Paxillin is a multidomain protein that localizes primarily to sites of cell adhesions, called focal adhesions, that link the extracellular matrix to actin cytoskeleton (reviewed in references 14, 33, 39, 40, and 41). The amino-terminal half of paxillin contains five LD repeats, each of which consists of eight amino acids beginning with an invariant leucine-aspartate sequence, for which the LD repeats are named. The LD motif mediates binding of focal adhesion kinase (FAK), vinculin, and E6 oncoprotein to paxillin and is implicated in paxillin-dependent localization of FAK and vinculin to focal adhesions (2, 41, 46). The carboxy terminus of paxillin contains four LIM domains and a double zinc finger motif that mediates binding to transcription factors and cytoskeletal proteins as well as tyrosine phosphatases (3, 34, 39). The LIM domain of paxillin also mediates dimerization of paxillin. Both LIM and LD domains are implicated in paxillin localization to focal contacts and its role in regulating cell spreading and motility (27, 46, 49). Paxillin localization within focal adhesions is required to mediate signals elicited following the interaction of integrins with extracellular matrix proteins and following activation of various growth factor receptors (19, 33).
Paxillin functions primarily as a molecular adaptor, providing multiple docking sites for an array of signaling and structural proteins at the plasma membrane. These range from the exchange factor PIX and p21-activated kinase (PAK) to the PAK structural proteins such as vinculin and actopaxin, which bind actin directly to regulators of actin cytoskeletal dynamics such as ARF, GAP, and protein kinase L (26, 33). In addition to focal adhesions, paxillin is found in cytoplasmic organelles, including Golgi (24). Paxillin association with poly(A) binding protein at the dense endoplasmic reticulum and in the edge of migrating cells (47) further supports its diverse localizations and functions. Paxillin also associates with numerous signaling molecules responsible for extensive posttranslational modifications of paxillin. These include adaptor molecules (p130Cas and CRK), kinases (FAK, Pyk2, PAK, and SRC), tyrosine phosphatases (PTP-PEST), and ARF-GAP proteins (16, 29, 45). Phosphorylation of paxillin is required for its ability to function as an adaptor protein implicated in cell spreading and cell migration through its effects on cellular cytoskeleton organization (28, 30, 49).
Disruption of the paxillin gene in mice results in early embryonic lethality (embryonic day E7 to E8), providing direct support for its critical role in regulating the development of mesodermally derived structures such as heart and somites (8). Cultured paxillin-null fibroblasts display abnormal focal adhesions, reduced cell migration, inefficient localization of focal adhesion kinase (FAK), and reduced fibronectin-induced phosphorylation of FAK, Cas, and mitogen-activated protein kinase (8). Similar changes in cell motility were observed in paxillin-null embryonic stem cells (45).
Paxillin is most abundant in smooth muscle tissue, where actin attachment to the extracellular matrix occurs via dense plaques (focal adhesion analogue) that enable efficient transduction during muscle contraction (33, 39, 40). Although changes in paxillin localization and expression have been reported to take place during development and mitosis and also in transformed cells, the mechanisms underlying such changes are not known. An increase in paxillin expression coincided with immortalization of mouse embryo fibroblasts (13). In contrast, a decrease in expression of paxillin was noted during mitosis of certain cell lines (48). Altered expression of paxillin was also observed during malignant conversion of mouse keratinocytes in breast cancer, as well as in small cell lung cancer samples (25, 32, 43). Changes in the pattern of paxillin expression could also be attributed to its localization, whose regulation is poorly understood.
The finding that RNF5 was a LIM-associated protein in C. elegans led us to test the hypothesis that RNF5 could cause ubiquitination of its human homologue, paxillin, based on the expectation that it would alter paxillin stability and/or localization. Here we demonstrate that RNF5 both associates with and ubiquitinates paxillin, resulting in altered paxillin localization and impaired cell motility.
MATERIALS AND METHODS
Cell culture and transfection.
Adenovirus-transformed human embryonic kidney (HEK) 293T cells, HeLa cells, and mouse fibroblast NIH 3T3 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% calf serum; paxillin-null and wild-type mouse embryo fibroblasts were maintained in DMEM supplemented with 10% fetal bovine serum. All cultures were supplemented with 100 U of penicillin and 100 U of streptomycin (Gibco-BRL) per ml and maintained in a 5% CO2 incubator at 37°C (8). Transfection was performed with the calcium phosphate precipitate technique for 293T cells and Lipofectamine Plus (Invitrogen) for HeLa and NIH 3T3 cells. The medium was changed 24 h after transfection, and cells were harvested 48 h later.
DNA constructs.
The full-length RNF5 cDNA and the two alternatively spliced forms (Δ17 and ΔRING) were PCR amplified from first-strand cDNA with C. elegans RNA as the template, a spliced leader SL2 primer (5′-GGTTTTAACCCAGTTACTCAAG-3′), and a gene-specific 3′ primer (5′-GACGAATAGAAGCCAGAAAAGCA-3′). PCR products were subcloned and sequenced. Flag-tagged full-length human RNF5 was constructed in the pEF-Flag vector. A RING mutant (C27A and C30A) was constructed with the aid of the QuikChange site-directed mutagenesis kit (Stratagene). RNF5 was deleted of its C-terminal domain via PCR-based cloning of the DNA fragment corresponding to amino acids 1 to 164 of RNF5 into pEF-Flag. The full-length paxillin-α cDNA was amplified by PCR and cloned into pcDNA3, pEF-hemagglutinin (HA), or pEF-HA-green fluorescent protein (GFP) vector. pGEX-2T-paxillin-α and pBabe-puro-Myc-paxillin-α were described previously (23, 47), and Myc-paxillin was subcloned into the pEF construct. LIM deletion mutants ΔLIM4 and ΔLIM1-4 and a fragment carrying the four LIM domains were generated by PCR and cloned into pEF-HA. The integrity of all constructs was verified by sequencing. The LIM domain protein Y105E8a.6 was identified with RNF5 as the bait in a yeast two-hybrid screen of a mixed-stage C. elegans cDNA library (kindly provided by Mark Vidal). RNF5 was cloned in plasmid pDBLeu (Life Technologies), and 107 yeast transformants were screened with Saccharomyces cerevisiae MaV203.
Antibodies and immunoprecipitations.
Cells were lysed (20 mM HEPES [pH 7.5], 150 mM NaCl, 0.5% NP-40, 1 mM EDTA, 1 mM dithiothreitol, and 10% glycerol containing 1 mM phenylmethylsulfonyl fluoride and protease inhibitors) and incubated on ice for 30 min. Total cell lysates were clarified by centrifugation at 15,000 rpm for 30 min. Supernatants were incubated overnight at 4°C with paxillin (PharMingen) or Flag (Sigma) antibodies. Antibodies against paxillin were purchased (Pharmingen) and used at a 1:1,000 dilution for Western blots and 1:400 dilution for immunohistochemistry. Antibodies to vinculin (Sigma), HA tag (UBI), and Flag tag (Sigma) were used according to the manufacturers' recommendations. Immunoprecipitation was performed by incubation for 90 min at 4°C with protein G-Sepharose (Sigma). After washing three times with lysis buffer, proteins were solubilized in 3 × Laemmli buffer and separated on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). For coimmunoprecipitation of endogenous RNF5 and paxillin, proteins were prepared with radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% NP-40, 0.5% deoxycholic acid, 0.1% SDS, 1 mM dithiothreitol, and 2 mM EDTA supplemented with 0.2 mM phenylmethylsulfonyl fluoride and a cocktail of protease inhibitors). Monoclonal antibodies against RNF5 were raised against the bacterial fusion protein glutathione S-transferase (GST)-RNF5. Positive clones were selected based on their inability to recognize GST and selective recognition of the RNF5 in immunoblots (dilution of 1:1000) and immunohistochemistry (dilution of 1:50) analysis.
In vitro binding assay.
Bacterially expressed and purified GST, GST-CeRNF5, GST-Δ17ΔRING, GST-hRNF5, GST-RINGm and GST-paxillin bound to glutathione beads were first incubated with 3 mg of bovine serum albumin in phosphate-buffered saline for 2 h, followed by incubation at 4°C for 2 h with 35S-labeled RNF5 or paxillin which was in vitro translated with the TNT coupled reticulocyte lysate system (Promega). Bead-bound material was subjected to three washes with 20 mM Tris [pH 7.5]-150 mM NaCl-1 mM EDTA-1 mM EGTA-0.5% NP-40-1 mM NaVO4-1 mM dithiothreitol supplemented with protease inhibitors before being subjected to separation by SDS-PAGE.
In vivo ubiquitination.
Cells were transfected with Flag-tagged RNF5 vector and HA-tagged ubiquitin to analyze ubiquitination as described previously (5, 6). Harvested cells were lysed by incubation with 1 volume of 2% SDS in TBS (10 mM Tris-HCl [pH 8.0]) at 95°C for 10 min. Nine volumes of 1% Triton X-100 and 2 mM EDTA in TBS were added, and lysates were incubated on ice for 30 min, followed by sonication (2 min). The solution was incubated for 30 min at 4°C with protein G beads (Invitrogen) and clarified by 30 min of centrifugation (14,000 rpm) at 4°C. The protein concentration was determined by the Bradford assay. For immunoprecipitation, 1 mg of protein was incubated with anti-Flag antibody at 4°C overnight before protein G beads were added for 2 h. The beads were washed twice with NaCl (1 M) in TBS supplemented with NP-40 (1%), β-mercaptoethanol (0.05%), and EDTA (1 mM). Proteins were loaded onto SDS-8 to 15% PAGE, followed by immunoblot analysis with the indicated antibodies and enhanced chemiluminescence (ECL) detection (Amersham).
In vitro ubiquitination.
Bacterially expressed and glutathione bead-purified GST-RNF5 (1 μg/10 μl) and GST-RINGm bound to glutathione beads were washed once with ubiquitination buffer (50 mM Tris-HCl [pH 8.0], 5 mM MgCl2, 0.5 mM dithiothreitol, 2 mM NaF, and 3 μM okadaic acid), GST-RNF5 or GST-RINGm beads were incubated for 30 min at 37°C with whole-cell extracts prepared from HeLa cells immunodepleted with RNF5 monoclonal antibodies, followed by extensive washes with phosphate-buffered saline containing NP-40 (0.25%), β-mercaptoethanol (0.05%), and EDTA (1 mM). The ubiquitination reaction was carried out in ubiquitination buffer supplemented with HA-ubiquitin (0.5 μg), 2 mM ATP, E1 (15 ng; Affinity Research, London, United Kingdom), and the purified E2 ubiquitin-conjugating enzyme UbcH5c for 60 min at 37°C. GST-RNF5 and GST-RINGm beads were then washed extensively with phosphate-buffered saline containing 0.5 M LiCl, 0.1% β-mercaptoethanol, 0.25% NP-40, and 1 mM EDTA before being eluted in Laemmli buffer, and separated by SDS-10% PAGE. [35S]paxillin was subjected to in vitro ubiquitination in the presence of GST-RNF5 or GST-mutant for 60 min at 37°C, and radiolabeled paxillin was separated by SDS-PAGE. The degree of [35S]paxillin ubiquitination was detected by autoradiography.
Protein stability assays.
To monitor changes in the stability of paxillin, pulse-chase analysis was performed with in vitro-translated 35S-labeled paxillin. Following its translation, paxillin was immunoprecipitated with antibodies against paxillin and incubated with RNF5 and ubiquitin components, as indicated in Results. Incubation of GST-RNF5 with cell extracts was performed as indicated above. Subsequently, RNF5 was washed extensively prior to the addition of paxillin and ubiquitin reaction components. The proteasome inhibitor MG132 was purchased from Peptide International.
Confocal microscopy.
Cells grown on 22-mm2 coverslips were fixed in freshly prepared 3% para-formaldehyde in phosphate-buffered saline ([pH 7.4) for 1 0 min at room temperature. The cells were then washed three times (5 min each) in phosphate-buffered saline, followed by permeabilization in 0.1% Triton X-100 in phosphate-buffered saline (pH 7.4) for 1 min and an additional three 5-min washes in phosphate-buffered saline. Cells were then incubated with 50 mM NH4Cl for 5 min following the washes and further incubation with phosphate-buffered saline supplemented with 5% bovine serum albumin for 30 min. The cells were incubated on 75-μl drops of antibodies (diluted in phosphate-buffered saline [pH 7.4] containing 0.2% bovine serum albumin as indicated in Results) for 1 h at room temperature in a humidity chamber. The cells were washed three times in phosphate-buffered saline (5 min each) before incubation with 75-μl drops of fluorescein-conjugated anti-rabbit immunoglobulin G (Molecular Probes) diluted (2 μg/ml) in phosphate-buffered saline (pH 7.4) containing 0.2% bovine serum albumin for 30 min at room temperature in a humidity chamber in the dark. The cells were rinsed three times in phosphate-buffered saline, followed by a rinse in 0.1% Triton X-100 in phosphate-buffered saline (pH 7.4) and then three additional rinses in phosphate-buffered saline. The coverslips were mounted on glass slides in Vectashield (Vector Laboratories).
Cell migration.
Cells were grown to confluence and then scratched with a pipette tip (8). Three wounds were made for each sample, and all were photographed at the zero time point and at subsequent time points. Assays were repeated at least three times.
RESULTS
RNF5 associates with paxillin in vivo.
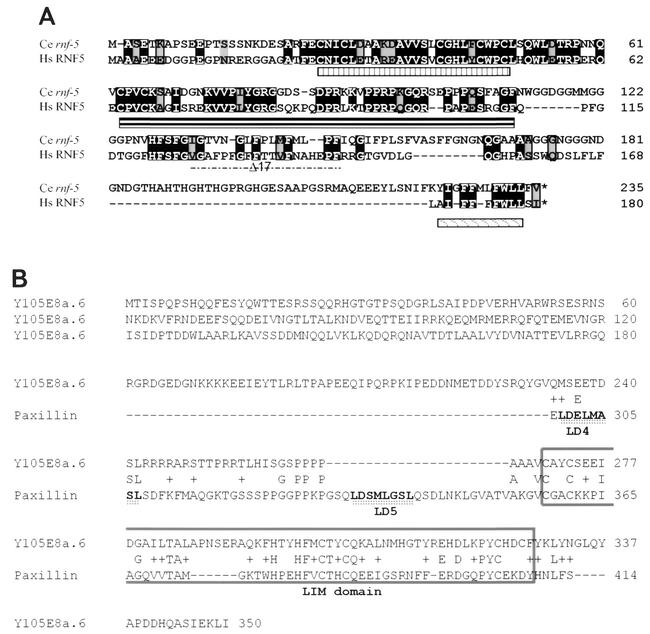
The human and C. elegans forms of RNF5 reveal conservation of the RING (implicated in E3 ligase activity), central (possible substrate recognition), and carboxyl-terminal (implicated in membrane binding) (22) domains (Fig. 1A). The search for RNF5-interacting proteins via the yeast two-hybrid system identified several membrane-associated and cytoskeleton-related proteins, including the putative C. elegans protein Y105E8A.V. The Y105E8A.V protein consists of a LIM domain and appears to localize primarily to body wall and vulval muscles in C. elegans. The expression of Y105E8A.V in muscles is restricted to dense bodies that represent focal adhesion-like attachment sites in C. elegans (Broday et al., unpublished data).
FIG. 1.
Alignment of C. elegans and human RNF5. (A) The amino acid sequences of human (Hs) and C. elegans (Ce) forms of RNF5 were aligned. Important domains in RNF5 are underlined (RING, central, and carboxyl-terminal domains). (B) Alignment of the C. elegans LIM-containing protein Y105E8a.6 and human paxillin. Homology between the two proteins was found in the LIM domain (highest in LIM1 and LIM4 of paxillin; boxed area) and also in the region between the LD4 and LD5 domains of paxillin.
The homology search for the human homologue of Y105E8A.V identified paxillin (Fig. 1B), a 68-kDa protein implicated in cytoskeleton organization, cell spreading, and motility that is localized primarily within focal adhesions (40, 41, 42), as seen with the C. elegans protein Y105E8A.V. To confirm that paxillin associates with C. elegans RNF5, we first performed a series of immunoprecipitations with C. elegans RNF5 and endogenously expressed paxillin. Transfection of Flag-tagged C. elegans RNF5 into 293T cells was followed by immunoprecipitation of endogenous paxillin and identified C. elegans RNF5 as a paxillin-associated protein (Fig. 2A).
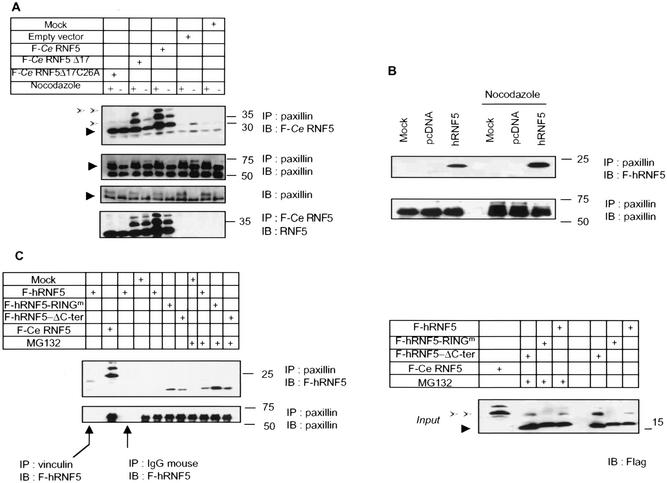
FIG. 2.
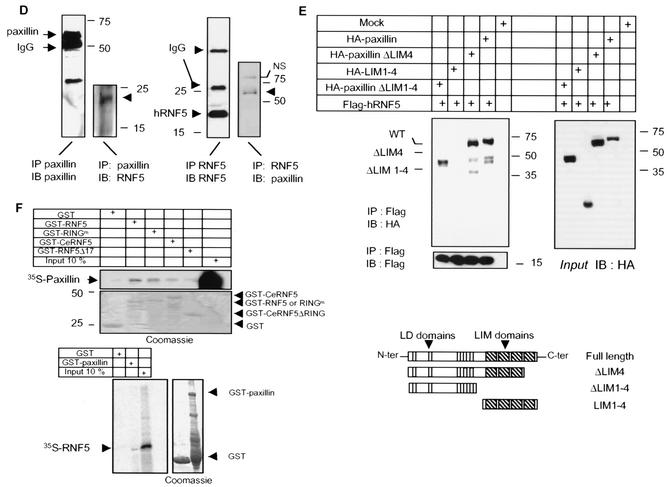
RNF5 association with paxillin. (A) C. elegans RNF5 associates with paxillin. Flag-tagged C. elegans RNF5 (F-Ce RNF5), a spliced variant lacking 17 amino acids, and the RING mutant C26A forms were transfected into 293T cells, and 24 h later the cells were subjected to treatment with nocodazole (100 ng/ml) or mock treatment, as indicated in the figure. Proteins were prepared 18 h later and subjected to immunoprecipitation (IP) with antibodies to endogenous paxillin. Immunoprecipitated material was analyzed via Western blots (IB) with the antibodies indicated in the figure. Arrowheads point to the corresponding protein. Empty arrows point to mono- and diubiquitin forms of RNF5. (B) Human RNF5 (hRNF5) binds to paxillin. Flag-tagged human RNF5 (F-hRNF5) was transfected into 293T cells that were mock or nocodazole treated. Proteins (1 mg) were subjected to immunoprecipitation with antibodies to paxillin, following Western blot analysis with the antibodies indicated in the figure. (C) Mutant forms of RNF5 exhibit stronger binding to paxillin than wild-type RNF5. Flag-tagged forms of RNF5 (human wild-type, RING mutant, and C-terminally deleted [ΔC-ter] forms and C. elegans RNF5) were transfected into 293T cells; 24 h later, proteins were prepared and subjected to immunoprecipitation with antibodies to paxillin, followed by immunoblot analysis with the antibodies indicated in the figure. The relative intensity of the ECL reaction was reduced compared with that in panel B to emphasize the difference in association of paxillin with the different forms of RNF5. Control reactions with vinculin antibodies and control IgG for immunoprecipitation followed by immunoblotting with Flag-RNF5 are shown. Expression of RNF5 forms (input) is shown on the right panel. (D) In vivo association of RNF5 and paxillin. Endogenous human RNF5 was immunoprecipitated with monoclonal antibodies raised against RNF5, followed by immunoblot analysis with the antibodies indicated in the figure. NS, nonspecific. (E) RNF5 associates with paxillin via the amino-terminal domain. Expression vectors containing the LIM or amino-terminal domain were coexpressed with RNF5 in 293T cells. Immunoprecipitations with the indicated antibodies identified an association with paxillin lacking the LIM domain (ΔLIM1-4) and containing only the amino-terminal domain. (F) In vitro association between paxillin and RNF5. GST-RNF5 (human wild-type and RING mutant forms and C. elegans wild-type and Δ17ΔRING forms) fusion proteins were incubated with 35S-labeled paxillin, and reciprocally, GST-paxillin was incubated with 35S-labeled RNF5. The amount of bound material shown reflects direct association that was followed by extensive high-salt washes.
An earlier report demonstrating a substantial decrease in paxillin levels during cell mitosis (48) led us to assess possible changes in the association between C. elegans RNF5 and paxillin in cells subjected to treatment with nocodazole, which efficiently drives cells to mitosis. Under normal growth conditions, immunoprecipitation of paxillin identified the native form of C. elegans RNF5 (26 kDa) as its associated protein. However, in mitotic cells, paxillin was also found in complex with modified forms (34 kDa and 40 kDa) of C. elegans RNF5. Similar to its association with wild-type C. elegans RNF5, paxillin also bound to a spliced variant of RNF5 lacking 17 amino acids (Δ17; Fig. 2A). Because RNF5 is a RING finger protein with putative E3 ligase activity, we also tested whether its association with paxillin requires an intact RING domain. Expression of C26A, a RING-mutated form of C. elegans RNF5 that is expected to lack E3 ligase activity, maintained its association with paxillin, suggesting that E3 ligase per se is not required for RNF5's association with paxillin (Fig. 2A).
In contrast to the three forms of wild-type and Δ17 C. elegans RNF5 that were found in complex with paxillin in nocodazole-treated cells, only the native form of RING mutant C. elegans RNF5 was found in complex with paxillin in untreated cells (Fig. 2A), further implicating RNF5 E3 ligase activity in the modified forms found to be associated with paxillin. These findings suggest that intact E3 ligase activity of C. elegans RNF5 is required for generating multiple C. elegans RNF5 forms, which may present mono- and diubiquitinated C. elegans RNF5 forms associated with paxillin. These findings also indicate that paxillin associates with modified forms of RNF5, as seen in cells that are in mitosis.
The human homologue of RNF5 is an 18-kDa protein that shows homology with the C. elegans RNF5 within the central regulatory domains (Fig. 1). Forced expression of the Flag-tagged human RNF5 homologue in 293T cells confirmed its association with endogenously expressed paxillin (Fig. 2B). C. elegans RNF5 exhibited greater association with paxillin than did human RNF5, which could be attributed to its different sequence (36% sequence identity to the human protein) or conformation or altered subcellular distribution (Fig. 2C). Larger amounts of RING mutant RNF5 were found in association with paxillin compared with wild-type RNF5 (Fig. 2C), suggesting that the E3 ligase activity of RNF5 may limit the amount of RNF5 associated with paxillin. Along these lines, the amount of RNF5 bound to paxillin increased in the presence of a proteasome inhibitor (Fig. 2C).
Importantly, we also observed the association of paxillin with endogenous RNF5. Using monoclonal antibodies to RNF5, we identified RNF5 as a paxillin-bound protein with either RNF5 or paxillin antibodies for the initial immunoprecipitation (Fig. 2D). Overall, a small fraction of paxillin was subject to association with RNF5, suggesting that modification of paxillin is required for this association. These data establish that both the human and C. elegans forms of RNF5 associate with paxillin in vivo. Also, interestingly, the C-terminal tail of RNF5 has been implicated in membrane association (22). Since RNF5 which lacks this domain exhibits greater association with paxillin than does the wild-type form (Fig. 2C), we conclude that localization of RNF5 to the plasma membrane may not be required for its association with paxillin. However, C-terminus-dependent localization is required for RNF5's effect on paxillin (see below).
Next we identified the site of RNF5 association on paxillin. The LIM domain is highly conserved between the C. elegans and human RNF5-associated proteins and consequently, we first assessed possible associations to LIM domains. Expression of paxillin in which either the fourth or all four LIM domains have been deleted retained the association with RNF5 (Fig. 2E). Similarly, expression of paxillin LIM domains (i.e., the C-terminal half of paxillin) was not sufficient for RNF5 association. To determine if the amino-terminal domain serves as such a docking site, we expressed the amino-terminal domain of paxillin, consisting of all five LD domains but no LIM domains (HA-paxillin ΔLIM1-4). Immunoprecipitation of RNF5 with Flag antibodies identified paxillin that lacks all four LIM domains. These results suggest that the amino-terminal domain is required for RNF5 association with paxillin (Fig. 2E). Although the C. elegans LIM-containing protein and human paxillin also show homology outside the LIM domain (Fig. 1B), we cannot exclude possible association based on those sequences or common secondary structures.
Incubation of 35S-labeled, in vitro-translated paxillin with bacterially expressed GST-RNF5 of human and C. elegans origin revealed a weak association between the two proteins (Fig. 2F). Reciprocal analysis confirmed the association between 35S-labeled, in vitro-translated RNF5 and GST-paxillin, albeit at a low affinity (Fig. 2F). The weak in vitro association, as opposed to the noticeable in vivo binding, suggests that the RNF5-paxillin interaction may be dependent on posttranslational modifications and/or the presence of an adaptor protein that is present in vivo.
RNF5 elicits ubiquitination of paxillin in vitro.
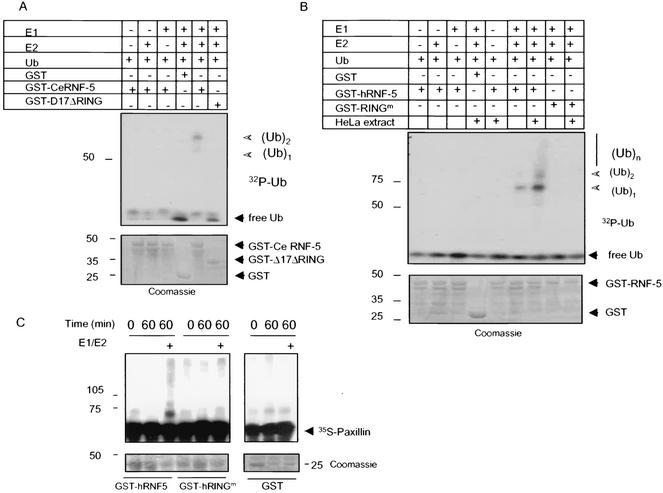
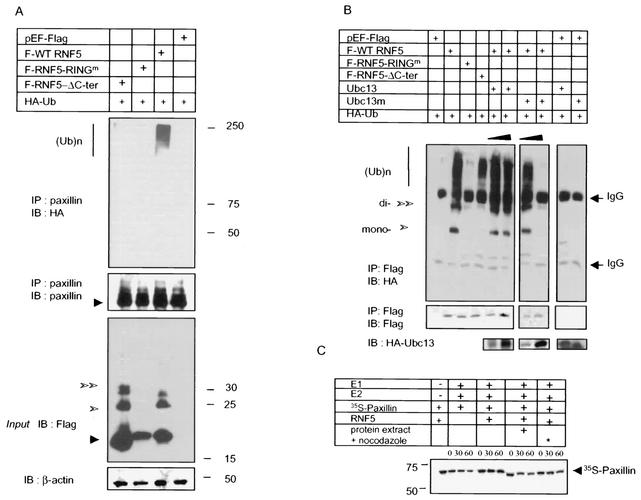
Not all RING finger proteins exhibit intrinsic E3 ligase activity. Therefore, we explored whether RNF5 elicits E3 ligase activity first with a bacterially expressed and purified form of C. elegans wild-type GST-RNF5. In vitro ubiquitination assays carried out in the presence of E1, E2, and [32P]ubiquitin revealed that the wild-type but not the RING mutant form of RNF5 exhibited limited E3 ligase activity, as reflected by the formation of mono- and diubiquitinated forms of RNF5 (Fig. 3A). When the same reaction was carried out with GST-RNF5 that had been incubated with cellular extracts (which were washed prior to the actual ubiquitination reaction), RNF5 was capable of generating polyubiquitin chains (Fig. 3B). These observations suggest that additional changes or components (i.e., posttranslational modification, association with partner E3 ligase, or a different set of E2's) elicit RNF5 E3 ligase activity in vitro. These results establish both that RNF5 exhibits E3 ligase activity and that the intact RING domain is required for this activity.
FIG. 3.
RNF5 E3 ligase activities and ubiquitination of paxillin in vitro. (A) E3 ligase activities of C. elegans RNF5 in vitro. In vitro ubiquitination assays were carried out with bacterially expressed and purified GST fused to full-length or Δ17C26(A) (17 amino acids within the RING domain mutated) C. elegans RNF5. The purified forms of E1, E2 (UbcH5c), and bacterially produced 32P-labeled ubiquitin (Ub) were added in the presence of ATP. GST-RNF5 bound to glutathione beads was washed before separation on SDS-PAGE and analysis via autoradiography. The positions of mono- and diubiquitin are indicated. The lower panel depicts Coomassie blue staining, reflecting the quantity of proteins used for the reaction. (B) E3 ligase activities of human RNF5 in vitro. The in vitro ubiquitination reaction was carried out as indicated above except that the human forms of wild-type and RING mutant RNF5 were used. As indicated in the figure, GST-hRNF5 was also incubated with HeLa cell extracts that were immunodepleted of RNF5 following extensive washes before adding E1, E2, 32P-ubiquitin, and ubiquitination buffer. Arrows point to the positions of the mono-, di-, and polyubiquitin forms. The lower panel depicts Coomassie blue staining, reflecting the quantity of proteins used for the reaction. (C) RNF5 mediates ubiquitination of paxillin in vitro. In vitro ubiquitination assays were performed with bacterially expressed and purified forms of GST-hRNF5 (wild type or RING mutant) and 35S-labeled in vitro-translated paxillin in the presence of E1 and E2 for the period indicated. Following the reaction, GST-hRNF5 was spun, and supernatant-containing [35S]paxillin was subjected to separation on SDS-PAGE. Shown is the autoradiograph of ubiquitinated paxillin. The lower panel depicts input of GST-hRNF5 (left panel) or GST (right panel).
RING finger proteins that possess E3 ligase activity target their associated proteins for ubiquitination (1, 21). To determine whether RNF5 targets the ubiquitination of paxillin in vitro, we used 35S-labeled, in vitro-translated paxillin as a substrate. The addition of a bacterially expressed GST-RNF5 in the presence of E1, E2, and ubiquitin resulted in the ubiquitination of paxillin, whereas the RING mutant form of RNF5 elicited paxillin ubiquitination similar to that seen with control GST (Fig. 3C). These data provide direct evidence for RNF5 E3 ligase activity and for its ability to induce the ubiquitination of paxillin in vitro.
RNF5 elicits ubiquitination of paxillin in vivo.
To assess whether RNF5 targets paxillin ubiquitination in vivo, HA-tagged ubiquitin was cotransfected with wild-type, RING mutant, or a C-terminally truncated form of RNF5. Of the three forms, only wild-type RNF5 caused extensive polyubiquitination of paxillin (Fig. 4A). Upon exposure to MG132, neither the ubiquitination nor the steady-state level of paxillin changed (data not shown), suggesting that paxillin is not destined for proteasomal degradation and that the intact RING of RNF5 is required for in vivo ubiquitination of paxillin. Forced expression of the C-terminally truncated form of RNF5 did not induce ubiquitination of paxillin, identifying the C-terminal tail of RNF5 as a requirement for this activity towards paxillin (Fig. 4A). The C-terminal tail of RNF5 consists of a membrane-anchoring motif, and consequently, the failure of C-terminally truncated RNF5 to cause ubiquitination of paxillin suggests that such ubiquitination may require anchoring of RNF5 to a cellular membrane.
FIG. 4.
RNF5 ubiquitinates paxillin in vivo without affecting its half-life. (A) RNF5 promotes paxillin ubiquitination in vivo. Wild-type (WT) and mutant forms of RNF5 (RING mutant and C-terminally deleted) were cotransfected with HA-tagged ubiquitin into 293T cells. Proteins were prepared 24 h after transfection and subjected to immunoprecipitation, followed by immunoblot analysis with the antibodies indicated in the figure. Sizes and polyubiquitin chains are marked on the side panels. Western blot analysis of RNF5 input is presented on the bottom panel. β-Actin blot was used to assure equal loading. (B) RNF5 E3 ligase in vivo is Ubc13 dependent. Wild-type and mutant forms of human RNF5 were cotransfected with HA-ubiquitin and wild-type or mutant forms of Ubc13 into 293T cells. Proteins prepared under denaturing conditions were subjected to immunoprecipitation with antibodies to Flag-tagged RNF5, followed by immunoblot with the antibodies indicated in the figure. The lower panel depicts expression of Ubc13. (C) RNF5 does not affect paxillin half-life in vitro. The half-life of paxillin was monitored in vitro with in vitro-translated 35S-labeled paxillin. Following its translation, paxillin was immunoprecipitated and incubated with E1, E2, and RNF5 as indicated in the figure. RNF5 was also incubated with protein extracts of mock-treated (+) and nocodazole-treated (∗) cells to enable posttranslational modifications that increase the efficiency of association with and ubiquitination of paxillin.
Analysis of RNF5 expression revealed that both the wild-type and C-terminally deleted forms but not the RING mutant form expressed a native as well as mono- and diubiquitinated forms (Fig. 4A). These observations suggest that, at least in part, RNF5's own ligase activity is required for the regulation of its own ubiquitination. Together, these observations suggest that while the ubiquitination of RNF5 depends on an intact RING domain independent of its C terminus, the ubiquitination of paxillin requires that both domains be intact.
To assess in vivo ubiquitination of RNF5, we monitored changes in the ubiquitination of wild-type and mutant forms of RNF5. The wild-type form of RNF5 exhibited an extensive degree of ubiquitination, although the C-terminally deleted form of RNF5 was also ubiquitinated, albeit to a lesser degree (Fig. 4B). Of interest, the polyubiquitination of RNF5 seen following its immunoprecipitation but not in direct Western blots suggests that a relatively small portion of total RNF5 is subjected to polyubiquitination (compare Fig. 4A and 4B). Based on the observation that RNF5 does not affect steady-state levels of paxillin despite polyubiquitination by RNF5, we assessed the possible formation of ubiquitin polychains that may have different topologies.
Ubc13 has been implicated in the formation of polyubiquitin chains of K63 topology that are not destined for proteasomal degradation (10). Accordingly, we considered the possible role of Ubc13 in RNF5's own ubiquitination. Forced expression of wild-type Ubc13 somewhat increased an already high level of RNF5 ubiquitination, while the mutant form of Ubc13 that was shown to serve as a potent dominant negative (11) decreased RNF5 ubiquitination, even to the complete elimination of it (Fig. 4B). These data suggest that RNF5 ubiquitination depends primarily on Ubc13, which is expected to form K63 polyubiquitin chains. Along these lines, expression of mutant Ubc13 also inhibited RNF5-mediated paxillin ubiquitination (data not shown), which is consistent with the mode of RNF5 polyubiquitination. Additional studies are required to establish the composition of polyubiquitin chains assembled on RNF5.
RNF5 does not affect paxillin half-life.
After establishing the RNF5-mediated ubiquitination of paxillin, we next assessed the effect of ubiquitination on paxillin stability. Cycloheximide chase analysis was carried out in HeLa cells cotransfected with HA-tagged paxillin and the wild-type and RING mutant forms of RNF5. The half-life of exogenous paxillin, estimated to be around 6 h, was not altered after expression of either RNF5 form in HeLa or NIH 3T3 cells (data not shown). These observations were further supported by an in vitro analysis of paxillin half-life. Incubation of 35S-labeled paxillin with RNF5 in the presence of cellular extracts (of normally growing or mitotic cells) did not alter the half-life of paxillin (Fig. 4C). These findings are consistent with the idea that RNF5's own and targeted ubiquitination is likely to be composed of K63 topology (Fig. 4B), which is unlikely to destine proteins for proteasomal degradation. Although these findings strongly suggest that ubiquitination of paxillin by RNF5 does not alter paxillin stability, we cannot exclude the remote possibility that a small subfraction of paxillin, masked by the high expression levels of paxillin in the cell, is affected by RNF5 at the level of stability.
RNF5 affects subcellular localization of paxillin.
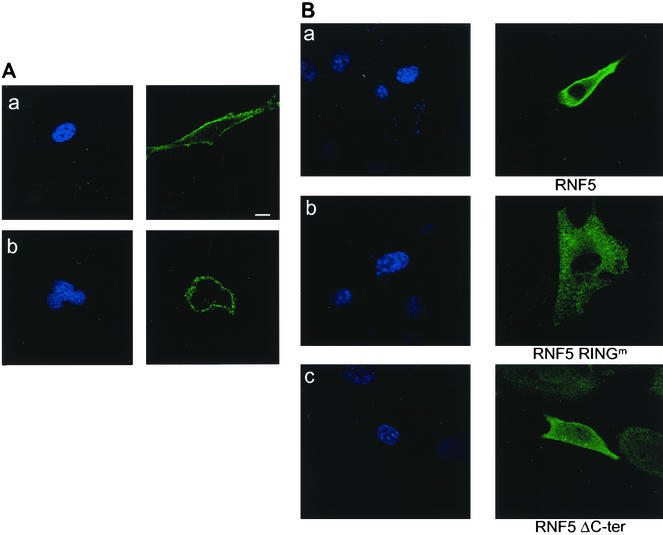
Using immunohistochemistry, we determined the cellular distribution of endogenous and Flag-tagged RNF5. Monoclonal antibodies that were raised against the bacterially expressed RNF5 detected a single moiety protein by Western blotting (Fig. 2D) and identified RNF5 staining along the cell membrane, with greater localization occurring within the membrane of both mouse- and human-derived cell lines (Fig. 5A, panels a and b, respectively). Forced expression of Flag-tagged RNF5 revealed that the wild-type form of RNF5 is found primarily in the plasma membrane, with some localization occurring within cytoplasmic organelles. This could be due to the higher amounts of the exogenously expressed RNF5 (Fig. 5B, panel a), yet resembles the analysis made with the monoclonal antibodies against the endogenous protein. The RING mutant form of RNF5 was found primarily within the cytoplasm and cytoplasmic organelles (Fig. 5B, panel b). RNF5 that lacked the C-terminal tail was no longer found within the plasma membrane, but was localized within cytoplasmic organelles and possibly in nuclei (Fig. 5A, panel c). These findings suggest that only RNF5 with its C-terminal tail is localized to plasma membrane and cytosolic organelles. The E3 ligase activity of RNF5 also appears to play a role in its subcellular distribution. Similarly, differences in RNF5 localization could also explain differences in binding and E3 ligase activity.
FIG. 5.
Cellular localization of endogenous and exogenously expressed RNF5. (A) Cellular localization of endogenous RNF5. NIH 3T3 (a) or HeLa (b) cells were subjected to immunocytochemistry-based confocal microscopic analysis with monoclonal antibodies raised against RNF5. 4′,6′-Diamidino-2-phenylindole (DAPI) staining is shown in the left panels. Scale bar. 10 μm. (B) Cellular localization of exogenously expressed RNF5. NIH 3T3 cells were transfected with Flag-tagged wild-type (a), RING mutant (b), and C-terminally deleted (c) forms of RNF5; 30 h later the cells were subjected to confocal microscopy-based immunocytochemistry analysis with antibodies to Flag. The left panel depicts DAPI staining. The picture represents multiple fields from independent experiments.
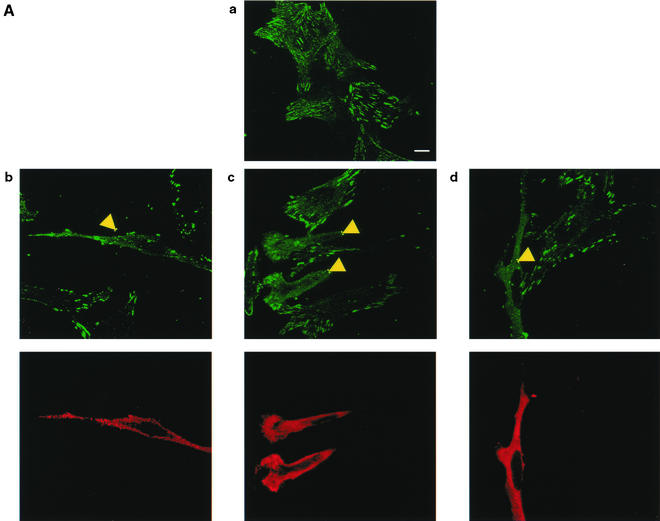
As RNF5 did not affect paxillin stability, we monitored possible changes in the localization of endogenous paxillin upon RNF5 expression. Immunostaining of cells with antibodies to endogenous paxillin revealed primary localization of paxillin within focal adhesions, with some cytoplasmic staining (Fig. 6A, panel a), as reported previously (24). Expression of RNF5 caused a noticeable decrease in the localization of paxillin within focal adhesions while increasing the localization of paxillin in the cytoplasm (Fig. 6A, panels b to d). These differences were clearly observed in the comparison of neighboring cells that expressed RNF5 and those that did not (Fig. 6A, panels b to d). These findings demonstrate that RNF5 expression alters paxillin subcellular localization and is reflected by impaired focal adhesion and increased cytoplasmic staining.
FIG. 6.
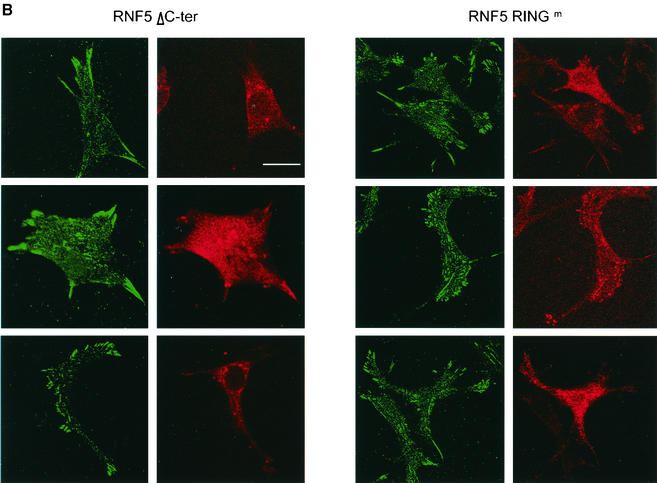
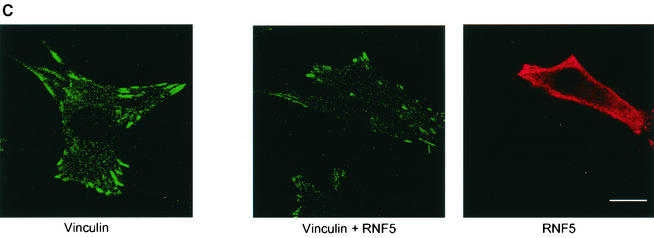
RNF5 expression affects cellular distribution of paxillin. (A) RNF5 alters paxillin localization. Localization of endogenous paxillin was monitored in RNF5 transfected cells. Panel a depicts staining of endogenous paxillin. Note the difference in paxillin localization in cells that express RNF5 (panels b to d) with those that do not (compare to the corresponding cells that express RNF5 [yellow arrows]). The figure represents multiple fields and data observed in five independent experiments. Yellow arrows point to cells which expressed RNF5 and exhibited altered localization of paxillin. Scale bar, 10 μm. (B) Mutant forms of RNF5 do not affect paxillin localization. NIH 3T3 cells were transfected with mutant Flag-RNF5 forms (RING mutant or C-terminally deleted forms), as indicated in the figure. Antibodies to Flag (RNF5) were recognized by red fluorescence, whereas antibodies to paxillin were recognized by green fluorescence. (C) RNF5 and vinculin localization. Analysis of vinculin was carried out on cells that were transfected or not with RNF5. Red staining depicts RNF5, whereas green reveals vinculin expression.
In contrast to the high efficiency of wild-type RNF5, expression of RING mutant and C-terminally truncated forms of RNF5 was less efficient at abrogating paxillin localization within the focal adhesions (Fig. 6B). These findings are in line with the observation that RNF5 which lacks ithe C-terminal domain is not capable of mediating paxillin ubiquitination, although it is capable of associating with paxillin. RNF5 was less efficient in abrogating the localization of vinculin within focal adhesions (Fig. 6C). Collectively, these findings point to a role for membrane anchoring and intact RING domains in RNF5's ability to effect paxillin ubiquitination and efficiently alter its cellular localization.
RNF5 inhibits cell motility.
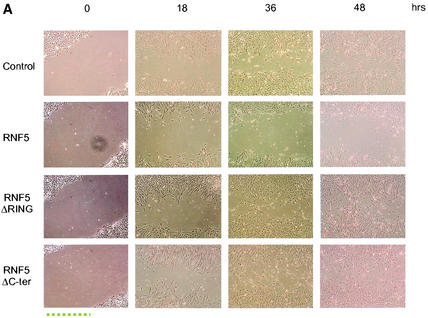
As the presence of paxillin within focal adhesions is required for cell motility and RNF5 is able to abolish paxillin localization within the focal adhesions, it is possible that RNF5 may be affecting cell motility. Accordingly, we performed the scratch-wound assay, monitoring the ability of cells to fill the gap formed upon the wound. Time lapse analysis was performed on NIH 3T3 cells that had been transfected with wild-type or mutant forms of RNF5. Analysis of cell motility revealed inhibition of gap filling of cells that expressed the wild-type form of RNF5. RNF5-expressing cells maintained a gap that was already filled by control cells or cells expressing mutant forms of RNF5 (Fig. 7A), indicating that RNF5 expression reduces cell motility.
FIG. 7.
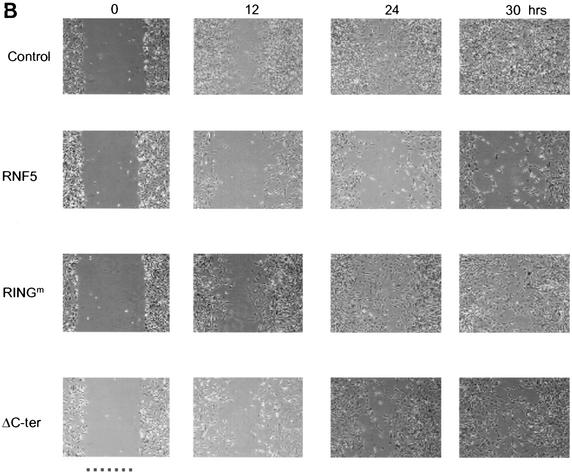
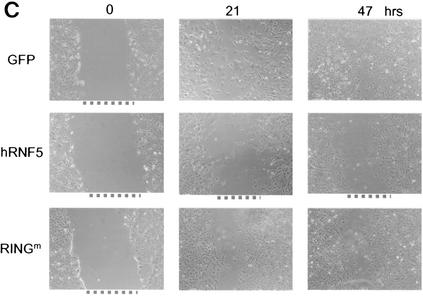
RNF5 inhibits migration of NIH 3T3 cells. (A) Cells were transfected with the RNF5 constructs indicated in the figure, and 48 h later cells were subjected to the wound assay. With a pipette tip, scratches were made in the centers of equally confluent cells. Time lapse photographs were made at the indicated time points, revealing the cells' ability to fill the wound (gap). Pictures shown represent multiple analyses. (B) The experiment was carried out as indicated for panel A with the exception that the plates were first coated with fibronectin (10 μg/ml). (C). Effect of RNF5 on cell motility in paxillin-null cells. Paxillin-null cells were subjected to transfection with the constructs indicated in the figure, and time lapse analysis was carried out to monitor changes in the migration of the cells as indicated above.
The role of paxillin in cell motility is often associated with cell contact with fibronectin. Using fibronectin-coated plates, we further evaluated the role of RNF5 in cell motility. The wild-type form of RNF5 was able to inhibit cell motility on the fibronectin-coated surface to a greater extent than its RING mutant and C-terminally deleted forms (Fig. 7B). These results further establish the role of RNF5 in cell motility.
The availability of paxillin-null cells prompted us to determine the possible effects of RNF5 on cell motility in the absence of paxillin, and thereby, we assessed the relative effect of paxillin on altered cell motility via RNF5. Although paxillin-null cells exhibit impaired cell motility (8, 32), forced expression of RNF5 in paxillin-null cells caused some additional decrease in cell motility. This was not evident after expression of the RING mutant form of RNF5 (Fig. 7C). These observations suggest that RNF5's effect on cell migration may also be directed against other cytoskeleton-related proteins that contain motifs similar to those found within the amino-terminal domain of paxillin.
DISCUSSION
The study presented here characterizes the RING finger protein RNF5. We demonstrate that RNF5 exhibits limited E3 ligase activities on its own in vitro and that its ability to elicit E3 ligase activities increases substantially after incubation with cellular extracts or expression in vivo. Based on these observations, we propose that RNF5 is an E3 ligase whose activities depend tightly on posttranslational modifications and possibly on its cooperation with other cellular proteins. Indeed, preliminary studies revealed the role of RNF5 phosphorylation as key in its E3 ligase activities (our unpublished observations). As RNF5 ubiquitination appears to depend on Ubc13, it is likely that the polyubiquitin chains formed on RNF5 are based on the K63 topology. Along these lines, the limited RNF5 E3 ligase activity evident in our in vitro reactions could also be attributed to the lack of the proper E2 and could provide an explanation for the ability of cell extracts that contain Ubc13 to also support the formation of a polyubiquitin chain in vitro.
Our study also identifies paxillin as a target for RNF5 E3 ligase activity; accordingly, RNF5 is the first E3 ligase shown to target paxillin ubiquitination. RNF5 and paxillin exhibit weak association with one another in vitro, yet the association is better seen in vivo, as revealed in a series of immunoprecipitations. These results suggest that posttranslational modifications of RNF5 and paxillin are important to their association. Our results cannot exclude the possibility that the interaction between RNF5 and paxillin is not mediated by an adaptor protein, perhaps similar to Csk in Skip1 targeting of p27 (7).
Mapping of RNF5 binding to the amino-terminal domain on paxillin suggests that modifications associated with this domain may be required for RNF5 function. For example, FAK and CRK play important roles in paxillin function through their phosphorylation within LD domains bearing phosphoacceptor sites. This activity has been implicated in paxillin functions in cytoskeleton organization and may be required for RNF5 association. Alternatively, RNF5 association may displace binding of other amino-terminal domain-associated proteins, thereby outcompeting the corresponding activity of paxillin.
Paxillin likely represents other cytoskeletal proteins that may be affected by RNF5. Preliminary experiments revealed an association between RNF5 and Hic5, a focal adhesion protein that shows extensive homology with paxillin (38) and has also been implicated in cell senescence (35). Therefore, it is plausible that RNF5 is one of the E3 ligases regulating the cellular distribution of proteins involved in cytoskeleton organization. Evidence for the effect of RNF5 on other possible cytoskeleton proteins also comes from paxillin-null cells, in which the expression of RNF5 resulted in inhibition of cell motility, albeit to a lower degree than in the presence of paxillin.
RNF5 does not appear to affect paxillin stability, but it does have a clear impact on its localization within focal adhesions. While causing paxillin to lose its specific localization within focal adhesions, RNF5 increases paxillin distribution throughout the cytoplasm. RNF5 requires its RING domain for altering paxillin localization, suggesting that paxillin ubiquitination by RNF5 is required for altering its cellular distribution. These observations are consistent with the finding that Ubc13 apparently plays an important role in RNF5's ubiquitination, as Ubc13-dependent polyubiquitin chains do not target degradation of the ubiquitinated substrate (11). These findings support RNF5 as an important regulator of paxillin localization and paxillin's ability to elicit its cell motility functions.
RNF5 also requires an intact C-terminal domain for its effect on paxillin. Although the C terminus of RNF5 is not required for paxillin association, it is required for RNF5 ubiquitination of paxillin and to abolish its localization within focal adhesions. These observations suggest that further modifications take place in the membrane-anchored form of RNF5, which may also explain the limited nature of the activities seen in vitro. Modifications within selective subcellular compartments are often observed and serve as efficient regulators of the degree and rate of changes seen, which in this case are limited to altered localization of paxillin.
Changes in paxillin localization are reflected in altered cell motility. Our study elucidates the biological implications of paxillin ubiquitination in that it links the well-recognized role of paxillin in cellular motility with its ubiquitination by RNF5, thereby providing mechanisms underlying the regulation of cell motility via the newly characterized E3 ligase RNF5. The effect of RNF5 on paxillin localization and consequently on cell motility suggests that RNF5 serves to limit cell migration. Indeed, preliminary analysis of RNF5 expression in human tumor cell lines revealed a decrease in RNF5 expression, which could provide the appropriate setting for efficient cell migration, a prerequisite for angiogenic/metastatic properties of tumor cells (our unpublished studies). The latter is supported by the finding that RNF5 is among the genes that are transcriptionally suppressed by the polycomb group protein EZH2 in advanced prostate tumors (44).
The effect of RNF5 on paxillin localization is expected to affect paxillin interaction with a number of proteins implicated in regulating cytoskeleton architecture and processes such as migration. For example, RNF5 may affect paxillin binding to FAK, which is important for focal adhesion turnover, spreading, migration, and survival (9, 14). RNF5 may also influence paxillin effects on Cas via the adapter protein Crk, thereby affecting Cas's role in cell migration (15). Although the role of these interactions is uncertain, it is possible that binding of RNF5 to paxillin could affect the subcellular localization or function of these proteins. On the other hand, RNF5 may also affect paxillin's interaction with negative regulators such as Csk and PTP-PEST (15, 34, 49). PTP-PEST has been shown to dephosphorylate Cas, and Csk negatively regulates Src PTKs (31). RNF5 association and ubiquitination of paxillin may impair paxillin's ability to sequester these proteins away from their targets, thus attenuating positive signals sent via Cas and Src.
The idea that RNF5 was originally isolated as a suppressor of a yeast secretory mutant, sec 15 (21), supports the role of RNF5 in localization of paxillin. Sec15 is involved in vesicular transport from the Golgi apparatus to the plasma membrane and is essential for exocytosis. The finding that overexpression of RNF5 can suppress this phenotype, together with our current observations, further emphasizes the role of RNF5 in cellular trafficking and exocytosis. Additionally, the growth defect of sec 15 is also suppressed by the overexpression of Rho3, which organizes the actin cytoskeleton and contributes to the paxillin link to actin filaments (21, 41).
Overall, our findings identifying RNF5 as an E3 ligase of paxillin and the effect of RNF5 on paxillin localization reveal a novel regulator of cytoskeleton regulatory proteins that in turn is important in cell motility regulation. The effects of RNF5 on paxillin resemble those of the C. elegans LIM protein Y105E8A.6, which is a newly identified LIM domain protein that is required for the formation of focal adhesion-like muscle attachment structures, also termed dense bodies, that provide a physical link between the muscles and the hypodermis (Broday et al., unpublished observations). Similar to focal adhesions, these structures contain integrin heterodimers as central structural units and cytoskeletal adaptor proteins such as vinculin, α-actinin, and talin. This parallel between the C. elegans and human RNF5 target proteins further supports the regulatory role of this newly characterized RING finger protein in cytoskeleton organization and motility.
Acknowledgments
We thank Lila Pirkalla and Gabriel Maulit for technical help and Z.Q. Pan and members of the Ronai laboratory for advice.
Microscopy was performed at the MSSM-Microscopy Shared Resource Facility, supported, in part, with funding from NIH-NCI shared resources grant 1 R24 CA095823-01. Support by NIH grant CA97105 to Z.R. is gratefully acknowledged.
REFERENCES
- 1.Borden, K. L. B. 2000. RING domains: master builders of molecular scaffolds? J. Biol. Chem. 295:1103-1112. [DOI] [PubMed] [Google Scholar]
- 2.Brown, M. C., M. S. Curtis, and C. E. Turner. 1998. Paxillin LD motifs may define a new family of protein recognition domains. Nat. Struct. Biol. 5:677-678. [DOI] [PubMed] [Google Scholar]
- 3.Brown, M. C., J. A. Perrotta, and C. E. Turner. 1998. Serine and threonine phosphorylation of the paxillin LIM domains regulates paxillin focal adhesion localization and cell adhesion to fibronectin. Mol. Biol. Cell 9:1803-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deshaies, R. J. 1999. SCF and Cullin/Ring H2-based ubiquitin ligases. Annu. Rev. Cell. Dev. Biol. 15:435-467. [DOI] [PubMed] [Google Scholar]
- 5.Fuchs, S. Y., B. Xie, V. Adler, V. A. Fried, R. J. Davis, and Z. Ronai. 1997. c-Jun NH2-terminal kinases target the ubiquitination of their associated transcription factors. J. Biol. Chem. 272:32163-32168. [DOI] [PubMed] [Google Scholar]
- 6.Fuchs, S. Y., L. Dolan, R. J. Davis, and Z. Ronai. 1996. Phosphorylation-dependent targeting of c-Jun ubiquitination by Jun N-kinase. Oncogene 13:1531-1535. [PubMed] [Google Scholar]
- 7.Ganoth, D., G. Bornstein, T. K. Ko, B. Larsen, M. Tyers, M. Pagano, and A. Hershko. 2001. The cell-cycle regulatory protein Cks1is required for SCF (Skp2)-mediated ubiquitinylation of p27. Nat. Cell Biol. 3:321-324. [DOI] [PubMed] [Google Scholar]
- 8.Hagel, M., E. L. George, A. Kim, R. Tamimi, S. L. Opitz, C. E. Turner, A. Imamoto, and S. M. Thomas. 2002. The adaptor protein paxillin is essential for normal development in the mouse and is a critical transducer of fibronectin signaling. Mol. Cell. Biol. 22:901-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hildebrand, J. D., M. D. Schaller, and J. T. Parsons. 1995. Paxillin, a tyrosine phosphorylated focal adhesion-associated protein binds to the carboxyl terminal domain of focal adhesion kinase. Mol. Biol. Cell 6:637-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hofmann, R. M., and C. M. Pickart. 1995. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell 96:645-653. [DOI] [PubMed] [Google Scholar]
- 11.Hofmann, R. M., and C. M. Pickart. 2001. In vitro assembly and recognition of lys-63 polyubiquitin chains. J. Biol. Chem. 276:27936-27943. [DOI] [PubMed] [Google Scholar]
- 12.Hu, G., S. Zhnag, M. Vidal, J. L. Baer, T. Xu, and E. R. Fearon. 1997. Mammalian homologs of seven in absentia regulate DCC via the ubiquitin-proteasome pathway. Genes Dev. 11:2701-2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ishino, K., Kaneyama, M. Shibanuma, and K. Nose. 2000. Specific decrease in the level of Hic-5, a focal adhesion protein, during immortalization of mouse embryonic fibroblasts, and its association with focal adhesion kinase. J. Cell. Biochem. 76:411-419. [DOI] [PubMed] [Google Scholar]
- 14.Jockusch, B. M., P. Bubeck, K. Giehl, M. Kroemker, J. Moschner, M. Rothkegel, M. Rudiger, K. Schluter, G. Stanke, and J. Winkler. 1995. The molecular architecture of focal adhesions. Annu. Rev. Cell Dev. Biol. 11:379-416. [DOI] [PubMed] [Google Scholar]
- 15.Klemke, R. L., J. Leng, R. Molander, P. C. Brooks, K. Vuori, and D. A. Cheresh. 1998. CAS/Crk coupling serves as a “molecular switch” for induction of cell migration. J. Cell Biol. 140:961-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kondo, A., S. Hashimoto, H. Yano, K. Nagayama, Y. Mazaki, and H. Sabe. 2000. A new paxillin-binding protein, PAG3/Papα/KIAA0400, bearing an ARF GTPase-activating protein activity is involved in paxillin recruitment to focal adhesions and cell migration. Mol. Biol. Cell 11:1315-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kyushiki, H., Y. Kuga, M. Suzuki, E. Takahashi, and M. Horie. 1997. Cloning, expression and mapping of a novel RING-finger gene (RNF5), a human homologue of a putative zinc-finger gene from Caenorhabditis elegans. Cytogenet. Cell Genet. 79:114-117. [DOI] [PubMed] [Google Scholar]
- 18.Levkowitz, G., H. Waterman, S. A. Ettenberg, M. Katz, A. Y. Tsygankov, I. Alroy, S. Lavi, K. Iwai, Y. Reiss, A. Ciechanover, S. Lipkowitz, and Y. Yarden. 1999. Ubiquitin ligase activity and tyrosine phosphorylation underlie suppression of growth factor signaling by c-Cbl/Sli-1. Mol. Cell 6:1029-1040. [DOI] [PubMed] [Google Scholar]
- 19.Lewis, J. M., and M. A. Schwartz. 1998. Integrins regulate the association and phosphorylation of paxillin by c-Abl. J. Biol. Chem. 273:14225-14230. [DOI] [PubMed] [Google Scholar]
- 20.Lorick, K. L., J. P. Jensen, S. Fang, A. M. Ong, S. Hatakeyama, and A. M. Weissman. 1999. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc. Natl. Acad. Sci. USA 96:11364-11369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsuda, N., and A. Nakano. 1998. RMA1, an Arabidopsis thaliana gene whose cDNA suppresses the yeast sec15 mutation, encodes a novel protein with a RING finger motif and a membrane anchor. Plant Cell Physiol. 39:545-554. [DOI] [PubMed] [Google Scholar]
- 22.Matsuda, N., T. Suzuki, K. Tanaka, and A. Nakano. 2001. Rma1, a novel type of RING finger protein conserved from Arabidopsis to human, is a membrane-bound ubiquitin ligase. J. Cell Sci. 114:1949-1957. [DOI] [PubMed] [Google Scholar]
- 23.Mazaki, Y., H. Uchida, O. Hino, S. Hashimoto, and H. Sabe. 1998. Paxillin isoforms in mice: lack of the γ isoform, and developmentally specific β isoform expression. J. Biol. Chem. 273:22435-22441. [DOI] [PubMed] [Google Scholar]
- 24.Mazaki, Y., S. Hashimoto, K. Okawa, A. Tsubouchi, K. Nakamura, R. Yagi, H. Yano, A. Kondo, A. Iwamatsu, A. Mizoguchi, and H. Sabe. 2001. An ADP-ribosylation factor GTPase-activating protein Git2-short/KIAA0148 is involved in subcellular localization of paxillin and actin cytoskeletal organization. Mol. Biol. Cell 12:645-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Newell, S. W., J. P. Perchellet, E. M. Perchellet, and E. T. Ulug. 1999. Alterations in focal adhesion kinase activity and associated proteins during malignant conversion of mouse keratinocytes. Mol. Carcinogenesis 25:73-83. [DOI] [PubMed] [Google Scholar]
- 26.Nikolopoulos, S. N., and C. E. Turner. 2000. Actopaxin, a new focal adhesion protein that binds paxillin LD motifs and actin and regulates cell adhesion. J. Cell Biol. 151:1435-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nikolopoulos, S. N., and C. E. Turner. 2001. Integrin-linked kinase (ILK) binding to paxillin LD1 motif regulates ILK localization to focal adhesions. J. Biol. Chem. 276:23499-23505. [DOI] [PubMed] [Google Scholar]
- 28.Nobes, C. D., and A. Hall. 1995. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell 81:53-62. [DOI] [PubMed] [Google Scholar]
- 29.Oh, E. S., H. Gu, T. M. Saxton, J. F. Timms, S. Hausdorff, E. U. Frevert, B. B. Kahn, T. Pawson, B. G. Neel, and S. M. Thomas. 1999. Regulation of early events in integrin signaling by protein tyrosine phosphatase SHP-2. Mol. Cell. Biol. 19:3205-3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Panetti, T. S. 2002. Tyrosine phosphorylation of paxillin, FAK, and p130CAS: effects on cell spreading and migration. Front. Biosci. 7:d143-d150. [DOI] [PubMed] [Google Scholar]
- 31.Sabe, H., A. Hata, M. Okada, H. Nakagawa, and H. Hanafusa. 1994. Analysis of the binding of the Src homology 2 domain of Csk to tyrosine-phosphorylated proteins in the suppression and mitotic activation of c-Src. Proc. Natl. Acad. Sci. USA 91:3984-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salgia, R., J. L. Li, D. S. Ewaniuk, Y. B. Wang, M. Sattler, W. C. Chen, W. Richards, E. Pisick, G. I. Shapiro, B. J. Rollins, L. B. Chen, J. D. Griffin, and D. J. Sugarbaker. 1999. Expression of the focal adhesion protein paxillin in lung cancer and its relation to cell motility. Oncogene 18:67-77. [DOI] [PubMed] [Google Scholar]
- 33.Schaller, M. D. 2001. Paxillin: a focal adhesion-associated adaptor protein. Oncogene 20:6459-6472. [DOI] [PubMed] [Google Scholar]
- 34.Shen, Y., G. Schneider, J. F. Cloutier, A. Veillette, and M. D. Schaller. 1998. Direct association of protein-tyrosine phosphatase PTP-PEST with paxillin. J. Biol. Chem. 273:6474-6481. [DOI] [PubMed] [Google Scholar]
- 35.Shibanuma, M., J. Mashimo, T. Kuroki, and K. Nose. 1994. Characterization of the TGF β 1-inducible hic-5 gene that encodes a putative novel zinc finger protein and its possible involvement in cellular senescence. J. Biol. Chem. 269:26767-26774. [PubMed] [Google Scholar]
- 36.Shimura, H., N. Hattori, S. Kubo, Y. Mizuno, S. Asakawa, S. Minoshima, N. Shimizu, K. Iwai, T. Chiba, K. Tanaka, and T. Suzuki. 2000. Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat. Genet. 25:302-305. [DOI] [PubMed] [Google Scholar]
- 37.Thien, C. B., and W. Y. Langdon. 2001. Cbl: many adaptations to regulate protein tyrosine kinases. Nat. Rev. Mol. Cell. Biol. 2:294-307. [DOI] [PubMed] [Google Scholar]
- 38.Thomas, S. M., M. Hagel, and C. E. Turner. 1999. Characterization of a focal adhesion protein, Hic-5, that shows extensive homology with paxillin. J. Cell Sci. 112:181-190. [DOI] [PubMed] [Google Scholar]
- 39.Turner, C. 2000. Paxillin interactions. J. Cell Sci. 23:4139-4140. [DOI] [PubMed] [Google Scholar]
- 40.Turner, C. 1998. Molecules in focus: paxillin. Int. J. Biochem. Cell. Biol. 98:1357-2725. [Google Scholar]
- 41.Turner, C. E. 2000. Paxillin and focal adhesion signaling. Nat. Cell. Biol. 2:E231-E236. [DOI] [PubMed] [Google Scholar]
- 42.Turner, C. E. 1994. Paxillin: a cytoskeletal target for tyrosine kinases. Bioessays 16:47-52. [DOI] [PubMed] [Google Scholar]
- 43.Vadlamudi, R., L. Adam, B. Tseng, L. Costa, and R. Kumar. 1999. Transcriptional up-regulation of paxillin expression by heregulin in human breast cancer cells. Cancer Res. 59:2843-2846. [PubMed] [Google Scholar]
- 44.Varambally, S., Dhanasekaran, S. M., Zhou, M., Barrette, T. R., Kumar-Sinha, S., Sanda, M. G., Ghosh, D., Pienta, K. J., Sewalt, R. G. A. B., Otte, A. P., Rubin, M. A., and A. M. Chinnaiyan. 2002. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 419:624-629. [DOI] [PubMed] [Google Scholar]
- 45.Wade, R., J. Bohl, and S. Vande Pol. 2002. Paxillin null embryonic stem cells are impaired in cell spreading and tyrosine phosphorylation of focal adhesion kinase. Oncogene 21:96-107. [DOI] [PubMed] [Google Scholar]
- 46.West, K. A., H. Zhang, M. C. Brown, S. N. Nikolopoulos, M. C. Riedy, A. F. Horwitz, and C. E. Turner. 2001. The LD4 motif of paxillin regulates cell spreading and motility through an interaction with paxillin kinase linker (protein kinase L). J. Cell Biol. 154:161-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Woods, A. J., M. S. Roberts, J. Choudhary, S. T. Barry, Y. Mazaki, H. Sabe, S. J. Morley, D. R. Critchley, and J. C. Norman. 2002. Paxillin associates with poly(A)-binding protein 1 at the dense endoplasmic reticulum and the leading edge of migrating cells. J. Biol. Chem. 277:6428-6437. [DOI] [PubMed] [Google Scholar]
- 48.Yamaguchi, R., Y. Mazaki, K. Hirota, S. Hashimoto, and H. Sabe. 1997. Mitosis specific serine phosphorylation and downregulation of one of the focal adhesion protein, paxillin. Oncogene 15:1753-1761. [DOI] [PubMed] [Google Scholar]
- 49.Yano, H., H. Uchida, T. Iwasaki, M. Mukai, H. Akedo, K. Nakamura, S. Hashimoto, and H. Sabe. 2000. Paxillin α and Crk-associated substrate exert opposing effects on cell migration and contact inhibition of growth through tyrosine phosphorylation. Proc. Natl. Acad. Sci. USA 97:9076-9081. [DOI] [PMC free article] [PubMed] [Google Scholar]