Abstract
Hepatitis B virus (HBV) includes an X gene (HBx gene) that plays a critical role in liver carcinogenesis. Because centrosome abnormalities are associated with genomic instability in most human cancer cells, we examined the effect of HBx on centrosomes. We found that HBx induced supernumerary centrosomes and multipolar spindles. This effect was independent of mutations in the p21 gene. Furthermore, the ability of HBV to induce supernumerary centrosomes was dependent on the presence of physiological HBx expression. We recently showed that HBx induces cytoplasmic sequestration of Crm1, a nuclear export receptor that binds to Ran GTPase, thereby inducing nuclear localization of NF-κB. Consistently, supernumerary centrosomes were observed in cells treated with a Crm1-specific inhibitor but not with an HBx mutant that lacked the ability to sequester Crm1 in the cytoplasm. Moreover, a fraction of Crm1 was found to be localized at the centrosomes. Immunocytochemical and ultrastructural examination of these supernumerary centrosomes revealed that inactivation of Crm1 was associated with abnormal centrioles. The presence of more than two centrosomes led to an increased frequency of defective mitoses and chromosome transmission errors. Based on this evidence, we suggest that Crm1 is actively involved in maintaining centrosome integrity and that HBx disrupts this process by inactivating Crm1 and thus contributes to HBV-mediated carcinogenesis.
Centrosomes are eukaryotic cellular structures that play a key role in cell division (5, 33). Centrosomes function as microtubule-organizing centers which control the number, polarity, and orientation of microtubules during interphase and nucleate microtubules to form bipolar spindles during mitosis (5, 33). Each centrosome contains two centrioles, which adhere to each other throughout the cell cycle and normally separate only once during the G1-to-S cell cycle transition, resulting in centrosome duplication (10, 30). Thus, intricate mechanisms exist to regulate centriole cohesion and separation, in which any unscheduled splitting of mother and daughter centrioles should be avoided.
Centrosome duplication is regulated by many intracellular events that are essential in maintaining genomic stability. A normal mitotic cell contains two and only two centrosomes, which ensure the formation of a bipolar spindle. Abnormal centrosome duplication is tightly linked to aneuploidy and is found in virtually every type of human cancer (25). Thus, disruption of the regulatory mechanisms that ensure the numeral integrity of centrosomes may be an early event in carcinogenesis. Consistently, mutation of tumor suppressor genes, such as p53, Brca1, Brca2, p21, Gadd45, and adenomatous polyposis coli has been shown to be associated with supernumerary centrosomes (6, 15, 17, 21, 22, 34, 39). Moreover, expression of oncogenic human papillomavirus type 16 E6 and E7 also leads to supernumerary centrosomes and mitotic defects (12; M. J. Difilipantonio, unpublished data).
Disruption of genes that regulate mitotic spindle assembly can also lead to abnormal mitosis and genomic instability. For example, Ran GTPase is an essential factor that regulates nucleocytoplasmic transport as well as mitotic spindle assembly (31, 36). Proteins that activate Ran, including RCC1, RanGAP1, and RanBP1, are also essential for Ran-mediated mitotic spindle assembly (8, 9). Saccharomyces cerevisiae with a disruption of SPI1, a yeast Ran homologue, is multiseptate, leading to mitotic hepaloidization and aneuploidy (26). The conditional inactivation of Crm1, a nuclear export receptor that binds to Ran, leads to disrupted higher-order chromosome organization and arrangements of centromeres or telomeres in S. cerevisiae (18). Overexpression of RanBP1 induces multipolar spindles (20). Multipolar spindles can cause aberrant mitoses, which may lead to chromosome imbalances and facilitate carcinogenesis.
Hepatocellular carcinoma is the fifth most prevalent malignant disease worldwide, and more than 85% of hepatocellular carcinoma cases are associated with hepatitis B virus (HBV) or hepatitis C virus (4, 7, 14, 28). HBV is a DNA tumor virus encoding the X oncoprotein (HBx) that contributes to liver carcinogenesis (14). HBx is essential for HBV replication in vivo (4) and induces liver cancer in transgenic mice and neoplastic transformation in cultured cells (24, 28, 40). HBx is frequently integrated into the cellular genome and expressed during the development of hepatocellular carcinoma (14, 28). Although HBx does not bind to DNA, it is a potent transcriptional coactivator, a property shared by many viral oncoproteins (28, 40). Several pleiotropic activities associated with HBx expression include the stimulation of mitogen-activated protein kinase and the tumor necrosis factor alpha and NF-κB signaling pathways (4, 23, 28, 40). Consequently, HBx may influence apoptosis by interacting with the NF-κB signaling cascade or p53, and it may stimulate cell proliferation through the activation of cyclin-dependent kinase activities (4, 23, 28, 40).
Previously, we and others demonstrated that HBx inactivates p53 and p53-mediated activation of p21 (1, 13, 23, 35). More recently, we showed that HBx contains a functional nuclear export signal motif utilizing the Crm1/Ran GTPase-mediated pathway. We further demonstrated that HBx can bind to and sequester Crm1 in the cytoplasm, thereby altering Crm1/Ran GTPase-dependent nuclear export of the NF-κB/IκBα complex. An export-defective mutant of HBx (HBx-NESM) failed to do so (16). Consistent with this finding, liver cells from HBV-infected chronic viral hepatitis carriers maintain a cytoplasmic sequestration of Crm1 (16). Recent evidence indicates that importin receptors α and β as well as their cofactors that bind to Ran also regulate mitotic spindle assembly. Thus, it is reasonable to speculate that the Crm1 export receptor may also control this process.
We hypothesize that HBx induces genomic instability by altering the fidelity of centrosome maintenance and duplication through acting on the Crm1-Ran complex. Here, we show that both HBx expression and Crm1-specific inhibition induce abnormal centriole synthesis and formation of multipolar spindles, which are associated with aneuploidy, whereas a nuclear export signal mutant of HBx lacks such activity. We also demonstrate, for the first time, aneuploidy in noncancerous hepatocytes from HBV-infected chronic liver disease patients. This study reveals that Crm1 plays a role in maintaining the fidelity of centrosome duplication and that HBx interferes with this function. The resultant genomic instability may contribute to liver cancer development associated with hepatitis B virus infection.
MATERIALS AND METHODS
Cell culture.
Telomerase (hTERT)-immortalized normal human fibroblast cells (NHF-hTERT) were a gift from Judith Campisi. These cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen), supplemented with 10% fetal bovine serum (FBS; Biofluids), penicillin and streptomycin (1×; 100 U/ml and 100 μg/ml, respectively), and glutamine (1×, 200 mM). HCT-116 (colon carcinoma) cells were cultured in Eagle's minimal essential medium (Invitrogen) supplemented with 10% FBS, penicillin-streptomycin (1×), and glutamine (1×). HepG2 cells (a hepatoma cell line) were maintained in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum, sodium pyruvate (1 mM), and nonessential amino acids (1×). Cells were incubated at 37°C in a humidified atmosphere with 5% CO2.
Adenovirus cloning and production.
Ad-HBx was constructed essentially as described previously (38). The Ad-HBxNESM construct (containing an HBx nuclear export signal mutant) was constructed by inserting the BglII-XbaI fragment of pGFP-HBxM2 (16) into the BamHI and XbaI sites of the pZEROTG-CMV adenoviral shuttle vector and then transferred to the adenovirus construct. Adenovirus production and titer determination were performed essentially as described previously (38).
HBx-dependent HBV DNA replication system.
To determine the cellular effects induced by a physiological level of HBx, we used a previously established HBx-dependent HBV DNA replication system (27). Analysis of HBV DNA replication was done as described previously (3, 27). Briefly, the payw1.2 plasmid (pHBV) contains a wild-type HBV genome and can replicate in HepG2 cells to produce viral particles, while the payw∗7 plasmid [pHBV(−HBx)] contains the same length of HBV genome without HBx expression, due to a point mutation at codon 7 of HBx, resulting in a termination signal. HepG2 cells were transfected with these plasmids by the Nucleofection protocol according to the manufacturer's instructions (Amaxa Biosystems; http://www.amaxa.com). The HBV DNA from cytoplasm-derived nucleocapsid particles as an indication of HBV DNA replication were isolated 4 days posttransfection and analyzed by the Southern blot technique as described previously (3, 27).
Indirect immunofluorescence assay and confocal analysis.
NHF-hTERT or HCT-116 cells were cultured on four-well chamber slides and either treated with leptomycin B (2, 10, or 25 nM) or infected with Ad-HBx (multiplicity of infection [MOI] of 6 for NHF-hTERT and an MOI of 10 for HCT116). We used this MOI based on the finding that the MOI of 6 gave rise to a moderate abundance (0.14%) of HBx transcripts (37), which was comparable to the amount of HBx transcripts (0.1%) expressed in HBV-positive hepatocellular carcinoma samples (Taro Yamashita, personal communication).
For centrosome analysis on HepG2 cells, exponentially growing cells were transfected with pHBV or HBV(−HBx) plasmids with the Nucleofection protocol (see detailed description in the section for analyzing HBV DNA replication). With this protocol, the transfection efficiency for HepG2 cells was over 64% (see manufacturer's data and data not shown). Following various times of incubation, cells were fixed with 4% paraformaldehyde for 10 min and methanol for 20 min and then incubated in phosphate-buffered saline. Samples were blocked with 10% normal donkey serum for 1 h at room temperature and stained with various primary antibodies for 1 h at 37°C, followed by either Alexa 488 fluorescein isothiocyanate-conjugated anti-mouse or -rabbit or Alexa 568 Texas Red-conjugated anti-mouse or -rabbit antibodies (Molecular Probes). Mouse monoclonal anti-γ-tubulin (GTU-88; Sigma) or rabbit polyclonal anti-γ-tubulin (Sigma) antibodies were used to detect centrosomes. Mouse monoclonal anti-α-tubulin (clone B-5-1-2; Sigma) was used to detect spindles. Mouse monoclonal anticentrin antibody (gift from J. Salisbury) was used to detect centrioles. For determining the localization of Crm1 at centrosomes, cells were extracted with 0.1% Triton X-100 and immediately fixed as described above. Nuclei were stained with 4′,6′-diamidino-2-phenylindole (DAPI). Conventional or confocal fluorescence microscope analysis was done essentially as described previously (16).
Cell cycle analysis.
NHF-hTERT cells were synchronized by serum starvation, infected with control virus (Ad-CMV) or HBx virus (Ad-HBx) at an MOI of 10 for 24 h alone or treated with 5 Gy of γ-radiation, and then released into medium containing 10% FBS and bromodeoxyuridine (65 μM). Cells were trypsinized, washed twice with phosphate-buffered saline, pelleted, and fixed with 70% ethanol overnight at 4°C. Samples were then stained with antibromodeoxyuridine-fluorescein isothiocyanate antibody, treated with 20 μl of RNase per ml for 30 min, counterstained with 60 μg of propidium iodide per ml for DNA content, and then subjected to FACScan analysis.
Western blot analysis.
Western blotting analysis was described previously (16). Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to nitrocellulose membranes (Invitrogen), blocked with 5% milk, and probed with appropriate antibodies, i.e., phospho-Raf (Ser 259), phospho-MEK1/2 (Ser217/Ser221), MEK1/2 antibody, phospho-ERK1/2 (Thr202/Tyr204), and ERK1/2 antibody from Cell Signaling Technology; DO-1 (p53 antibody), anti-WAF1, anti-cyclin E, anti-cyclin A, and anti-Cdk2 from Santa Cruz, and then detected by chemiluminescence (ECL kit; Amersham).
Interphase fluorescence in situ hybridization analysis.
Archived frozen tissues were allowed to thaw briefly on ice and “touched” to glass slides (Plus Gold slides; Thomas Scientific) and then fixed in sequential 70%, 90%, and 100% ethanol for 3 min each. Samples were allowed to air dry and treated with pepsin (Sigma) for various predetermined incubation times. Fluorescence in situ hybridization detection was performed as previously described (39) with specific bacterial artificial chromosome clones for chromosomes 1q (243 M13) and 6p (175A4). Fluorescence-labeled DNA probes were hybridized to denatured slides and incubated in a humidified chamber at 37°C overnight. Nuclei were stained with DAPI and mounted with 1,4-phenylenediamine (Anti-fade; Sigma).
RESULTS
Induction of centrosome amplification and aberrant mitosis by HBx.
The ability of HBx to disrupt the fidelity of centrosome number was determined by expressing HBx in human cell lines. To ensure efficient expression of the HBx gene, we constructed replication-defective adenoviral vectors encoding either wild-type HBx (Ad-HBx) or a nuclear export signal-mutated HBx (L98A, G99A, and L100A) (Ad-NESM). Efficient expression of wild-type HBx and the HBx NESM mutant were verified by Western blot analysis with anti-HBx antibodies (data not shown). However, we were not able to immunocytochemically detect HBx expression at the single-cell level with the amount of Ad-HBx used.
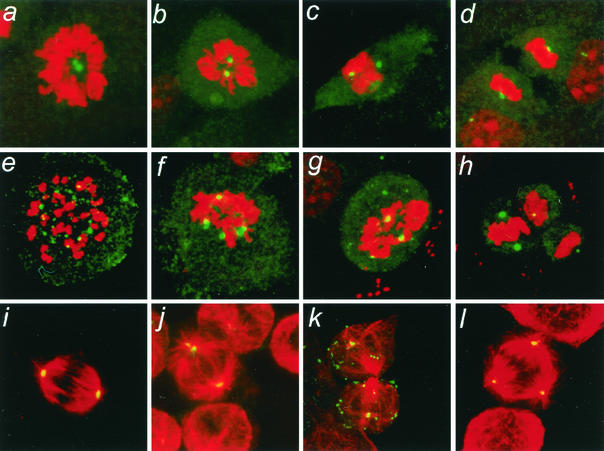
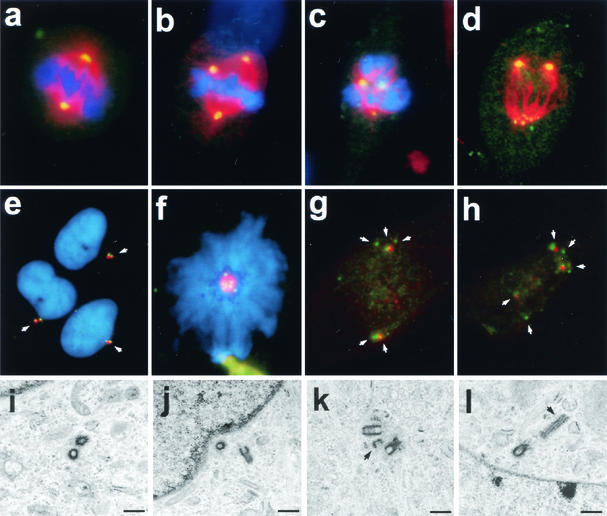
To determine whether wild-type HBx affects centrosome numbers, centrosomes were visualized immunocytochemically in an hTERT-immortalized normal human fibroblast line (NHF-hTERT) with a rabbit polyclonal anti-γ-tubulin antibody. Uninfected NHF-hTERT cells (data not shown) and cells infected with control adenovirus (Ad-CMV) contained the typical one or two γ-tubulin-positive foci (indicative of centrosomes) in interphase (data not shown) and at various stages of mitosis (Fig. 1a to d, Table 1). Two centrosomes in each normal mitotic cell ensured the formation of a bipolar spindle (Fig. 1i). In contrast, Ad-HBx-infected cells contained more than two γ-tubulin-positive foci in 7% of interphase and 39% of mitotic cells (Fig. 1e to h, Table 1). Some of the Ad-HBx-infected mitotic cells contained more than 10 γ-tubulin-positive foci (Fig. 1e). Many of the M-phase γ-tubulin-positive foci in both Ad-CMV- and Ad-HBx-infected cells were juxtaposed to condensed chromatin (Fig. 1c, d, f, and g). Double immunostaining for α- and γ-tubulin revealed the formation of multipolar spindles (Fig. 1l).
FIG. 1.
Induction of abnormal spindle pole formations by HBx. NHF-hTERT cells were infected with adenovirus encoding HBx (Ad-HBx) (panels e to h and j to l) or a control adenovirus (Ad-CMV) (panels a to d and i). Centrosomes were visualized by staining cells with rabbit polyclonal anti-γ-tubulin antibody (green), followed by propidium iodide for staining chromatin (red) (panels a to h). For analysis of mitotic spindles, cells were coimmunostained with rabbit polyclonal anti-γ-tubulin antibody (green) and mouse monoclonal anti-α-tubulin antibody (red) (panels i to l). Representative images are shown. Magnification, ×630.
TABLE 1.
Induction of centrosome amplification in NHF cells by wild-type HBx but not by a nuclear export signal mutanta
| Construct | Interphase
|
Mitosis
|
||||
|---|---|---|---|---|---|---|
| No. of centrosomes
|
% of cells with centrosome amplification | No. of centrosomes
|
% of cells with centrosome amplification | |||
| 1-2 | ≥3 | 1-2 | ≥3 | |||
| Ad-CMV | 974 | 13 | 1.3 | 86 | 7 | 7.5 |
| Ad-HBx | 971 | 71 | 6.8 | 115 | 75 | 39.3 |
| Ad-NESM | 537 | 6 | 1.1 | 185 | 16 | 8.0 |
Mitotic indices: Ad-CMV, 6.1% (53 of 865); Ad-HBx, 10.9% (108 of 995).
Many HBx-expressing mitotic cells exhibited abnormal mitotic spindle assembly, as evidenced by the presence of more than two spindle poles (Fig. 1j to l) and/or disarrayed microtubules (Fig. 1k). Aberrant mitotic cells were often observed in Ad-HBx-infected samples (Fig. 1g and h), and some appeared to display two sets of spindle poles in a single mitotic cell, as if they had attempted to divide twice (Fig. 1h), possibly as a result of unequal chromosome segregation due to multiple centrosomes. Increased numbers of centrioles, as determined by an anticentrin monoclonal antibody, were observed in NHF-hTERT cells infected with Ad-HBx (data not shown). It was also noted that supernumerary centrosomes in NHF-hTERT cells induced by HBx were more pronounced in mitosis than in interphase (Table 1), implying that HBx may preferentially affect mitotic components of the centrosome duplication cycle.
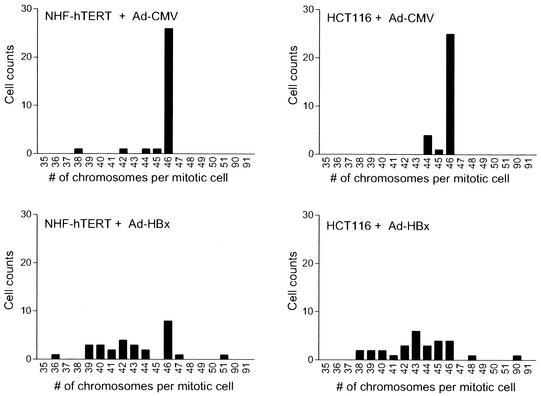
Multipolar spindles during mitosis may lead to unequal chromosome segregation, resulting in aneuploidy. Consistent with this prediction, metaphases from about 73% of HBx-infected NHF-hTERT cells and 90% of HBx-infected HCT116 cells displayed a chromosome loss or gain, compared with only 13% and 17%, respectively, when infected with control Ad-CMV virus (Fig. 2). These results indicate that HBx is able to directly induce genomic instability in both normal fibroblasts and diploid colon epithelial tumor cells.
FIG. 2.
Destabilization of chromosomes by HBx. To determine the fidelity of chromosome segregation, cells were infected with control (Ad-CMV) or AD-HBx adenovirus and incubated for 48 h. Cells were treated with 0.56% KCl for 5 min and fixed with methanol-glacial acetic acid (3:1). Mitotic spreads were prepared and stained with Giemsa. Mitotic cells were randomly selected, and the number of chromosomes per mitotic cell was counted. A total of 30 mitotic cells were analyzed for each condition.
HBx-expressing cells have normal γ-radiation-induced cell cycle checkpoints.
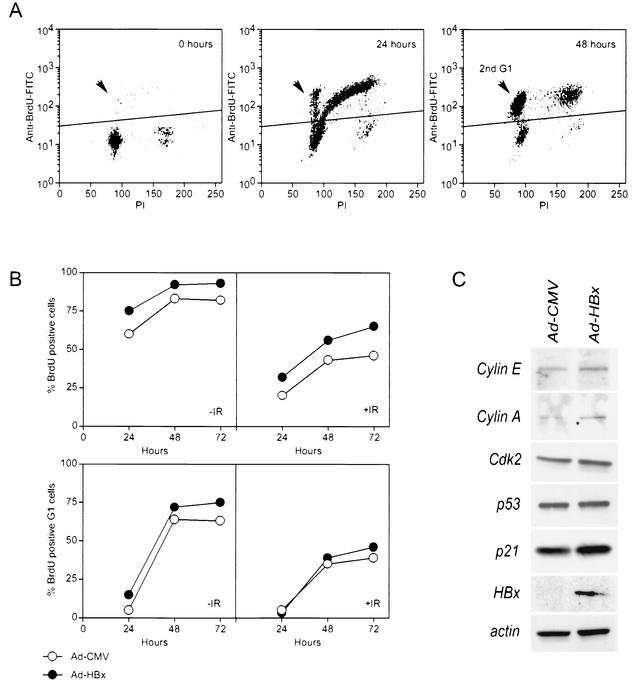
Faithful centrosome duplication relies on normal cell cycle progression (5). HBx expression has been shown to stimulate cell proliferation, possibly through the activation of cyclin-dependent kinases (2) and inactivation of cell cycle checkpoint proteins, including p53 and p21 (35). To determine whether the HBx-induced centrosome abnormalities could be due to interference with cell cycle checkpoints, Ad-HBx-infected NHF-hTERT cells treated with γ-radiation or untreated were analyzed by fluorescence-activated cell sorting. NHF-hTERT cells were synchronized in G0/G1 by contact inhibition and serum starvation (0.1% FBS) for 48 h. The quiescent cells were infected with Ad-CMV or Ad-HBx (MOI = 10) and incubated for an additional 24 h in low serum. Cell cycle reentrance was achieved by subculturing to low density and adding 10% FBS. While greater than 95% of serum-starved cells were in G0/G1, over 60% of the cells had progressed past the first G1-S checkpoint 24 h after release. About 10% of these cells had reached the second G1 phase in this time period, indicating that they had proceeded through the subsequent G2-M checkpoint (Fig. 3). By 48 h, greater than 80% of the cells had bypassed the first G1-S checkpoint, and at least 60% had bypassed the subsequent G2-M checkpoint (Fig. 3A).
FIG. 3.
HBx weakly stimulates cell cycle progression without induction of any measurable changes in γ-radiation-induced cell cycle checkpoints or expression of various cell cycle regulators. NHF-hTERT cells were synchronized by serum starvation in the presence of control (Ad-CMV) or HBx (Ad-HBx) adenovirus. Cells untreated or treated with 5 Gy of γ-radiation were released into the cell cycle in medium containing 10% FBS and bromodeoxyuridine (BrdU). Cells were harvested at 0, 24, 48, and 72 h and subjected to FACScan analysis. (A) Representative FACScan profiles from uninfected and unirradiated (IR) samples. Events above the horizontal lines indicate bromodeoxyuridine-positive cells that were included in the analysis to examine the effect of HBx in panel B. Arrows indicate bromodeoxyuridine-positive G1 events, which represent cells that divided successfully and entered a second G1. (C) Unsynchronized NHF-hTERT cells were infected with control or HBx adenovirus and incubated for 24 h. Cell lysates were subjected to Western blot analysis with antibodies specific to cyclin E, cyclin A, cdk2, p53, p21, HBx, and β-actin.
Consistent with previous observations (2), HBx slightly stimulated cell proliferation, as evidenced by the higher percentages of bromodeoxyuridine-positive cells and an increase in mitotic index (Fig. 3B, Table 1). Moreover, the amount of Ad-HBx used to observe multipolar spindles only minimally activated mitogen-activated protein kinase with no alteration of the level of several cell cycle-dependent proteins (data not shown). However, the induction of a G1 or G2 arrest after γ-irradiation was apparently normal in Ad-HBx-infected cells (Fig. 3B). Furthermore, there was no significant change in the levels of cyclin E, cyclin A, Cdk2, p53, and p21 upon HBx expression (Fig. 3C). These experiments demonstrate that while HBx stimulates cell growth, DNA damage-induced cell cycle checkpoints remain intact.
HBx induces supernumerary centrosomes in p53- and p21-deficient cells.
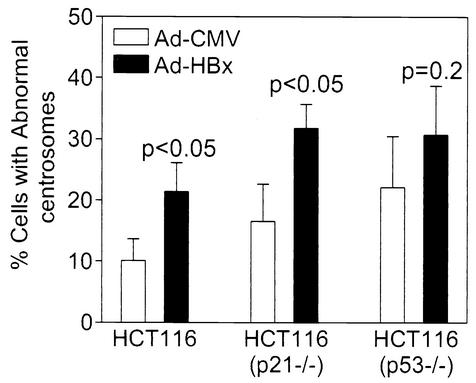
Loss of p53 or p21 can lead to supernumerary centrosomes (6, 17). To test whether induction of supernumerary centrosomes by HBx depends on inactivation of the p53-dependent pathway, wild-type, p53−/−, or p21−/− HCT116 cells were infected with Ad-CMV or Ad-HBx at an MOI of 10. About 10% of wild-type, 15% of p21−/−, and 20% of p53−/− HCT116 cells contained three or more centrosomes when infected with Ad-CMV (Fig. 4). Ad-HBx infection resulted in a statistically significant increase in cells that contained three or more centrosomes in both wild-type and p21−/− cells (P < 0.05), demonstrating that p21 is not required for HBx-induced supernumerary centrosomes. Although Ad-HBx further enhanced centrosome abnormality in p53−/− cells, the difference was statistically insignificant (P = 0.2). Thus, the involvement of p53 in HBx-mediated abnormal centrosomes requires further investigation.
FIG. 4.
HBx induces supernumerary centrosomes in p53- and p21-deficient cells. Cells were infected with control (Ad-CMV) or HBx-containing (Ad-HBx) adenovirus for 24 h and then analyzed essentially as described for Fig. 1. Data were obtained from three independent experiments, and at least 150 cells were analyzed in each experiment. A Student's t test was used to compare Ad-CMV with Ad-HBx, and P values are indicated.
HBV-induced supernumerary centrosomes require functional HBx.
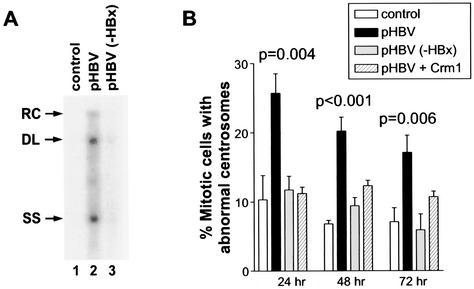
To determine whether the induction of supernumerary centrosomes by ectopic expression of HBx has any physiological relevance, we analyzed centrosomes in a hepatoma cell line (HepG2) transiently transfected with replication-competent wild-type HBV DNA (pHBV) or an HBx-deficient HBV DNA [pHBV(−HBx)]. Consistent with published data (27), pHBV but not pHBV(−HBx) replicated efficiently in HepG2 cells (Fig. 5A). Because HBx preferentially induced supernumerary centrosomes in mitotic cells, we focused our analysis on mitotic HepG2 cells. In control mitotic cells, a total of approximately 10, 7, and 7% of cells with abnormal centrosomes were observed at 24, 48, and 72 h, respectively, after transfection (Fig. 5B). There was a significant increase in mitotic cells with abnormal centrosomes at all three time points in pHBV-transfected cells, but not in pHBV(−HBx)-transfected cells (Fig. 5B). These analyses indicate that induction of supernumerary centrosomes in HepG2 cells by HBV depends on a physiological expression of HBx during HBV replication.
FIG. 5.
Induction of supernumerary centrosomes in HepG2 cells by HBV DNA in an HBx-dependent manner. (A) HBV replication is dependent on HBx expression. Consistent with previously published data (27), high levels of HBV replicative forms (cytoplasmic nucleocapsid particles) can be detected in HepG2 cells transiently transfected with the wild-type HBV construct (pHBV), but not with an HBx-devoid HBV construct, pHBV(−HBx), for 4 days. The relaxed circular (RC), double-stranded linear (DL), and single-stranded (SS) DNA species are indicated by arrows on the left. (B) HepG2 cells were transfected with the pHBV or pHBV(−HBx) construct and incubated for 24, 48, or 72 h. Cells were fixed and analyzed for centrosomes as described for Fig. 1. At least 100 mitotic HepG2 cells were analyzed for each condition in each experiment, and more than two centrosomes in each mitotic cell were considered abnormal centrosomes. Where error bars are shown, data were obtained from three independent experiments. A Student's t test was used to compare the experimental groups to the control, and statistically significant P values are indicated.
Leptomycin B mimics HBx induction of aberrant centriole replication and multipolar spindles.
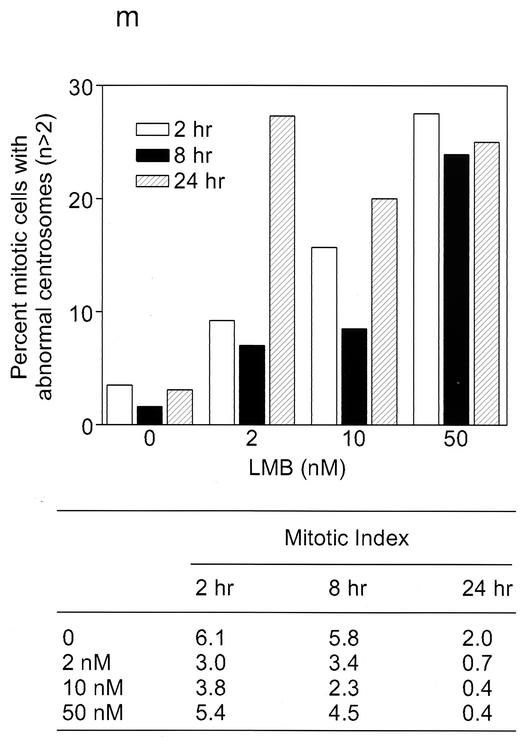
Recently, we showed that wild-type HBx but not a nuclear export signal mutant colocalizes with and sequesters Crm1 in the cytoplasm, thereby inducing NF-κB/IκB nuclear localization (16). Therefore, we next examined whether an adenovirus vector encoding a nuclear export signal mutant of HBx (Ad-NESM) could induce supernumerary centrosomes in NHF cells. Table 1 shows that while wild-type HBx efficiently induced supernumerary centrosomes and abnormal spindles, Ad-NESM was totally devoid of such activity (Table 1). These data raise the possibility that HBx-mediated alteration of Crm1 localization, and hence its activity, may give rise to the observed supernumerary centrosomes and abnormal mitoses. Consistently, induction of supernumerary centrosomes in HepG2 cells by pHBV was effectively blocked by overexpression of Crm1 (Fig. 5B). To further assess whether Crm1 has a role in controlling centrosome number, we treated NHF-hTERT cells with a Crm1-specific inhibitor, leptomycin B. Leptomycin B specifically bound to Crm1, thereby inactivating the Crm1-dependent nuclear export pathway. Treatment of NHF-hTERT cells with leptomycin B resulted in a dose-dependent increase in the percentage of mitotic cells with supernumerary centrosomes (Fig. 6b to d). The effect was observed as early as 2 h.
FIG. 6.
Aberrant centriole replication and formation of multipolar spindles induced by Crm1-specific inhibitor leptomycin B. (a to h) Untreated (a and e) and 50 nM leptomycin B-treated (b to d and f to h) NHF-hTERT cells were fixed at 2 h and then coimmunostained with antibodies specific to either γ-tubulin (green) and α-tubulin (red) (a to d) or γ-tubulin (red) and centrin (green) (e to h). Chromatin was stained with DAPI (blue). A representative single cell image is shown in panels g and h. Magnification, ×630. Arrows indicate centrioles. (i to l) Electron micrographs of thin sections of untreated NHF-hTERT cells with normal centriole pairs (i and j) and leptomycin B-treated cells with abnormal centrioles (arrowheads) (k and l). Bar, 0.5 μm. Three cells with abnormal centrioles were observed among a total of 50 cells surveyed. (m) Mitotic NHF-hTERT cells with abnormal centrosomes (each cell contained more than two centrosomes) were quantified in the absence and presence of various concentrations of leptomycin B (LMB) and at different time points. At least 200 mitotic cells were analyzed. The mitotic index (percent mitotic cells) was also recorded.
We also used a monoclonal anticentrin antibody to determine the status of centrioles after treatment with leptomycin B. Untreated cells always displayed paired centrin signals, which colocalized with γ-tubulin (Fig. 6e), indicating the presence of a single centrosome. In contrast, leptomycin B-treated cells often contained more than four centrin signals (Fig. 6f to h). The centrin signals were frequently well separated, and some were not even associated with γ-tubulin (Fig. 6g to h). Many leptomycin B-treated mitotic cells contained three or more γ-tubulin-positive foci, some of which were associated with either a single or three more centrin signals. These results imply that disruption of a Crm1-dependent process may result in abnormal centriole replication, which in turn leads to abnormal spindles. Electron microscope analysis revealed the presence of extra minicentrioles that were separated from their mother centrioles (Fig. 6k) or abnormally elongated centrioles (Fig. 6l) in leptomycin B-treated NHF-hTERT cells. The effect of leptomycin B did not appear to be the result of a continued centrosome cycle in the presence of cell cycle arrest, because minimal mitotic arrest was observed in cells treated with leptomycin B at early time points (Fig. 6m).
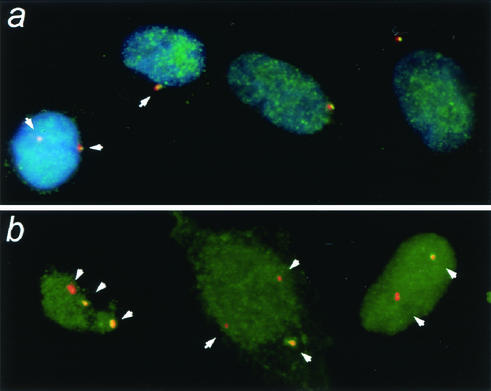
Immunocytochemistry analysis with a polyclonal anti-Crm1 antibody indicated that greater than 90% of Crm1 was localized in the nucleus of NHF cells (data not shown). To test whether a small fraction of Crm1 protein could be found at the centrosomes, we incubated cells with Triton X-100 to remove soluble Crm1 prior to fixation and immunocytochemistry analysis. With this approach, we found a fraction of Crm1 colocalizing with every centrosome in the cells (Fig. 7a). Similarly, the GFP-Crm1 signal, when transiently expressed, was found to be localized with centrosomes (data not shown). In contrast, after treatment with leptomycin B, we often observed cells with centrosomes that lacked any detectable Crm1 signal, particularly in cells with more than two centrosomes (Fig. 7b). Thus, it appears that Crm1 acts directly at the centrosome and that amplified centrosomes are free of its actions.
FIG. 7.
Fraction of Crm1 protein is located with centrosomes and is disrupted in leptomycin B-treated cells. (a) Untreated NHF-hTERT cells; (b) NHF-hTERT cells treated with 10 nM leptomycin B for 2 h. Cells were briefly incubated with Triton X-100 to remove soluble proteins as described in Materials and Methods, fixed, and coimmunostained with polyclonal anti-Crm1 antibodies (green) and monoclonal anti-γ-tubulin antibody (red). Representative cells are shown. Magnification, ×630.
Increased frequency of aneuploidy in HBV-positive liver tissues.
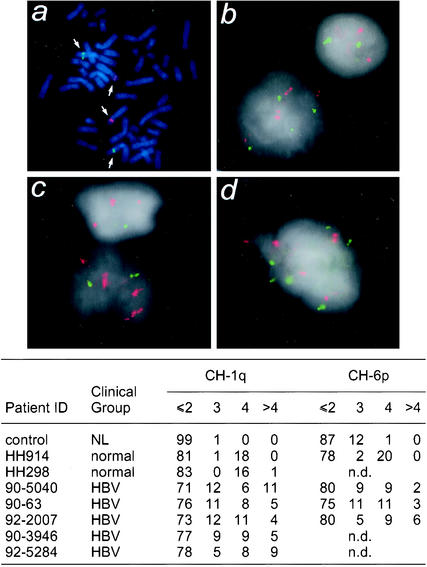
Due to the observed defects in centrosome number and mitotic spindles, we used interphase fluorescence in situ hybridization to identify changes in chromosome number in liver cells from patients chronically infected with HBV. Because the gain of a hybridization signal can be quantitated with more confidence than a loss, we selected chromosomes 1 and 6 as targets due to their preferential gain in HBV-related hepatocellular carcinoma. Cultured lymphocytes from a healthy individual (NL) were used to verify the efficiency of probe hybridization to mitotic spreads (Fig. 8a) and interphase cells (Fig. 8b). Touch preparations of liver tissues from two healthy donors and five chronically HBV-infected patients were included for this analysis. Interphase cells with strong hybridization signals in areas with minimal background from connective tissue were randomly selected, and the number of fluorescent signals (red for chromosome 6 and green for chromosome 1) in 100 cells was quantified for each sample.
FIG. 8.
Increased frequency in gains of both chromosomes 1 and 6 associated with chronically hepatitis B virus-infected liver samples. Metaphase spreads of normal lymphocytes were used as a control to determine the efficiency of fluorescence in situ hybridization and to show that the bacterial artificial chromosome probes hybridized to desired chromosomes 1q (green) and 6p (red) (a). Touch preparations from two normal livers (b) and five HBV-positive liver samples (c and d) were then hybridized with the bacterial artificial chromosome clones. A total of 100 cells were analyzed for each sample, and the number of copies of chromosomes 1q and 6p per cell was determined and is summarized in the table. n.d., not determined.
In this analysis, a normal diploid cell (2N) would contain two fluorescent signals and a tetraploid cell (4N) would contain four fluorescent signals. Cells containing more than four fluorescent signals were scored as a gain. A cell containing three fluorescent signals may also be aneuploid or tetraploid, with a fourth signal being obscured. Almost all of the NL cells and >80% of the hepatocytes from two normal liver donors were 2N, with the remainder being primarily 4N. Aneuploidy was rarely (1%) detected in these cells. In contrast, an increased frequency of aneuploidy (>4 copies of fluorescent signals) was detected in all five HBV samples (2 to 11%), with some cells containing more than 10 copies of chromosome 1 or 6. These data indicate that aneuploidy for chromosomes 1 and 6 occurs in HBV-infected liver cells prior to the development of hepatocellular carcinoma.
DISCUSSION
We have shown that the hepatitis B virus-encoded HBx gene is able to induce supernumerary centrosomes and multipolar spindles. This is reminiscent of the activity by the oncogenic papillomavirus-encoded E6 and E7 proteins (12). Because most human cancers show centrosome abnormalities (25) and E7-induced abnormal centrosome synthesis is an early event in the evolving malignant phenotype (11), abnormal amplification of centrosomes and structural alteration of centrosomes may be early events during human carcinogenesis. Our results support this model, as we showed that oncogenic HBx can induce multipolar spindles and aneuploidy. Moreover, aneuploidy can be detected in livers chronically infected with HBV, a preneoplastic condition predisposing individuals to hepatocellular carcinoma. It should be noted that we have not demonstrated abnormal centrosomes in HBV-positive liver tissues due to technical difficulties. The connection between the observed aneuploidy in premalignant liver tissues and an in vitro effect of HBV-induced multipolar spindles is only correlative and requires further investigation.
Our studies revealed a novel role for the Crm1 nuclear export receptor in regulating the fidelity of mitotic centrosome maintenance. We show that wild-type HBx, but not a nuclear export-deficient mutant, was able to induce centrosome abnormalities in mitotic cells, as illustrated by abnormal centrioles and the formation of multipolar spindles. Similarly, a Crm1-specific inhibitor, leptomycin B, resembled HBx in its ability to induce such centrosome abnormalities. Furthermore, colocalization of Crm1 and γ-tubulin in the pericentriolar matrix appeared to be disrupted by leptomycin B treatment. Our results are consistent with the model that Crm1 may act either alone or together with Ran and other nuclear export signal-containing cellular factors to maintain the fidelity of centrosome duplication.
Although it has been demonstrated that the centrosome is capable of duplicating itself independent of cell cycle transition (10), it can only duplicate once per cell cycle under normal conditions. This process is initiated by splitting of mother and daughter centrioles, most likely through phosphorylation by cyclin-dependent kinases (10). This unique ability implies that there are intricate mechanisms to inhibit unscheduled centriole synthesis and splitting throughout the cell cycle except at the G1 to S transition. Nucleophosmin was identified recently as a candidate to inhibit centriole splitting in the G0/G1 phase (32). It is plausible that different inhibitors may be responsible for preventing unwanted centriole synthesis or splitting in newly duplicated centrosomes. Our data showed that inactivation of Crm1 can lead to abnormal centriole synthesis and multipolar mitoses. Consistently, electron microscope analysis indicated that many leptomycin B-treated cells contain extra minicentrioles, as if they had been synthesized prematurely and split from their mother centrioles prior to completion of synthesis. Moreover, many of the centrioles in leptomycin B-treated cells did not appear to contain γ-tubulin, a part of the pericentriolar matrix. Taken together, these data imply that Crm1 plays an essential role in preventing extra centriole synthesis and possibly splitting and unscheduled centrosome duplication, thus ensuring the fidelity of chromosome segregation.
Because Crm1 is involved in nuclear transport of many cellular proteins, including cell cycle regulators and transcription factors, it is possible that HBx or leptomycin B can induce multipolar spindles by disrupting Crm1-dependent transport of cellular proteins involved in transduction of cell cycle signals or gene expression. However, recent studies have revealed a direct role of most of the Ran-associated proteins in spindle assembly that is independent of protein transport (9). For example, Ran is a typical GTPase that cycles between the GTP-bound and GDP-bound states, principally through the actions of cytoplasmic RanBP1 and nuclear chromatin-bound RCC1 (9). In addition to its role in nucleocytoplasmic transport, Ran and its GTPase-related proteins (including RanGAP1, RCC1, and RanBP1) are also essential for microtubule assembly and the formation of normal mitotic spindles (9). These two independent cellular processes may both rely on Ran and Ran-associated proteins.
It is known that when bound to Ran-GTP, importin α and β do not bind to nuclear localization signal proteins. On the contrary, the Crm1 export receptor only has high affinity for the nuclear export signal proteins when bound to Ran-GTP. These features led to the hypotheses that spindle assembly may be initiated either by the formation of a complex of Crm1-Ran-nuclear export signal proteins or by the dissociation of import receptors and Ran from their nuclear localization signal proteins (19). Recent studies indicate that importin α and β can negatively regulate nuclear localization signal-containing proteins such as TPX2 and NuMA to modulate microtubule assembly (19, 29). In light of the recent findings as well as the data presented here, we propose the following working model. The formation of functional bipolar spindles requires both a positive regulatory signal for microtubule nucleation by the Ran-GTP/importin/nuclear localization signal complex and the inhibition of centriole splitting by the Ran-GTP/Crm1/nuclear export signal complex. Analogous to nuclear localization signal-containing proteins, such as TPX2 and NuMA, that participate in microtubule nucleation, we speculate that there are specific nuclear export signal-containing proteins that may be responsible for Crm1-mediated negative regulation of centrosome synthesis. Together, these two steps ensure the fidelity of bipolar spindle assembly, and the loss of its control may lead to chromosomal instability, aneuploidy, and tumorigenesis.
Acknowledgments
We are grateful to Judy Campisi for the NHF-hTERT cells, Bert Vogelstein for the p21- and p53-deficient HCT116 cells, Jeffery Salisbury for the anticentrin antibody, Gerald Grosveld for the anti-Crm1 rabbit polyclonal antibody, and Robert Schneider for the pHBV and pHBV(−HBx) constructs. We thank Patrizia Lavia and Taro Yamashita for stimulatory discussion and sharing unpublished data. We thank Curtis Harris and Eric Huang for critical reading of the manuscript and Susan Garfield and Alice Uy for technical support. We also thank Dorothea Dudek for editorial support and Karen MacPherson for bibliographic assistance.
This work was supported by the Intramural Research Program from the Center for Cancer Research, National Cancer Institute, and in part by NIH/NCI grant CA90522 to K.F.
REFERENCES
- 1.Arbuthnot, P., A. Capovilla, and M. Kew. 2000. Putative role of hepatitis B virus X protein in hepatocarcinogenesis: effects on apoptosis, DNA repair, mitogen-activated protein kinase and JAK/STAT pathways. J. Gastroenterol. Hepatol. 15:357-368. [DOI] [PubMed] [Google Scholar]
- 2.Benn, J., and R. J. Schneider. 1995. Hepatitis B virus HBx protein deregulates cell cycle checkpoint controls. Proc. Natl. Acad. Sci USA 92:11215-11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bouchard, M. J., L. H. Wang, and R. J. Schneider. 2001. Calcium signaling by HBx protein in hepatitis B virus DNA replication. Science 294:2376-2378. [DOI] [PubMed] [Google Scholar]
- 4.Brechot, C., D. Gozuacik, Y. Murakami, and P. Paterlini-Brechot. 2000. Molecular bases for the development of hepatitis B virus (HBV)-related hepatocellular carcinoma (hepatocellular carcinoma). Semin. Cancer Biol. 10:211-231. [DOI] [PubMed] [Google Scholar]
- 5.Brinkley, B. R. 2001. Managing the centrosome numbers game: from chaos to stability in cancer cell division. Trends Cell Biol. 11:18-21. [DOI] [PubMed] [Google Scholar]
- 6.Bunz, F., A. Dutriaux, C. Lengauer, T. Waldman, S. Zhou, J. P. Brown, J. M. Sedivy, K. W. Kinzler, and B. Vogelstein. 1998. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science 282:1497-1501. [DOI] [PubMed] [Google Scholar]
- 7.Butel, J. S. 2000. Viral carcinogenesis: revelation of molecular mechanisms and etiology of human disease. Carcinogenesis 21:405-426. [DOI] [PubMed] [Google Scholar]
- 8.Carazo-Salas, R. E., G. Guarguaglini, O. J. Gruss, A. Segref, E. Karsenti, and I. W. Mattaj. 1999. Generation of GTP-bound Ran by RCC1 is required for chromatin-induced mitotic spindle formation. Nature 400:178-181. [DOI] [PubMed] [Google Scholar]
- 9.Dasso, M. 2001. Running on Ran: nuclear transport and the mitotic spindle. Cell 104:321-324. [DOI] [PubMed] [Google Scholar]
- 10.Doxsey, S. 2001. Re-evaluating centrosome function. Nat. Rev. Mol. Cell Biol. 2:688-698. [DOI] [PubMed] [Google Scholar]
- 11.Duensing, S., A. Duensing, C. P. Crum, and K. Munger. 2001. Human papillomavirus type 16 E7 oncoprotein-induced abnormal centrosome synthesis is an early event in the evolving malignant phenotype. Cancer Res. 61:2356-2360. [PubMed] [Google Scholar]
- 12.Duensing, S., L. Y. Lee, A. Duensing, J. Basile, S. Piboonniyom, S. Gonzalez, C. P. Crum, and K. Munger. 2000. The human papillomavirus type 16 E6 and E7 oncoproteins cooperate to induce mitotic defects and genomic instability by uncoupling centrosome duplication from the cell division cycle. Proc. Natl. Acad. Sci. USA 97:10002-10007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elmore, L. W., A. R. Hancock, S. F. Chang, X. W. Wang, S. Chang, C. P. Callahan, D. A. Geller, H. Will, and C. C. Harris. 1997. Hepatitis B virus X protein and p53 tumor suppressor interactions in the modulation of apoptosis. Proc. Natl. Acad. Sci. USA 94:14707-14712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feitelson, M. A. 1999. Hepatitis B virus in hepatocarcinogenesis. J. Cell Physiol. 181:188-202. [DOI] [PubMed] [Google Scholar]
- 15.Fodde, R., J. Kuipers, C. Rosenberg, R. Smits, M. Kielman, C. Gaspar, J. H. van Es, C. Breukel, J. Wiegant, R. H. Giles, and H. Clevers. 2001. Mutations in the APC tumour suppressor gene cause chromosomal instability. Nat. Cell Biol. 3:433-438. [DOI] [PubMed] [Google Scholar]
- 16.Forgues, M., A. J. Marrogi, E. A. Spillare, C. G. Wu, M. Yoshida, and X. W. Wang. 2001. Interaction of the hepatitis b virus x protein with the Crm1-dependent nuclear export pathway. J. Biol. Chem. 276:22797-22803. [DOI] [PubMed] [Google Scholar]
- 17.Fukasawa, K., T. Choi, R. Kuriyama, S. Rulong, and G. F. Vande Woude. 1996. Abnormal centrosome amplification in the absence of p53. Science 271:1744-1747. [DOI] [PubMed] [Google Scholar]
- 18.Funabiki, H., I. Hagan, S. Uzawa, and M. Yanagida. 1993. Cell cycle-dependent specific positioning and clustering of centromeres and telomeres in fission yeast. J. Cell Biol. 121:961-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gruss, O. J., R. E. Carazo-Salas, C. A. Schatz, G. Guarguaglini, J. Kast, M. Wilm, N. Le Bot, I. Vernos, E. Karsenti, and I. W. Mattaj. 2001. Ran induces spindle assembly by reversing the inhibitory effect of importin alpha on TPX2 activity. Cell 104:83-93. [DOI] [PubMed] [Google Scholar]
- 20.Guarguaglini, G., L. Renzi, F. D'Ottavio, B. Di Fiore, M. Casenghi, E. Cundari, and P. Lavia. 2000. Regulated Ran-binding protein 1 activity is required for organization and function of the mitotic spindle in mammalian cells in vivo. Cell Growth Differ. 11:455-465. [PubMed] [Google Scholar]
- 21.Hollander, M. C., M. S. Sheikh, D. V. Bulavin, K. Lundgren, L. Augeri-Henmueller, R. Shehee, T. A. Molinaro, K. E. Kim, E. Tolosa, J. D. Ashwell, M. P. Rosenberg, Q. Zhan, P. M. Fernandez-Salguero, W. F. Morgan, C. X. Deng, and A. J. Fornace, Jr. 1999. Genomic instability in Gadd45a-deficient mice. Nat. Genet. 23:176-184. [DOI] [PubMed] [Google Scholar]
- 22.Kaplan, K. B., A. A. Burds, J. R. Swedlow, S. S. Bekir, P. K. Sorger, and I. S. Nathke. 2001. A role for the adenomatous polyposis coli protein in chromosome segregation. Nat. Cell Biol. 3:429-432. [DOI] [PubMed] [Google Scholar]
- 23.Kew, M. C. 1997. Increasing evidence that hepatitis B virus X gene protein and p53 protein may interact in the pathogenesis of hepatocellular carcinoma. Hepatology 25:1037-1038. [DOI] [PubMed] [Google Scholar]
- 24.Koike, K. 1995. Hepatitis B virus HBx gene and hepatocarcinogenesis. Intervirology 38:134-142. [DOI] [PubMed] [Google Scholar]
- 25.Lingle, W. L., and J. L. Salisbury. 2000. The role of the centrosome in the development of malignant tumors. Curr. Top. Dev. Biol. 49:313-329. [DOI] [PubMed] [Google Scholar]
- 26.Matsumoto, T., and D. Beach. 1991. Premature initiation of mitosis in yeast lacking RCC1 or an interacting GTPase. Cell 66:347-360. [DOI] [PubMed] [Google Scholar]
- 27.Melegari, M., P. P. Scaglioni, and J. R. Wands. 1998. Cloning and characterization of a novel hepatitis B virus x binding protein that inhibits viral replication. J. Virol. 72:1737-1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murakami, S. 2001. Hepatitis B virus X protein: a multifunctional viral regulator. J. Gastroenterol. 36:651-660. [DOI] [PubMed] [Google Scholar]
- 29.Nachury, M. V., T. J. Maresca, W. C. Salmon, C. M. Waterman-Storer, R. Heald, and K. Weis. 2001. Importin beta is a mitotic target of the small GTPase Ran in spindle assembly. Cell 104:95-106. [DOI] [PubMed] [Google Scholar]
- 30.Nigg, E. A. 2002. Centrosome aberrations: cause or consequence of cancer progression? Nat. Rev. Cancer 2:815-825. [DOI] [PubMed] [Google Scholar]
- 31.Ohba, T., M. Nakamura, H. Nishitani, and T. Nishimoto. 1999. Self-organization of microtubule asters induced in Xenopus egg extracts by GTP-bound Ran. Science 284:1356-1358. [DOI] [PubMed] [Google Scholar]
- 32.Okuda, M., H. F. Horn, P. Tarapore, Y. Tokuyama, A. G. Smulian, P. K. Chan, E. S. Knudsen, I. A. Hofmann, J. D. Snyder, K. E. Bove, and K. Fukasawa. 2000. Nucleophosmin/B23 is a target of CDK2/cyclin E in centrosome duplication. Cell 103:127-140. [DOI] [PubMed] [Google Scholar]
- 33.Sluder, G., and E. H. Hinchcliffe. 1999. Control of centrosome reproduction: the right number at the right time. Biol. Cell 91:413-427. [PubMed] [Google Scholar]
- 34.Tutt, A., A. Gabriel, D. Bertwistle, F. Connor, H. Paterson, J. Peacock, G. Ross, and A. Ashworth. 1999. Absence of Brca2 causes genome instability by chromosome breakage and loss associated with centrosome amplification. Curr. Biol. 9:1107-1110. [DOI] [PubMed] [Google Scholar]
- 35.Wang, X. W., M. K. Gibson, W. Vermeulen, H. Yeh, K. Forrester, H. W. Sturzbecher, J. H. J. Hoeijmakers, and C. C. Harris. 1995. Abrogation of p53-induced apoptosis by the hepatitis B virus X gene. Cancer Res. 55:6012-6016. [PubMed] [Google Scholar]
- 36.Wilde, A., and Y. Zheng. 1999. Stimulation of microtubule aster formation and spindle assembly by the small GTPase Ran. Science 284:1359-1362. [DOI] [PubMed] [Google Scholar]
- 37.Wu, C. G., M. Forgues, S. Siddique, J. Farnsworth, K. Valerie, and X. W. Wang. 2002. SAGE transcript profiles of normal primary human hepatocytes expressing oncogenic hepatitis B virus X protein. FASEB J. 16:1665-1667. [DOI] [PubMed] [Google Scholar]
- 38.Wu, C. G., D. M. Salvay, M. Forgues, K. Valerie, J. Farnsworth, R. S. Markin, and X. W. Wang. 2001. Distinctive gene expression profiles associ-ated with hepatitis B virus x protein. Oncogene 20:3674-3682. [DOI] [PubMed] [Google Scholar]
- 39.Xu, X., Z. Weaver, S. P. Linke, C. Li, J. Gotay, X. W. Wang, C. C. Harris, T. Ried, and C. X. Deng. 1999. Centrosome amplification and a defective G2-M cell cycle checkpoint induce genetic instability in BRCA1 exon 11 isoform-deficient cells. Mol. Cell 3:389-395. [DOI] [PubMed] [Google Scholar]
- 40.Yeh, C. T. 2000. Hepatitis B virus X protein: searching for a role in hepatocarcinogenesis. J. Gastroenterol. Hepatol. 15:339-341. [DOI] [PubMed] [Google Scholar]