Abstract
In Lactobacillus johnsonii strain NCC533, two prophages were integrated into tRNA genes and one was disrupted by integration. In a survey, the prophages were restricted to strains sharing an essentially identical restriction pattern. Microarray analysis showed that the prophage DNA represents about 50% of the NCC533 strain-specific DNA.
In many bacteria, genome sequencing identified prophages as a prominent part of the laterally acquired DNA. About two-thirds of the sequenced γ-proteobacteria and low-GC-content gram-positive bacteria contained one—many, even multiple—prophage (9). Further observations suggested an important role of prophages for bacterial genomes. Many prophages from bacterial pathogens encoded important virulence factors (4). In several pathogenic bacteria, sequencing of multiple strains identified prophages as a major contributor to the strain-specific DNA (1, 16). Much less is known about the role of prophages in the genomes from nonpathogenic bacteria. This is also true for the economically important lactic acid bacteria, which are otherwise well investigated for their phages (6). This group contains bacterial starters like Lactococcus lactis, which is used in the food industry; Lactobacillus commensals of mucosal surfaces (e.g., gut and vagina), which are explored for their probiotic (i.e., health promoting) activities; and important human pathogens like Streptococcus pyogenes, a paradigm for the role of prophages in the evolution of pathogenic bacteria (1, 2, 14, 17). To fill this gap, we provided partial genome sequences for prophages from the gut commensal Lactobacillus johnsonii (12). The completion of the genome sequencing of the L. johnsonii strain NCC533 (D. Pridmore et al., unpublished data) now allows a more detailed analysis of these prophages. In the present report, we investigated the integration and distribution of these prophages in different strains from L. johnsonii.
Prophage integration sites.
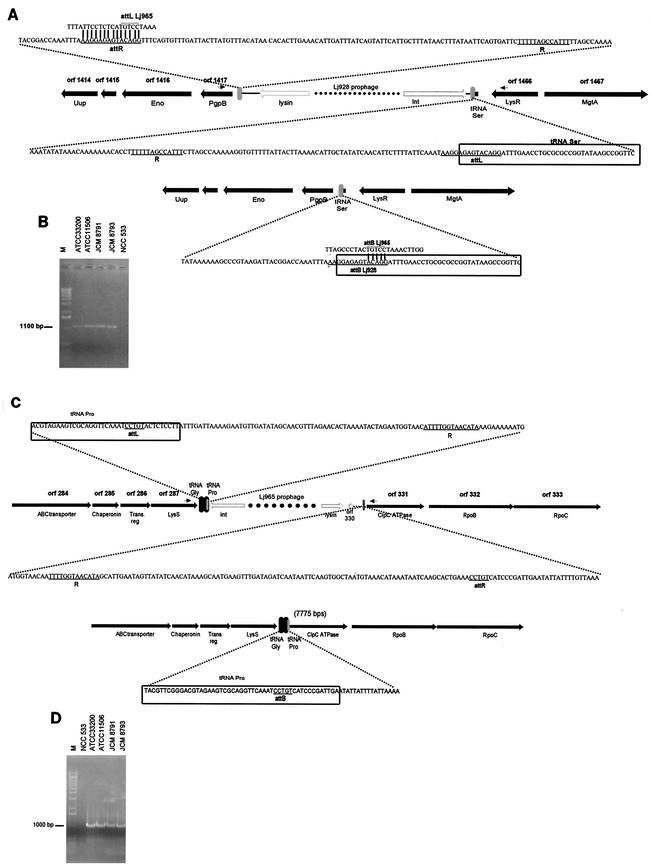
Prophage Lj928 is located between an anonymous gene (i.e., open reading frame [ORF] 1417, or Ljo_1417, on the NCC533 genome map) on one side and a tRNASer gene followed by a transcriptional regulator (lysR) gene on the other side (Fig. 1A). Lj928 is flanked by a 14-bp and a 13-bp repeat. Most L. johnsonii strains from our collection amplified a 1.1-kb PCR product when primers were placed in the bacterial genes that bracket the prophage in NCC533 (Fig. 1B). This PCR product contained the 14-bp sequence that overlaps the 3′ end of a tRNASer gene but did not contain the 13-bp sequence. The sequences flanking the deduced attB were identical to those to the left and the right, respectively, of the 14-bp repeats, thus identifying them as the likely attL and attR sites (Fig. 1A). The data suggest a Campbell-like integration of prophage Lj928 into a tRNASer gene that is functionally reconstituted upon prophage integration.
FIG.1.
The integration sites of prophages Lj928 and Lj965. (A) Bacterial genes flanking the Lj928 prophage. The predicted host genes are depicted by black arrows and are annotated with their NCC533 genome ORF number and a short description. The outmost prophage lysin and integrase genes are depicted by white arrows. A gray vertical bar represents the deduced attachment sites attL and attR; their nucleotide sequences are provided in an enlarged insert. The core sequence of the attL and attR sites are underlined, as is the 13-bp repeat R. The corresponding sequence of the attB site from a L. johnsonii strain lacking Lj928 is provided below. The boxed sequence indicates part of the tRNA gene. The small arrows provide the locations of the primers for PCR. (B) PCR amplifications of the attB site for Lj928 from the indicated L. johnsonii strains. Lane M, 1-kb DNA ladder (Gibco BRL). Primers placed within Lj928 served as positive control for NCC533 DNA (data not shown). (C) The bacterial DNA flanking the Lj965 prophage integration site. See panel A for annotations and note the different orientation of the two prophages on the bacterial chromosome. (D) PCR of the attB site from the indicated L. johnsonii strains. The accession numbers for the flanking bacterial DNA sequences are AY320376 to AY320379.
Prophage Lj965 is located between a lysyl-tRNA synthetase (lysS) and two tRNA genes (tRNAGly and tRNAPro) on one side and a bacterial clpC ATPase gene on the other side (Fig. 1C). Lj965 is flanked by a 13-bp repeat. By using primers placed in the bacterial genes flanking the prophage, we amplified a 1-kb-long DNA segment from most L. johnsonii strains (Fig. 1D). However, we did not detect the 13-bp sequence when the PCR product was sequenced. As in the case of Lj928, the 13-bp repeat is thus part of the phage DNA. The longest conserved DNA sequence between both prophage-flanking sites and the PCR product was a 5-bp repeat in the 3′ end of the tRNAPro gene. The sequences to the right and left of this deduced attB were identical to those abutting the likely attL and attR sites. Four observations are noteworthy: first, the short 5-bp attachment core sequence deduced for Lj965 is part of the 14-bp core sequence deduced for Lj928 (Fig. 1A). Second, the integrases of both prophages share 90% amino acid identity. Third, the region around the deduced Lj965 attL—but not the attR site—shared sequence identity with the entire 14-bp core sequence of Lj928 (Fig. 1A). Fourth, while Lj928 reconstituted a tRNASer gene, Lj965 integration changed the acceptor stem of the tRNAPro gene by complementing it via a tRNASer-specific sequence.
Upon prophage induction, one would expect an excised and circularized phage genome as well as a PCR product with primers running out of the int and lys genes. However, we observed no such PCR product for either prophage with total NCC533 DNA.
Distribution of prophages.
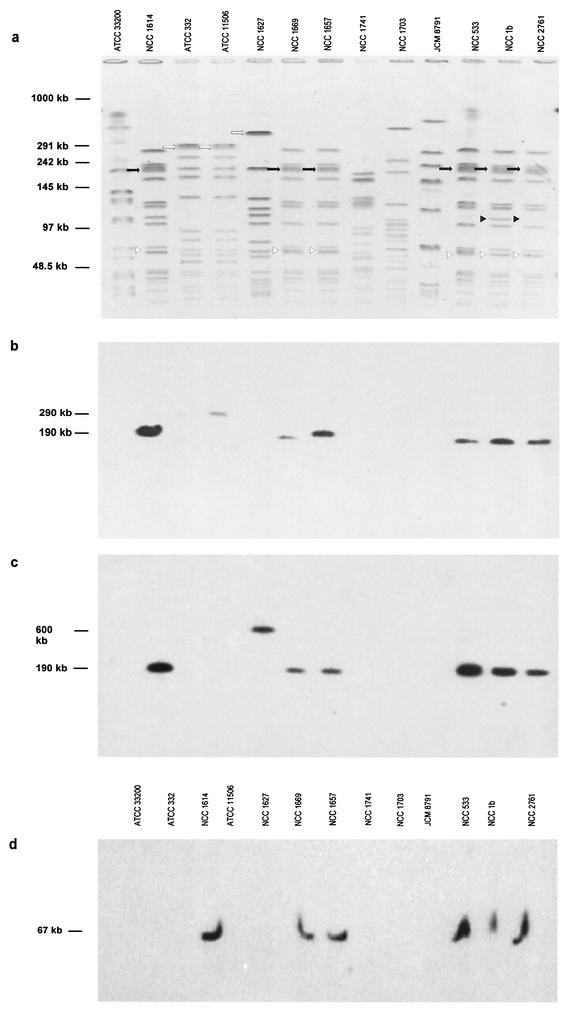
Thirteen L. johnsonii strains, including a human blood isolate, an ATCC strain that is the type strain for the species, as well as isolates from mouse, pig, calf, human and chicken feces, from calf stomach, and from dairy products were studied for their restriction patterns. Agarose-embedded bacterial cells were treated with 50 U of SmaI (20). Pulsed-field gel electrophoresis (PFGE) (Fig. 2a) was performed by a contour-clamped homogenous electric field mode in a CHEF-DRII apparatus (Bio-Rad, Richmond, Calif.; 20 h at 6 V/cm; ramping pulse time from 1 to 20 s). Two strains showed a pattern identical to that of NCC533, three strains differed by a single restriction band, and six strains displayed a clearly distinct SmaI pattern. Southern hybridization showed that strains with similar or identical SmaI patterns carried both prophages, while we detected neither prophage in all strains with different SmaI patterns (Fig. 2b to d). A weak 290-kb hybridization signal was also obtained in strain ATCC 11506 with the first Lj965 probe, while the second Lj965 probe yielded a strong hybridization signal at 600 kb in strain NCC1627 (Fig. 2b and c). Although the Lj965 and Lj928 prophages may be restricted to NCC533-like strains, it appears that individual genes from Lj965 are also found in molecularly distinct L. johnsonii strains.
FIG.2.
Pulsed-field gels and hybridization. (a) PFGE of SmaI-digested DNA from the indicated L. johnsonii strains stained with ethidium bromide (negative of photo). The molecular size of the DNA molecular markers is given. Corresponding Southern hybridization with radiolabeled PCR probes from Ljo_294 to Ljo_295 of prophage Lj965 (b), Ljo_292 of prophage Lj965 (c), and Ljo_1456 and Ljo_1455 of prophage Lj928 (d). The restriction fragments containing prophages Lj965 and Lj928 are indicated with a black arrow and white triangle, respectively. The black triangle indicates a distinct restriction band in NCC533-like strains. Bands cross-hybridizing with specific Lj965 probes are indicated with white arrows.
Since the prophages in the five NCC533-related strains are found on SmaI fragments of the same size as those in strain NCC533, they are probably located at the same sites in all of those strains. PCR across the attL sites of both prophages concurred with this suggestion (data not shown). Since we lack evidence that either of these prophages has ever been excised from a bacterial chromosome after having integrated, it is plausible to anticipate that the integrations in strain NCC533 occurred after it had diverged from the other L. johnsonii strains that were tested for the presence of the prophages. Sequence comparisons between the six strains that carry both prophages could give an estimate of how much evolutionary time has passed since they diverged and thus a minimum estimate of how long prophages can remain stably integrated. Preliminary random sequencing of strains sharing the same PFGE profile with NCC533 revealed less than 1% base pair differences (data not shown), thus suggesting that they were all derived from a relatively recent common ancestor.
Half of the L. johnsonii strain-specific DNA is prophage DNA.
During the first round of NCC533 sequencing, a tiling program was designed to select a set of 1,750 pUC18 clones (median insert length, 1.3 kb) that covered, with minimal overlap, 85% of the NCC533 genome. Nonclonable DNA segments and well-conserved DNA sequences coding for tRNA, rRNA, and ribosomal proteins were not represented. PCR amplicons were prepared for all selected clones by using the universal vector primers (Robo Amp 42000; MWG-Biotech) and then spotted in duplicates on a nylon Hybond N+ membrane (Amersham, Dübendorf, Germany), which also contained negative controls. Genomic DNA from NCC533 and the eight L. johnsonii strains indicated in Fig. 3 was digested with SalI and labeled with [32P]dCTP by randomly primed DNA labeling (Boehringer Mannheim GmbH). Prehybridization and hybridization of the filters were carried out at 42°C in 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate at pH 7), 5 mM EDTA, 0.1% sodium dodecyl sulfate (SDS), 2.5% skimmed milk, and 50% formamide. Following 18 h of hybridization, the membrane was rinsed twice (20 min) at room temperature in 2× SSC-0.1% SDS, twice (20 min) at 60°C in 0.1% SSC-0.1% SDS, and exposed to X-OMAT autoradiography film (Eastman Kodak Co., Rochester, N.Y.).
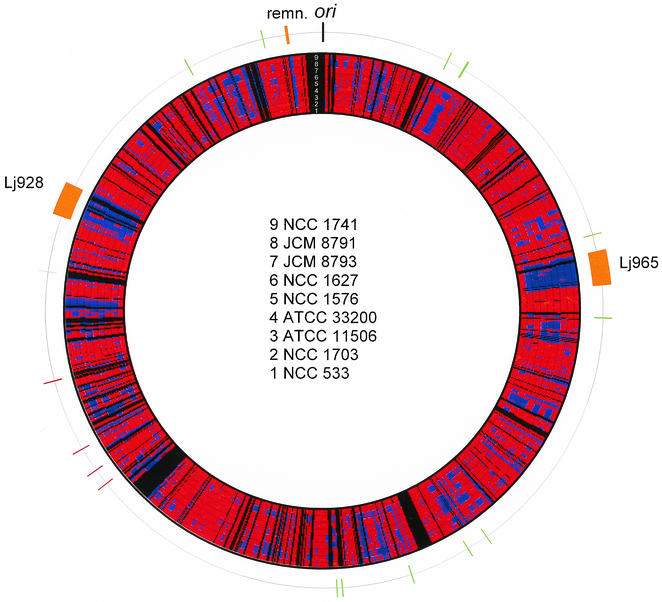
FIG. 3.
Microarray analysis. Graphical representation of the genome comparison of eight different L. johnsonii strains with the NCC533 genome (innermost ring) derived from DNA microarray hybridization experiments. The numbers in the concentric rings at the top delineate the eight different L. johnsonii strains identified in the center of the figure. The red and blue ring segments, respectively, indicate genome regions identified as present or absent in the investigated strains when projected on the NCC533 genome map. Black areas indicate NCC533 genome regions that were not represented on the arrays used in these experiments. The outermost ring shows the location of the likely origin of replication (black), two prophages and a likely mobile DNA remnant (orange), transposase (green), and integrase genes (red) in the genome of L. johnsonii NCC533.
The autoradiographies were scanned and analyzed with Imagene 3.0 (BioDiscovery, Inc.) software. As a positive control, the membrane was hybridized against homologous NCC533 DNA. Ninety-five percent of a total of 3,791 spots could be evaluated. Sixty-one percent of these spots were conserved over all the strains investigated, while 32% were shared with only part of the other L. johnsonii strains. Under the chosen hybridization conditions, 7% were NCC533-specific spots. From the lengths of the PCR products loaded on the microarrays, we calculated that the NCC533-specific DNA consists of 140 kb; the total genome size is about 2 Mb. The two prophages represent 79 kb of DNA, i.e., 4% of the NCC533 genome. Having analyzed the prophage sequences that were found in a patchwise fashion in the other test strains, we estimated that 65 kb of the NCC533-specific DNA is provided by prophage DNA. However, this figure is approximate because overlapping segments of the target DNA made a precise calculation difficult.
A color-coded projection of the microarray hybridization results from the eight L. johnsonii strains on the NCC533 genome is presented in Fig. 3. Two large blue segments of NC533 strain-specific DNA were represented by prophages Lj965 and Lj928. Numerous smaller strain-specific DNA segments were observed; two relatively prominent segments were detected at the 9- and 12 o'clock map positions from NCC533 (Fig. 3). The second segment is a likely mobile DNA element—it is associated with an integrase gene (Ljo_1773), accompanied by a phage-like repressor (Ljo_1779), and flanked by a 13-bp repeat separated by 5.7 kb (data not shown). PCR analysis revealed that all strains with a distinct PFGE pattern from NCC533 lacked the 5.7-kb segment and displayed a single 13-bp and identical flanking sequences (data not shown). This observation suggests that a Campbell-type integration process has introduced this mobile DNA element, possibly as a prophage that subsequently suffered massive DNA loss. None of the other NCC533-specific genome regions was associated with transposases or integrases (Fig. 3). Three of the four integrases not linked to mobile DNA in NCC533 resembled XerC,D-like recombinases from L. gasseri (89 to 94% amino acid identity) but were only distantly related to each other.
Conclusions.
With respect to prophage-host interaction, our investigations revealed a number of interesting parallels between L. johnsonii and S. pyogenes, a lactic acid bacterium that has been intensively studied for this relationship (1, 2, 14, 17). As the two host bacteria are phylogenetically related, it might not be surprising that the prophages from both species were closely related in their genetic organization (11, 12). In addition, prophages are a quantitatively important element of mobile DNA in the two bacterial genomes and, in both species, prophages constitute an important part of the strain-specific DNA (17). Furthermore, genes that were not linked to any known phage functions were transcribed from the prophages, thus suggesting potential lysogenic conversion genes (e.g., superantigens in S. pyogenes [5] and tRNA in L. johnsonii [Ventura et al., submitted]). Finally, some S. pyogenes prophages also integrated into tRNA genes (15), but this is a characteristic shared with many prophages (8). However, the lack of reconstitution of an intact tRNA gene by Lj965 sequences is unusual. This host gene inactivation by prophage integration (negative lysogenic conversion) might be the consequence of Lj965 using a secondary attachment site for integration. Interestingly, both NCC533 prophages carry tRNA genes that might compensate for disruptive integration into tRNA genes. Lj965 carries four tRNA genes but, notably, not a tRNAPro gene.
In several important animal (E. coli and Salmonella) (10, 16) and plant (Xyllela) bacterial pathogens (18) as well as in the investigated Lactobacillus commensal, prophage DNA turned out to represent a major part of the DNA that distinguishes strains in a species (17, 18). Since many prophages from nonpathogenic bacteria also encode extra genes that are transcribed in the lysogenic state (3, 19), prophage DNA is probably a major potential target for selection forces that influence the strain composition in a given environment. It was postulated that specific prophage combinations can contribute specific constellations of lysogenic conversion genes (13) that are of selective value to a given strain under a specific ecological situation regardless of whether the bacterium is a pathogen, a commensal, or a free-living organism (7). Epidemiological data suggested selection of S. pyogenes clones with specific prophage constellation over the last decades (1). The data from L. johnsonii also suggest dynamic prophage-host interaction. The residence time of the two investigated prophages cannot be long, since they were only detected in strains sharing an essentially identical PFGE pattern.
Acknowledgments
We thank the Swiss National Science foundation for the financial support of Marco Ventura and Carlos Canchaya (Research grant 5002 to 057832).
We also thank Anne Bruttin for reading the manuscript.
REFERENCES
- 1.Banks, D. J., S. B. Beres, and J. M. Musser. 2002. The fundamental contribution of phages to GAS evolution, genome diversification and strain emergence. Trends Microbiol. 10:515-521. [DOI] [PubMed] [Google Scholar]
- 2.Beres, S. B., G. L. Sylva, K. D. Barbian, B. Lei, J. S. Hoff, N. D. Mammarella, M. Y. Liu, J. C. Smoot, S. F. Porcella, L. D. Parkins, D. S. Campbell, T. M. Smith, J. K. McCormick, D. Y. Leung, P. M. Schlievert, and J. M. Musser. 2002. Genome sequence of a serotype M3 strain of group A Streptococcus: phage-encoded toxins, the high-virulence phenotype, and clone emergence. Proc. Natl. Acad. Sci. USA 99:10078-10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyce, J. D., B. E. Davidson, and A. J. Hillier. 1995. Identification of prophage genes expressed in lysogens of the Lactococcus lactis bacteriophage BK5-T. Appl. Environ. Microbiol. 61:4099-4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyd, E. F., and H. Brüssow. 2002. Common themes among bacteriophage-encoded virulence factors and diversity among the bacteriophages involved. Trends Microbiol. 10:521-529. [DOI] [PubMed] [Google Scholar]
- 5.Broudy, T. B., V. Pancholi, and V. A. Fischetti. 2002. The in vitro interaction of Streptococcus pyogenes with human pharyngeal cells induces a phage-encoded extracellular DNase. Infect. Immun. 70:2805-2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brüssow, H. 2001. Phages of dairy bacteria. Annu. Rev. Microbiol. 55:283-303. [DOI] [PubMed] [Google Scholar]
- 7.Brüssow, H., and R. W. Hendrix. 2002. Phage genomics: small is beautiful. Cell 108:13-16. [DOI] [PubMed] [Google Scholar]
- 8.Campbell, A. M. 1992. Chromosomal insertion sites for phages and plasmids. J. Bacteriol. 174:7495-7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Canchaya, C., C. Proux, G. Fournous, A. Bruttin, and H. Brüssow. 2003. Prophage genomics. Microbiol. Mol. Biol. Rev. 67:238-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng, W., S. R. Liou, G. Plunkett III, G. F. Mayhew, D. J. Rose, V. Burland, V. Kodoyianni, D. C. Schwartz, and F. R. Blattner. 2003. Comparative genomics of Salmonella enterica serovar Typhi strains Ty2 and CT18. J. Bacteriol. 185:2330-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desiere, F., W. M. McShan, D. van Sinderen, J. J. Ferretti, and H. Brüssow. 2001. Comparative genomics reveals close genetic relationships between phages from dairy bacteria and pathogenic Streptococci: evolutionary implications for prophage-host interactions. Virology 288:325-341. [DOI] [PubMed] [Google Scholar]
- 12.Desiere, F., R. D. Pridmore, and H. Brüssow. 2000. Comparative genomics of the late gene cluster from Lactobacillus phages. Virology 275:294-305. [DOI] [PubMed] [Google Scholar]
- 13.Figueroa-Bossi, N., S. Uzzau, D. Maloriol, and L. Bossi. 2001. Variable assortment of prophages provides a transferable repertoire of pathogenic determinants in Salmonella. Mol. Microbiol. 39:260-271. [DOI] [PubMed] [Google Scholar]
- 14.Ikebe, T., A. Wada, Y. Inagaki, K. Sugama, R. Suzuki, D. Tanaka, A. Tamaru, Y. Fujinaga, Y. Abe, Y. Shimizu, and H. Watanabe. 2002. Dissemination of the phage-associated novel superantigen gene speL in recent invasive and noninvasive Streptococcus pyogenes M3/T3 isolates in Japan. Infect. Immun. 70:3227-3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McShan, W. M., Y. F. Tang, and J. J. Ferretti. 1997. Bacteriophage T12 of Streptococcus pyogenes integrates into the gene encoding a serine tRNA. Mol. Microbiol. 23:719-728. [DOI] [PubMed] [Google Scholar]
- 16.Ohnishi, M., K. Kurokawa, and T. Hayashi. 2001. Diversification of Escherichia coli genomes: are bacteriophages the major contributors? Trends Microbiol. 9:481-485. [DOI] [PubMed] [Google Scholar]
- 17.Smoot, J. C., K. D. Barbian, J. J. Van Gompel, L. M. Smoot, M. S. Chaussee, G. L. Sylva, D. E. Sturdevant, S. M. Ricklefs, S. F. Porcella, L. D. Parkins, S. B. Beres, D. S. Campbell, T. M. Smith, Q. Zhang, V. Kapur, J. A. Daly, L. G. Veasy, and J. M. Musser. 2002. Genome sequence and comparative microarray analysis of serotype M18 group A Streptococcus strains associated with acute rheumatic fever outbreaks. Proc. Natl. Acad. Sci. USA 99:4668-4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Sluys, M. A., M. C. de Oliveira, C. B. Monteiro-Vitorello, C. Y. Miyaki, L. R. Furlan, L. E. Camargo, A. C. da Silva, D. H. Moon, M. A. Takita, E. G. Lemos, M. A. Machado, M. I. Ferro, F. R. da Silva, M. H. Goldman, G. H. Goldman, M. V. Lemos, H. El Dorry, S. M. Tsai, H. Carrer, D. M. Carraro, R. C. de Oliveira, L. R. Nunes, W. J. Siqueira, L. L. Coutinho, E. T. Kimura, E. S. Ferro, R. Harakava, E. E. Kuramae, C. L. Marino, E. Giglioti, I. L. Abreu, L. M. Alves, A. M. do Amaral, G. S. Baia, S. R. Blanco, M. S. Brito, F. S. Cannavan, A. V. Celestino, A. F. da Cunha, R. C. Fenille, J. A. Ferro, E. F. Formighieri, L. T. Kishi, S. G. Leoni, A. R. Oliveira, V. E. Rosa, Jr., F. T. Sassaki, J. A. Sena, A. A. de Souza, D. Truffi, F. Tsukumo, G. M. Yanai, L. G. Zaros, E. L. Civerolo, A. J. Simpson, N. F. Almeida, Jr., J. C. Setubal, and J. P. Kitajima. 2003. Comparative analyses of the complete genome sequences of Pierce's disease and citrus variegated chlorosis strains of Xylella fastidiosa. J. Bacteriol. 185:1018-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ventura, M., A. Bruttin, C. Canchaya, and H. Brüssow. 2002. Transcription analysis of Streptococcus thermophilus phages in the lysogenic state. Virology 302:21-32. [DOI] [PubMed] [Google Scholar]
- 20.Walker, D. C., and T. R. Klaenhammer. 1994. Isolation of a novel IS3 group insertion element and construction of an integration vector for Lactobacillus spp. J. Bacteriol. 176:5330-5340. [DOI] [PMC free article] [PubMed] [Google Scholar]