Abstract
Histidine kinases DivJ and PleC initiate signal transduction pathways that regulate an early cell division cycle step and the gain of motility later in the Caulobacter crescentus cell cycle, respectively. The essential single-domain response regulator DivK functions downstream of these kinases to catalyze phosphotransfer from DivJ and PleC. We have used a yeast two-hybrid screen to investigate the molecular basis of DivJ and PleC interaction with DivK and to identify other His-Asp signal transduction proteins that interact with DivK. The only His-Asp proteins identified in the two-hybrid screen were five members of the histidine kinase superfamily. The finding that most of the kinase clones isolated correspond to either DivJ or PleC supports the previous conclusion that DivJ and PleC are cognate DivK kinases. A 66-amino-acid sequence common to all cloned DivJ and PleC fragments contains the conserved helix 1, helix 2 sequence that forms a four-helix bundle in histidine kinases required for dimerization, autophosphorylation and phosphotransfer. We present results that indicate that the four-helix bundle subdomain is not only necessary for binding of the response regulator but also sufficient for in vivo recognition specificity between DivK and its cognate histidine kinases. The other three kinases identified in this study correspond to DivL, an essential tyrosine kinase belonging to the same kinase subfamily as DivJ and PleC, and the two previously uncharacterized, soluble histidine kinases CckN and CckO. We discuss the significance of these results as they relate to kinase response regulator recognition specificity and the fidelity of phosphotransfer in signal transduction pathways.
Asymmetric cell division in the alpha proteobacterium Caulobacter crescentus produces a mother stalked cell and a new motile swarmer cell. The mother cell initiates chromosome replication immediately after cell separation. During the ensuing S phase, the cell elaborates the flagellum and other polar surface structures required to construct the incipient swarmer cell. The new progeny swarmer cell produced at division enters a presynthetic gap (G1 phase) during which the cell loses motility and elaborates a polar stalk at the point of flagellum attachment before initiating chromosome replication and entering the cell division cycle. The sequence of developmental events is closely coordinated with cell cycle progression (2, 10, 24). Genetic analysis of conditional cell division cycle mutants has provided evidence that successive steps in the cell division cycle provide checkpoints for the execution of developmental events, including flagellum biosynthesis, gain of motility, and stalk formation (9; reviewed in reference 22).
Differentiation in C. crescentus is controlled and coordinated by multiple signal transduction pathways mediated by members of the His-Asp family of proteins (10, 22, 24). The C. crescentus genome encodes 61 histidine protein kinases (23), and genetic analysis has shown that at least 4 of them initiate signal transduction pathways that regulate cell cycle progression and differentiation. These include the transmembrane kinases DivJ (38), PleC (35), and DivL (38), which belong to kinase subfamily 1. This subfamily contains the majority of the C. crescentus histidine kinases (24). The fourth kinase involved in cell cycle regulation, CckA, is a hybrid histidine kinase (12) in subfamily 4. There is only limited sequence similarity in the conserved catalytic domains of the subfamily 1 and 4 kinases (24).
Pseudoreversion analyses of mutants blocked in polar morphogenesis or cell division identified two signal transduction pathways initiated by DivJ and PleC, both of which are mediated by the essential single-domain response regulator DivK (32, 35). The pathway initiated by DivJ (25) controls an early step in the cell division cycle. The second signal transduction pathway, which is initiated by the PleC kinase, regulates cell motility late in the cell division cycle and the swarmer cell to stalked cell transition after cell separation (32, 35). A target of the DivJ → DivK cell division pathway and the PleC → DivK motility pathway was identified as the essential response regulator CtrA (29, 37) on the basis of suppression of the DivJ and PleC phenotypes by a conditional sokA allele of ctrA (31, 37). Recent work suggests that a target of the DivK-mediated signal transduction pathway regulating the early cell division cycle step is the ClpX/P-dependent turnover of CtrA (11).
The genetic analyses described above suggested that DivJ and PleC are cognate DivK kinases and that DivK participates in at least two different signal transduction pathways that are initiated in response to different cell cycle checkpoints (24). Biochemical experiments support this conclusion. The purified catalytic domains of the two kinases are autophosphorylated in the presence of ATP and transfer the phosphoryl group to DivK (8). The catalytic fragments of DivJ and PleC also catalyze dephosphorylation of phospho-DivK in a purified system (8). Experiments measuring DivK phosphorylation in intact cells (36) and by isolated membranes (T. Hofmeister and A. N., unpublished observation) indicate that DivK phosphorylation depends primarily on the DivJ kinase.
The function of DivK in programming polar morphogenesis and cell cycle progression in response to DivJ and PleC raises the questions of how the specificity of DivK interaction with these kinases is determined and the identity of other His-Asp signal transduction proteins that might interact with DivK. To address these issues, we have used DivK as the bait protein in a yeast two-hybrid screen of random DNA fragments of the C. crescentus genome. Although candidates for interaction include the 61 histidine kinases encoded by the C. crescentus genome (23), only 5 histidine kinases were identified in the screen and many of the isolated clones encoded fragments of either DivJ or PleC. This result supports our previous conclusion that DivJ and PleC are the cognate kinases regulating DivK activity (7). Of particular interest is the identification of a conserved 66-amino-acid sequence shared by all of the interacting fragments of the DivJ and PleC kinases. We present results indicating that this conserved α-helical sequence, which is expected to form a four-helix bundle in the kinase structure necessary for dimerization and phosphotransfer (33), is sufficient in vivo for recognition of the cognate response regulator. We discuss this conclusion and three additional histidine kinases identified in the two-hybrid screen.
MATERIALS AND METHODS
Media and growth conditions.
The Caulobacter strains, yeast strains, and plasmids used in this study are described in Table 1. Yeast cells were grown in yeast peptone dextrose or synthetic complete (SC) medium as described by Miller and Rose (19). The C. crescentus strains were derived from strain CB15 (ATCC 19089), and cultures were grown in PYE medium at 30°C (28). Spectinomycin (50 μg/ml), kanamycin (50 μg/ml), gentamicin (2 μg/ml), and tetracycline (2 μg/ml) were added when appropriate.
TABLE 1.
Stains and plasmids used in this study
| Strain or plasmid | Genotype or description | Source or construction |
|---|---|---|
| Caulobacter strains | ||
| CB15 | Wild type | ATCC 19089 |
| PC1131 | CB15/pRK2L1 | This study |
| PC3017 | divJ356::Ω | 31 |
| PC3026 | cckO302::GmrdivJ356::Ω (Spcr) | φCr30(PC3332) × PC3017 |
| PC3327 | cckN+cckN301::Kanr Gmr | CB15::pNOR702 (Materials and Methods) |
| PC3331 | cckO+cckO302::Gmr KanrsacB | CB15::pNOR1302 (Materials and Methods) |
| PC3332 | cckO303::Gmr | Sucrose selection of PC3331 |
| PC3335 | cckO303::Gmr | φCr30(PC3332) × CB15 |
| PC3356 | cckN301::Kanr | φCr30(PC3327) × CB15 |
| Yeast strains | ||
| YE001 | MATatrp1-901 leu2-3 leu2-112 ura3-52 his3-200 gal4Δ gal80Δ LYS2::GAL1-HIS3 GAL2-ADE2 met3::GAL7-lacZ | Same as PJ69-4A; 14 |
| YE002 | YE001/pNOR001 | pGBDU-divK+; this study |
| Plasmids | ||
| pAROGmr | Derivative of pARO181 (ATCC 77125; 27); Kanr was replaced with a Gm resistance-encoding aacC1 cassette (30) | This study |
| pNPTS128 | Kanr derivative of pLITMUS28 with sacB and oriT | 1 |
| pGBDU-C3 | Bait vector | 14 |
| pGAD-C1-, 2-, 3 | Prey vector | 14 |
Construction of C. crescentus yeast two-hybrid libraries.
Plasmid libraries containing the wild-type DNA were constructed as described by James et al. (14), with minor modifications. The purified DNA from C. crescentus strain CB15 was partially digested with three restriction enzymes, AciI, HinP1I, and MspI (New England Biolabs), separately. Each of these three enzymes cuts the GC-rich Caulobacter chromosome very frequently (on average, once in less than 100 bp), rendering the resulting partial digests extremely random. Aliquots of digested DNA samples were examined by Southern hybridization analysis to ascertain that partial and complete digestions were evenly distributed among restriction fragments, by using the 32P-labeled 501-bp SstI-BamHI fragment containing divK as a probe. DNA fragments in the range of 0.5 to 2 kb from each of the three enzyme digests were separately purified from agarose gel and then combined and fused to the GAL4-activating domain at the ClaI site in the pGAD vectors in three frames (14), generating libraries, CB-C1, CB-C2, and CB-C3. Electrophoretic examination of 16 random transformants from each library has shown that 90% of the clones contained insert DNA (data not shown).
Two-hybrid screen with DivK as the bait protein.
The bait plasmid containing the entire divK gene was constructed by creating the ClaI site at the 5′ end of the divK open reading frame, taking advantage of a preexisting ClaI half site, GAT. DivK, encoded by the 387-bp ClaI-BamHI fragment (see the divK DNA sequence [GenBank accession no. U13765]), was fused in frame to the GAL4 DNA binding domain in the vector pGBDU-C3 (14) to generate plasmid pNOR1001. pNOR1001 was transformed into yeast strain PJ69-4A (renamed YE001 for this study) (Table 1), which contains three reporter genes, GAL1-HIS3, GAL2-ADE2, and GAL7-lacZ (14), generating strain YE002. The library DNAs in three different frames (CB-C1, CB-C2, and CB-C3) were then independently transformed into YE002. Approximately 4 × 106 transformants were screened as described by Miller and Rose (18), except that the retesting of Ade+ prey plasmids was omitted. The omission may have resulted in the further examination of some of the false-positive clones.
Analysis of interacting clones.
Prey plasmid DNAs isolated from the yeast cells were transformed into Escherichia coli strain DH5α (Bethesda Research Laboratories) for further analysis. Identities of interacting clones were determined either by DNA sequencing or by Southern hybridization analyses. The 5′ nucleotide sequences of 127 clones were sequenced from the upstream end of the 5′ GAL4-insert fusion junction. Among 76 clones that encode either DivJ or PleC, 28 clones were also sequenced from the 3′ end to determine the end points of the respective polypeptide fragments. Among the sequenced clones, 32 represented unique hits but 28 of them were not in-frame fusions to open reading frames in the Caulobacter database (23) and were presumably false positives. The four single-hit in-frame fusions, which are not discussed in Results, are in the CC1471, CC0013, CC3043, and CC0766 genes. Several of these clones encode very short peptides. The probes used for Southern hybridization analyses were 32P-labeled DNA fragments containing the core catalytic (H-box) domain of either divJ (1,067-bp NotI-XhoI fragment; see the DNA sequence with GenBank accession no. M98873) or pleC (1,030-bp BamHI-ClaI fragment; see the DNA sequence with GenBank accession no. M91449).
Disruption of cckN and cckO.
Since cckN is located adjacent to divJ on the chromosome (23), the 1.5-kb BamHI-EcoRI fragment containing cckN (CC1062 [23]) was subcloned from a clone containing divJ and the upstream sequence (25). cckN was disrupted by deleting the 718-bp NcoI-NcoI fragment, which removed most of the gene, and replacing it with the 1.3-kb Kanr-encoding fragment (from pKM18-5; obtained from S. Inouye). The disrupted construct was cloned into the suicide vector pAROGm (Table 1; a Gmr derivative of pARO181 [27]) to give pNOR702 and integrated into the chromosome, resulting in a strain containing both wild-type cckN and cckN::Kanr (strain PC3327; Table 1). Strain PC3356 containing only cckN::Kanr was obtained by phage Cr30-mediated transduction (20).
The 1,866-bp BglII-BamHI fragment covering the C-terminal two-thirds of cckO (CC0026 [23]) was amplified from CB15 genomic DNA and cloned as an EcoRI-BamHI fragment in the suicide vector pNPTS128 (Table 1) (1) to give plasmid pNOR1301. The internal 681-bp EcoRV-NotI fragment of cckO was then deleted from pNOR1301 and replaced with the Gmr cassette (30), generating pNOR1302. The deletion removed the linker region connecting the kinase catalytic domain and the response regulator domain. Plasmid pNOR1302 was integrated into the CB15 genome to give PC3331, followed by sucrose selection (15) to obtain a cckO::Gmr replacement strain (PC3332) that had lost the plasmid sequence. Isogenic cckO::Gmr strain PC3335 was constructed by transduction of the cckO::Gmr-encoding allele of PC3332 into wild-type strain CB15 (Table 1). A ca. 3-kb-long DNA fragment presumed to contain the full-length cckO (CC0026) gene and the promoter region was also amplified from genomic DNA and cloned into pRK2L1 (21) to give pNOR1306. The cckN::Kanr and cckO::Gmr disruption mutations were confirmed by PCR amplification of the chromosome region containing the deletion-insertion and comparing them to those obtained from the wild-type strain CB15 DNA.
Flow cytometry (fluorescence-activated cell sorter [FACS] analysis).
Early exponential cultures (optical density at 650 nm of ca. 0.1 to 0.3) were treated with rifampin (Sigma) at a final concentration of 10 μg/ml for 3 h at 30°C to allow completion of chromosome replication but to prevent reinitiation of DNA synthesis. Cells were then harvested, stained with Sytox green nucleic acid stain (Molecular Probes, Eugene, Oreg.), and analyzed with a FACSort flow cytometer as previously described (31).
RESULTS
Yeast two-hybrid screen for proteins interacting with DivK.
Previous genetic and biochemical results (7, 32) have provided evidence that DivJ and PleC are cognate histidine kinases of response regulator DivK. To examine these interactions and to screen for other His-Asp proteins that interact with DivK by using the yeast two-hybrid assay, we constructed prey libraries of small, random fragments of C. crescentus DNA in the three reading frames in the pGAD shuttle vectors (see Materials and Methods and reference 14). As the bait clone, the divK gene was fused in frame to the DNA binding domain of GAL4 in plasmid pGBDU-C3 (14). Approximately 4 × 106 yeast transformants harboring both the bait and library DNAs were screened first for activation of ADE2 and then for activation of HIS3 in a secondary screen (18). A total of 182 interacting clones were identified. DNA sequence analysis of 88 clones from the region upstream of the 5′ fusion junction demonstrated that 23 clones contained inserts that encode PleC polypeptides and 38 contained inserts that encode DivJ polypeptides. The remaining 94 clones were examined by Southern blot analysis with 32P-labeled DNA probes containing either divJ or pleC restriction fragments (data not shown; see Materials and Methods). This analysis identified an additional 26 PleC clones and 35 DivJ clones. Clones that were not unambiguously identified as divJ or pleC by the Southern hybridization analysis were further analyzed by DNA sequencing.
The majority of the isolates examined by the two procedures just outlined correspond to either PleC (49 clones) or DivJ (73 clones) clones. An additional eight clones encoded polypeptide fragments of the tyrosine kinase DivL (38). We also identified 11 clones encoding fragments of either the histidine kinase CckN (3 clones) or CckO (8 clones). CckN (CC1062) and CckO (CC0026) are in the C. crescentus database (23), but they have not been previously studied (see below). None of the four remaining in-frame clones (see Materials and Methods) identified a predicted His-Asp signal transduction protein, including a possible phosphotransferase predicted to be downstream of DivK (37). It is also noteworthy that no DivK clones were recovered in this screen, although DivK forms dimers in vitro (6).
A second, smaller-scale screen with the same divK bait clone that was carried out as a control in another screen identified an additional 39 interacting clones. The majority of these clones again corresponded to either PleC (19 clones) or DivJ (16 clones). One DivL and one CckN clone were also isolated. These results confirmed our findings from the larger screen described above. They are consistent with the conclusion that only a small subset of the histidine kinases in C. crescentus recognize the DivK bait protein in the yeast two-hybrid screen.
DivJ, PleC, and DivL clones that interact with DivK contain the conserved helix 1, helix 2 sequence.
The DivJ and PleC fragments, a subset of which are shown in Fig. 1, form an overlapping array of sequences. The 76 PleC and DivJ clones examined by DNA sequencing have in common the translated H-box sequence, which contains the His residue that is the site of autophosphorylation and phosphotransfer in these histidine kinases (data not shown; reviewed in reference 4). Alignment of these common sequences showed that they are similar to the helix 1, helix 2 sequence of the homodimeric core domain of the E. coli EnvZ protein and other histidine kinases (Fig. 2) (33). The helix 1, helix 2 sequence of EnvZ forms a four-helix bundle that is responsible for dimerization of the histidine kinase and contains the site of autophosphorylation and phosphotransfer (33). Computer analysis of the PleC and DivJ fragments identified in the yeast two-hybrid screen supports the idea that the conserved sequence containing the H box also forms a four-helix bundle, although the exact location of helix 2 relative to helix 1 differs slightly from that of EnvZ (Fig. 2; analysis not shown).
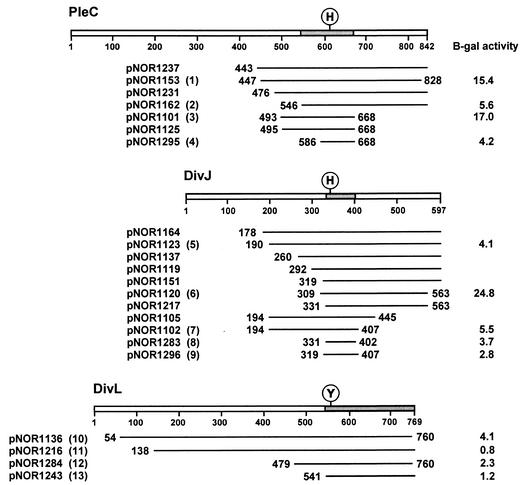
FIG. 1.
Polypeptide fragments of kinases DivJ, PleC, and DivL that interact with DivK in the yeast two-hybrid screen. An alignment of translated inserts in representative DivJ, PleC, and DivL clones is shown. The positions of the N- and C-terminal residues are indicated for each fragment, except those that extended to the C-terminal end of the proteins. Locations of the conserved sites of phosphorylation, histidine (H) in DivJ and PleC and tyrosine (Y) in DivL, are shown. Shaded areas of each kinase represent overlapping sequences common to the respective DivJ, PleC, and DivL clones. The numbers in parentheses following plasmid names identify the clones tested in the spot test shown in Fig. 3. β-Galactosidase (B-gal) assays (17) were performed on the cultures of the same clones that were grown in SC medium lacking leucine and uracil to mid-exponential phase. At least three independent cultures of each strain were assayed. Including the clones shown here, the fusion joints of 76 clones were sequenced. Among 34 pleC clones sequenced, the most frequent fusion points were either at amino acid 456 (7 clones) or between amino acids 492 and 495 (21 clones). Of the 42 DivJ clones sequenced, a total of 21 DivJ fragments started between residues 190 and 196 and 6 started between residues 328 and 331. The C-terminal ends of most clones were not sequenced.
FIG. 2.
Amino acid sequence alignment of the helix 1, helix 2 sequences in histidine kinases. The amino acid sequences of conserved DivJ, PleC, CckN, and DivL fragments identified in the yeast two-hybrid screen with DivK as the bait protein (shaded) are aligned with the core helix 1, helix 2 domain of the E. coli EnvZ protein. CckA and CckO are included for comparison, although cckA clones were not isolated in the screen and cckO clones did not encode the helix 1, helix 2 sequence (see text). Locations of helixes 1 and 2 in EnvZ were determined as described previously (33). Residues conserved in both PleC and DivJ are in bold.
Experiments with the purified catalytic domains of PleC, DivJ, and DivL have shown that DivL is relatively inefficient at catalyzing phosphotransfer to DivK (38). Given this observation, the interaction between DivL and DivK in the two-hybrid assay was unexpected. As described above, however, we isolated a total of nine divL clones in the two screens. All of the cloned fragments contain the conserved helix 1, helix 2 sequence (Fig. 1 and 2) and the Tyr-550 residue, which is the site of phosphorylation in the DivL kinase (38). The N-terminal end of the shortest cloned DivL fragment, the 228-amino-acid insert encoded by pNOR1243, lies just upstream of the conserved helix 1 sequence and extends to the end of the protein (Fig. 2). We conclude from these results that DivK also interacts with the four-helix bundle of the DivL kinase, as proposed for DivJ and PleC.
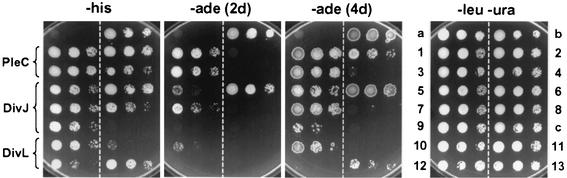
We have used spot tests and β-galactosidase assays of representative PleC, DivJ, and DivL clones to assess their interaction with the DivK bait protein. There is generally a good correlation between the growth of these hybrid clones on the spot tests (Fig. 3) and the enzyme activities (Fig. 1; see also Fig. 4). Although all of the clones interacted with DivK in the His− plate assay, two of the DivL clones, pNOR1216 and pNOR1284, displayed the interaction only at one of the lower dilutions (Fig. 3). The more stringent assay on the Ade− plate (after 2 days) confirmed this result and indicates that the interaction of the four DivL clones with DivK is weaker than that of many of the PleC and DivJ clones (Fig. 3). The results of the β-galactosidase assays suggest a similar conclusion (Fig. 1). A weaker interaction of the DivL kinase with DivK could account for the smaller number of DivL clones isolated in our screen.
FIG. 3.
Interactions of DivK with DivJ, PleC, and DivL in the yeast two-hybrid assay. Yeast strains containing both bait and prey plasmids were grown overnight at 30°C in SC medium lacking uracil and leucine. Cultures were serially diluted 10-fold and spotted onto SC plates lacking leucine and uracil (−leu −ura), histidine (−his), or adenine (−ade). All of the strains tested contained the divK bait plasmid and interacting clones, except a, b, and c. a, divK bait plus empty prey vector; b, pGDB-KAR9 plus pGAD-BIM1 (18); c, pNOR1196 plus empty bait vector. The rationale behind the method is that stronger interactions between bait and prey polypeptides result in higher enzymatic activities for the synthesis of histidine and adenine and thus permit more rapid growth on the selective His− and Ade− plates. The numbered clones correspond to those indicated in parentheses in Fig. 1. Before being photographed, the Leu− Ura−, His−, and Ade− plates were incubated for 2 days (2d), and the Ade− plate was incubated for 4 days (4d), at 30°C.
FIG. 4.
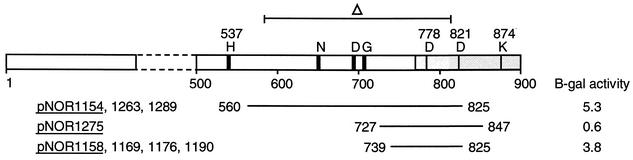
Alignment of CckO clones. Alignment of translated inserts in cckO clones with the translated CckO sequence (CC0026). The unshaded area represents the kinase domain, and the shaded area represents the receiver domain, of the hybrid kinase. The predicted sites of phosphorylation at H537, D821, other conserved motifs in the kinase domain (N, D, and G boxes [reviewed in reference 5]), and conserved residues in the receiver domain are shown. β-Galactosidase (B-gal) assays on each of the CckO clones underlined were performed as described in the legend to Fig. 1.
The four-helix bundle domains of the DivJ and PleC kinases are sufficient for recognition of DivK.
Among the 17 divJ and 11 pleC clones that were sequenced from both the 5′ and 3′ ends, the sizes of the translated inserts ranged from 72 to 420 amino acids for DivJ and from 174 to 400 amino acids for PleC. The isolation of a DivJ fragment of only 72 amino acids (pNOR1283) and a PleC fragment of 174 amino acids (pNOR1125; Fig. 1) indicates that the conserved 66-residue helix 1, helix 2 sequence may be sufficient, as well as necessary, for recognition of DivK. To examine this possibility in more detail, we constructed two additional prey clones with inserts encoding PleC (amino acid residues 586 to 668; plasmid pNOR1295) or DivJ (residues 319 to 407; plasmid pNOR1296) that contain primarily the conserved helix 1, helix 2 sequence (Fig. 1). The pNOR1295 and pNOR1296 clones, like the 72-amino-acid DivJ fragment encoded by the pNOR1283 clone, interacted with DivK, as determined by spot tests (Fig. 3) and β-galactosidase activity assays (Fig. 1). These short PleC and DivJ fragments displayed interactions with DivK that are similar to or stronger than those observed between DivK and the DivL kinase fragments. Although the interactions of these PleC and DivJ clones are weaker than those observed for the longer PleC clones pNOR1153 and pNOR1101 and the DivJ clone pNOR1120, they are comparable to those observed for other, longer clones, including the PleC clone pNOR1162 and the DivJ clone pNOR1123. Thus, the strength of interaction between the various kinase fragments and DivK is not strictly related to the length of the sequences flanking the helix 1, helix 2 motif. These findings support the conclusion that the recognition specificity of the kinases for DivK is confined to the four-helix bundle domain.
Genetic characterization of cckN and cckO.
CckN is a cytoplasmic kinase that is closely related in sequence to DivJ (Fig. 2). The four CckN (CC1062) clones identified by their interaction with DivK encode all or most of the kinase sequence, including the core dimerization domain. The cckN gene is divergently transcribed from divJ on the C. crescentus genome. This gene organization and the sequence similarity of DivJ, PleC, and CckN (Fig. 2) suggested that CckN might belong to a network of kinases controlling DivK activity. To determine if a cckN mutant displayed a defective cell cycle or a developmental phenotype similar to those caused by mutations in either divJ or pleC, we constructed a Kanr disruption allele of cckN and replaced the genomic cckN+ allele to give cckN::Kanr knockout strain PC3356 (see Materials and Methods) (Table 1).
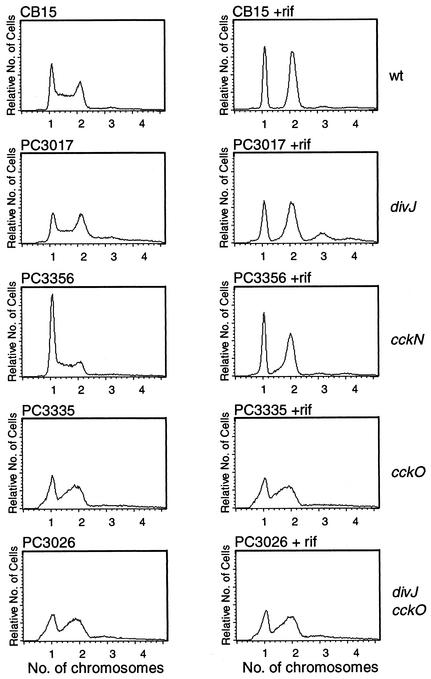
Our ability to construct the disruption strain indicates that cckN is not essential. Moreover, we were unable to detect mutant phenotypes, e.g., no obvious defects in cell division, motility, or stalk formation, as determined by light microscopy (data not shown). We also examined cell cycle regulation in the cckN disruption mutant by FACS analysis and observed a significant and reproducible increase in the number of cells with one chromosome, suggesting a possible G1 defect in DNA replication (see Fig. 5).
FIG. 5.
FACS analysis of divJ, cckN, and cckO null mutants. Cultures were grown in PYE medium supplemented with appropriate antibiotics to optical densities at 650 nm of ca. 0.1 to 0.3, and a portion of each culture was further incubated with rifampin (rif) for 3 h at 30°C before harvesting. Samples were prepared for flow cytometry as described in Materials and Methods, and DNA content (chromosome number) was measured. Normally, swarmer cells (G1 phase) contain one chromosome per cell and predivisional cells (G2 phase) contain two chromosomes per cell (3). wt, wild type.
The second novel kinase identified in this screen was CckO (CC0026), a cytoplasmic, hybrid kinase of 900 amino acids with a C-terminal receiver domain. Surprisingly, none of the eight CckO clones identified in the two-hybrid screen contain the conserved helix 1, helix 2 sequence (Fig. 4). Instead, all of the isolated clones contain the same 87-amino-acid sequence that includes the kinase response regulator linker region along with the N-terminal half of the CckO receiver domain, including the presumptive site of phosphorylation (Asp-821; Fig. 4). The isolation of eight independent cckO clones argues that the observed CckO-DivK interaction is not fortuitous. To confirm this interaction, we have reconstituted an isogenic set of interacting strains by retransforming the parent yeast strain containing the DivK bait plasmid (YE002; Table 1) with the eight cckO prey plasmids isolated from E. coli. The levels of two-hybrid interaction in these strains tested in a plate spot test (data not shown) and β-galactosidase assays (Fig. 4) were comparable to those observed for many of the divJ, pleC, and divL clones shown in Fig. 1 and 3.
To examine the possible function of cckO, we constructed a cckO::Gmr knockout strain. We were able to obtain the disruption construct in the absence of a complementing wild-type cckO sequence (see Materials and Methods). This indicates that cckO is not essential for colony formation on plates under the conditions used in this selection. Surprisingly, however, cckO::Gmr knockout strain PC3335 (see Materials and Methods) (Table 1) grew very poorly in liquid medium and exhibited a significant increase in the number of predivisional cells and short filaments (data not shown). FACS analysis of strain PC3335 revealed that a large number of cells contain greater than 1n but smaller than 2n chromosomes both before and after incubation for 3 h in the presence of rifampin (Fig. 5). This suggests that a defect in DNA synthesis in the mutant prevents or slows the completion of DNA replication. Double-knockout mutant strain PC3026, containing both cckO::Gmr and divJ::Ω, exhibited the same profile as PC3335, while in divJ knockout strain PC3017 (31), DNA replication was completed in the presence of rifampin (Fig. 5), as reported previously (36). This suggests that the cckO mutant block is epistatic to the divJ mutant block.
DISCUSSION
The yeast two-hybrid screen reported here identified five members of the histidine kinase superfamily in C. crescentus that interact with the essential response regulator DivK. These kinases include PleC, DivJ, and DivL, which have been previously characterized both genetically and biochemically (22), and the two novel kinases CckN and CckO. DivK plays a role in at least two signal transduction pathways regulating cell differentiation. The first of these pathways is initiated by the histidine kinase DivJ and controls a cell division step early in the cell cycle (10, 25), while the second pathway is initiated by the kinase PleC and controls motility in late predivisional cells (31, 32). The observation that two-thirds of the independently isolated clones in our screen correspond to either DivJ or PleC (Fig. 1) supports the previous conclusion (7, 32) that these kinases are cognates of DivK. In addition, all of the interacting DivJ and PleC clones, as well as the DivL and CckN clones, share a conserved α-helical sequence of 66 residues. We suggest below that this kinase sequence motif, which is expected to form a four-helix bundle that is the site of autophosphorylation, phosphotransfer, and kinase dimerization (26, 33, 34), is sufficient in vivo for recognition specificity between kinases and their cognate response regulators.
The failure to isolate clones identifying certain His-Asp proteins in our screen may be significant. These include proteins with sequence homology to a histidine phosphotransferase that would function downstream of DivK in a signal transduction pathway regulating CtrA activity (37). Clones encoding DivK and the essential kinase CckA were also not recovered. DivK clones may not have been isolated either because the DivK-DivK interaction does not occur in vivo or because the interaction could not be detected under our assay conditions. The failure to identify CckA, which is required for CtrA phosphorylation (10), is not surprising. The core dimerization domain sequence of this subfamily IV kinase (24) diverges from that of the histidine kinases shown in this study to interact with DivK (Fig. 2).
Response regulator interactions with the four-helix bundle subdomain of kinases and phosphotransferases.
The four-helix bundle appears to be a general structural feature of histidine kinases and phosphotransferases. As shown in studies of the E. coli histidine kinase EnvZ (33) and the Bacillus subtilis phosphotransferase Spo0B (34, 39), it plays an essential role in interaction with the respective cognate response regulators. Structural studies have shown that Spo0F of B. subtilis interacts with α1 and α2′ of the two Spo0B protomers, as well as two α-β loops in the C-terminal domain of the phosphotransferase (39). Nuclear magnetic resonance titration experiments indicate that OmpR interacts with multiple residues on both helixes 1 and 2 of the four-helix bundle of EnvZ (33). These structural studies suggest that an interaction between the response regulator DivK and its cognate histidine kinases in the two-hybrid assay described here would also occur with the four-helix bundle dimerization domain.
A genetic approach has not previously been used to address the question of whether the four-helix bundle of histidine kinases alone is sufficient to confer recognition specificity for cognate response regulators. The screen for C. crescentus proteins that interact with the response regulator DivK described here suggests that kinase response regulator interactions can be studied with the yeast two-hybrid assay. Our results indicate that the four-helix bundle sequence is both necessary and sufficient to confer specificity of recognition between this response regulator and its cognate histidine kinases in vivo. This screen of a genomic library of prey clone inserts also suggests that the specificity of the interaction of the core kinase dimerization domain with the cognate response regulator may be sufficient to exclude unacceptable levels of cross talk between noncognate kinase response regulator pairs, an issue explored previously in studies of His-Asp signal transduction in B. subtilis (39). An indication of the specificity of kinase response regulator interactions was the identification of only five protein kinases in the yeast two-hybrid screen, despite the fact that C. crescentus is predicted to encode 61 histidine kinases that could potentially interact with DivK (23).
On the basis of the finding that all of the interacting DivJ and PleC clones contain the helix 1, helix 2 sequence (as shown in the alignment in Fig. 2), we concluded that the specificity of the DivK-kinase interaction is confined to this conserved 66-amino-acid sequence. The ability of the newly constructed DivJ and PleC prey clones that encode only the helix 1, helix 2 sequence to interact with DivK in the two-hybrid assay supports this conclusion (Fig. 1). However, the extents of PleC-DivK and DivJ-DivK interactions for individual clones measured by growth on spot test plates and assays of β-galactosidase activity differed (Fig. 1 and 3). Although the strength of the interactions seemed to be influenced by the presence of sequences flanking the helix 1, helix 2 sequence, it did not depend on the length of these sequences.
Relatively few DivL and CckN clones were isolated, but all of them contain the conserved helix 1, helix 2 sequence. These α-helical sequences in DivJ, PleC, and CckN are very similar to one another. The cytoplasmic domains of the three kinases cluster as a group in a PILEUP analysis of the 29 subfamily 1 kinases, while DivL and CckO, also members of subfamily 1, are not adjacent to these kinases in the analysis (data not shown) (24).
Interactions of DivK with DivL, CckN, and CckO.
Several clones encoding the polypeptides of DivL kinase (38) were recovered in the screen (Fig. 1). Because phosphotransfer experiments have previously shown that DivL is much more efficient in phosphotransfer to the response regulator CtrA than to DivK (38), the interaction between DivK and DivL was unexpected. The nine DivL clones isolated contain a presumptive four-helix bundle sequence, however, and we propose that this structure subdomain also functions in DivL as the site of dimerization and interaction with DivK. The affinity of DivK-DivL interactions estimated for DivL clones is somewhat less than that for most of the PleC and DivJ clones. It should also be noted that the amino acid sequence in this subdomain in DivL diverges to some extent from those in PleC and DivJ, particularly in presumptive helix 2 (Fig. 2).
Genes encoding the two remaining kinases identified in this screen, CckN and CckO, were reported in the C. crescentus genome sequencing project (23). As discussed above, the soluble CckN kinase is similar in sequence to DivJ and PleC around the phosphotransfer domain (Fig. 2). The cckN gene, like divJ and pleC, is not essential, but unlike divJ and pleC, the cckN disruption mutant displays no detectable morphological phenotype. The only indication that this kinase may play a role in the cell division cycle was the accumulation of cckN mutant cells arrested in G1 (Fig. 5).
The cckO gene is also not essential, as judged by the successful isolation of a disruption in this gene. The cckO::Gmr mutant strain did, however, display a severe growth phenotype in liquid medium, and it also showed an increase in the number of predivisional cells, as well as an increase in the number of short filaments (data not shown). Consistent with the latter observation were the results of FACS analysis of cckO mutant cells, which displayed a distribution profile suggesting that the mutant is defective in the completion of DNA replication (Fig. 5). The interaction of the hybrid kinase CckO with DivK in the clones isolated is unusual. None of the clones contained the four-helix bundle sequences; instead, all contained the kinase response regulator linker region and the N-terminal portion of the receiver module. This unexpected interaction is under investigation.
Stability of kinase response regulator interactions.
One question raised by the present studies is whether the yeast two-hybrid system, which is not typically used for detection of catalytic interactions, might be of general use in the identification of cognate kinases or phosphotransferases for orphan response regulators. Our failure to identify any kinases in a yeast two-hybrid screen with the response regulator CtrA (29) or PleD (8) as the bait protein (unpublished results) suggests that many kinase response regulator interactions may not be stable enough for detection in a genomewide screen. The transmitter (catalytic) domain of the NtrB kinase and its cognate response regulator NtrC have been shown to interact in a pairwise combination in the yeast two-hybrid system (16), but neither of these proteins was used as bait to screen a prey library in that study.
The stability of DivK interactions with its cognate kinases may explain the spatial colocalization of DivK with the DivJ and PleC kinases at the C. crescentus cell poles: DivK colocalizes to the stalked pole with the DivJ kinase and to the flagellated pole with the PleC kinase (13). We propose that the localization of DivK may depend directly on the subcellular targeting of this soluble response regulator to the cell poles via its interaction with the four-helix bundles of the localized DivJ and PleC kinases (31, 36). Structural studies of DivK have shown that the metal ion coordination required for activity of the response regulator is unorthodox (6). These results have suggested that the required active-site geometry of DivK is achieved by its interaction with the cognate kinase. It is thus tempting to speculate that the observed colocalization of DivK with PleC or DivJ represents enzymatically active DivK-kinase complexes at the cell poles.
Acknowledgments
This work was supported by Public Health Service grant GM58794 from the National Institutes of Health.
We are most grateful to Rita Miller and other members of Mark Rose's laboratory for teaching us all of the techniques used for yeast two-hybrid analysis. We thank Leslie Kulick for help in two-hybrid screening, Ping Hu and Adam Niedzwiecki for β-galactosidase assays, and Christina DeCoste for performing FACS analysis. We are grateful to Christine Hirvonen for helpful discussions of alignments of helix 1, helix 2 sequences in C. crescentus kinases and to Stephen Sciochetti for critically reading the manuscript.
REFERENCES
- 1.Alley, M. R. 2001. The highly conserved domain of the Caulobacter McpA chemoreceptor is required for its polar localization. Mol. Microbiol. 40:1335-1343. [DOI] [PubMed] [Google Scholar]
- 2.Brun, V. Y., and R. Janakiraman. 2000. The dimorphic life cycle of Caulobacter and stalked bacteria, p. 297-317. In Y. Brun and L. Shimkets (ed.), Prokaryotic development. ASM Press, Washington, D.C.
- 3.Degnen, S., and A. Newton. 1972. Chromosome replication during development in Caulobacter crescentus. J. Mol. Biol. 64:671-680. [DOI] [PubMed] [Google Scholar]
- 4.Dutta, R., L. Qin, and M. Inouye. 1999. Histidine kinases: diversity of domain organization. Mol. Microbiol. 34:633-640. [DOI] [PubMed] [Google Scholar]
- 5.Grebe, T., and J. Stock. 1999. The histidine protein kinase superfamily. Adv. Microb. Physiol. 41:139-227. [DOI] [PubMed] [Google Scholar]
- 6.Guillet, V., N. Ohta, S. Cabantous, A. Newton, and J.-P. Samama. 2002. Crystallographic and biochemical studies of DivK reveal novel features of an essential response regulator in Caulobacter crescentus. J. Biol. Chem. 277:42003-42010. [DOI] [PubMed] [Google Scholar]
- 7.Hecht, G., T. Lane, N. Ohta, J. Sommer, and A. Newton. 1995. An essential single domain response regulator required for normal cell division and differentiation in Caulobacter crescentus. EMBO J. 14:3915-3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hecht, G., and A. Newton. 1995. Identification of a novel response regulator required for the swarmer to stalked cell transition in Caulobacter crescentus. J. Bacteriol. 177:6223-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huguenel, E. D., and A. Newton. 1982. Localization of surface structures during procaryotic differentiation: role of cell division in Caulobacter crescentus. Differentiation 21:71-78. [DOI] [PubMed] [Google Scholar]
- 10.Hung, D., H. McAdams, and L. Shapiro. 2000. Regulation of the Caulobacter cell cycle, p. 361-378. In Y. V. Brun and L. Shimkets (ed.), Prokaryotic development. ASM Press, Washington, D.C.
- 11.Hung, D. Y., and L. Shapiro. 2002. A signal transduction protein cues proteolytic events critical to Caulobacter cell cycle progression. Proc. Natl. Acad. Sci. USA 99:13160-13165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacobs, C., I. Domian, J. Maddock, and L. Shapiro. 1999. Cell cycle-dependent polar localization of an essential bacterial histidine kinase that controls DNA replication and cell division. Cell 97:111-120. [DOI] [PubMed] [Google Scholar]
- 13.Jacobs, C., D. Hung, and L. Shapiro. 2001. Dynamic localization of cytoplasmic signal transduction response regulator controls morphogenesis during the Caulobacter cell cycle. Proc. Natl. Acad. Sci. USA 98:4095-4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.James, P., J. Halladay, and E. A. Craig. 1996. Genome libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144:1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jenal, U., and T. Fuchs. 1998. An essential protease involved in bacterial cell-cycle control. EMBO J. 17:5658-5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martinez-Argudo, I., J. Martin-Nieto, P. Salinas, R. Maldonado, M. Drummond, and A. Contreras. 2001. Two-hybrid analysis of domain interactions involving NtrB and NtrC two-component regulators. Mol. Microbiol. 40:169-178. [DOI] [PubMed] [Google Scholar]
- 17.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 18.Miller, R. K., S. C. Cheng, and M. D. Rose. 2000. Bim1p/Yeb1p mediates the Kar9p-dependent cortical attachment of cytoplasmic microtubules. Mol. Biol. Cell 11:2949-2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller, R. K., and M. D. Rose. 1998. Kar9p is a novel cortical protein required for cytoplasmic microtubule orientation in yeast. J. Cell Biol. 140:377-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Minnich, S. A., N. Ohta, N. Taylor, and A. Newton. 1988. Role of the 25-, 27-, and 29-kilodalton flagellins in Caulobacter crescentus cell motility: method for construction of deletion and Tn5 insertion mutants by gene replacement. J. Bacteriol. 170:3953-3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mullin, D., and A. Newton. 1989. Ntr-like promoters and upstream regulatory sequence ftr are required for transcription of a developmentally regulated Caulobacter crescentus flagellar gene. J. Bacteriol. 171:3218-3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Newton, A., and N. Ohta. 2003. Role of multiple sensor kinases in cell cycle progression and differentiation in Caulobacter crescentus, p. 377-396. In M. Inouye and R. Dutta (ed.), Histidine kinases in signal transduction. Academic Press, Inc., New York, N.Y.
- 23.Nierman, W. C., T. V. Feldblyum, M. T. Laub, I. T. Paulsen, K. E. Nelson, J. Eisen, J. F. Heidelberg, M. R. K. Alley, N. Ohta, J. R. Maddock, I. Potcka, W. C. Nelson, A. Newton, S. Stephens, N. D. Phadke, B. Ely, R. T. DeBoy, R. J. Dodson, A. S. Durkin, M. L. Gwinn, D. H. Haft, J. F. Kolonay, J. Smit, M. B. Craven, H. Khouri, J. Shetty, K. Berry, T. Utterback, K. Tran, A. Wolf, J. Vamathevan, M. Ermolaeva, O. White, S. L. Salzberg, J. C. Venter, L. Shapiro, and C. M. Fraser. 2001. Complete genome sequence of Caulobacter crescentus. Proc. Natl. Acad. Sci. USA 98:4136-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohta, N., T. Grebe, and A. Newton. 2000. Signal transduction and cell cycle checkpoints in developmental regulation of Caulobacter, p. 341-359. In Y. Brun and L. Shimkets (ed.), Prokaryotic development. ASM Press, Washington, D.C.
- 25.Ohta, N., T. Lane, E. G. Ninfa, J. M. Sommer, and A. Newton. 1992. A histidine protein kinase homologue required for regulation of bacterial cell division and differentiation. Proc. Natl. Acad. Sci. USA 89:10297-10301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park, H., S. K. Saha, and M. Inouye. 1998. Two-domain reconstitution of a functional protein histidine kinase. Proc. Natl. Acad. Sci. USA 95:6728-6732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parke, D. 1990. Construction of mobilizable vectors derived from plasmids RP4, pUC18 and pUC19. Gene 93:135-137. [DOI] [PubMed] [Google Scholar]
- 28.Poindexter, J. S. 1964. Biological properties and classification of the Caulobacter group. Bacteriol. Rev. 28:231-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quon, K., G. Marczynski, and L. Shapiro. 1996. Cell cycle control by an essential bacterial two-component signal transduction protein. Cell 84:83-93. [DOI] [PubMed] [Google Scholar]
- 30.Schweizer, H. D. 1993. Small broad-host-range gentamycin resistance gene cassettes for site-specific insertion and deletion mutagenesis. BioTechniques 15:831-834. [PubMed] [Google Scholar]
- 31.Sciochetti, S. A., T. Lane, N. Ohta, and A. Newton. 2002. Protein sequences and cellular factors required for polar localization of a histidine kinase in Caulobacter crescentus. J. Bacteriol. 184:6037-6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sommer, J. M., and A. Newton. 1991. Pseudoreversion analysis indicates a direct role of cell division genes in polar morphogenesis and differentiation in Caulobacter crescentus. Genetics 129:623-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tomomori, C., T. Tanaka, R. Dutta, H. Park, S. K. Saha, Y. Zhu, R. Ishiima, D. Lui, K. I. Tong, H. Kurokawa, H. Qian, M. Inouye, and M. Ikura. 1999. Solution structure of the homodimeric core domain of Escherichia coli histidine kinase EnvZ. Nat. Struct. Biol. 6:729-734. [DOI] [PubMed] [Google Scholar]
- 34.Varughese, K. I., Madhusudan, X. Z. Zhou, J. M. Whiteley, and J. A. Hoch. 1998. Formation of a novel four-helix bundle and molecular recognition sites by dimerization of a response regulator phosphotransferase. Mol. Cell 2:485-493. [DOI] [PubMed] [Google Scholar]
- 35.Wang, S. P., P. L. Sharma, P. V. Schoenlein, and B. Ely. 1993. A histidine protein kinase is involved in polar organelle development in Caulobacter crescentus. Proc. Natl. Acad. Sci. USA 90:630-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wheeler, R. T., and L. Shapiro. 1999. Differential localization of two histidine kinases controlling bacterial cell differentiation. Mol. Cell 4:683-694. [DOI] [PubMed] [Google Scholar]
- 37.Wu, J., N. Ohta, and A. Newton. 1998. An essential, multicomponent signal transduction pathway required for cell cycle regulation in Caulobacter. Proc. Natl. Acad. Sci. USA 95:1443-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu, J., N. Ohta, J.-L. Zhao, and A. Newton. 1999. A novel bacterial tyrosine kinase essential for cell division and differentiation. Proc. Natl. Acad. Sci. USA 96:13068-13073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zapf, J., U. Sen, Madhusudan, and J. A. Hoch. 2000. A transient interaction between two phosphorelay proteins trapped in a crystal lattice reveals the mechanism of molecular recognition and phosphotransfer in signal transduction. Structure 8:851-862. [DOI] [PubMed] [Google Scholar]