Abstract
MinD binds to phospholipid vesicles in the presence of ATP and is released by MinE, which stimulates the MinD ATPase. Membrane binding requires a short conserved C-terminal region, which has the potential to form an amphipathic helix. This finding has led to a model in which the binding of ATP regulates the formation or accessibility of this helix, which then embeds in the membrane bilayer. To test this model, we replaced each of the four hydrophobic residues within this potential helix with tryptophan or a charged residue. Introduction of a negatively charged amino acid decreased membrane binding of MinD and its ability to activate MinC. In contrast, mutants with tryptophan substitutions retained the ability to bind to the membrane and activate MinC. Fluorescence emission spectroscopy analysis of the tryptophan mutants F263W, L264W, and L267W confirmed that these tryptophan residues did insert into the hydrophobic interior of the bilayer. We conclude that membrane binding by MinD involves penetration of the hydrophobic residues within the C-terminal amphipathic helix into the hydrophobic interior of the bilayer.
The min system spatially regulates the placement of the septum in Escherichia coli through an oscillation process in which the Min proteins rapidly switch between the ends of the cell (8, 9, 11, 22, 23). This oscillatory behavior of the Min proteins prevents the formation of Z rings near the cell poles while allowing a Z ring to form at the middle of the cell (1, 20). The min locus encodes three proteins (5), MinC, MinD, and MinE, all of which oscillate. MinD and MinE are the essential components of the oscillatory system, and their movements are coupled (23). MinC is an antagonist of FtsZ assembly (14) and is positioned in the cell by the action of the MinDE oscillator through interaction with MinD (11, 22).
The oscillatory behavior of the Min proteins, as well as the dependency patterns, was determined with functional green fluorescent protein (GFP) fusions. Expression of each of the Min proteins in the absence of the others revealed that MinD bound uniformly to the membrane whereas MinC and MinE were cytoplasmic (11, 22, 23). MinE was recruited to the membrane by MinD and also induced oscillation of MinD between the poles (8, 9). MinC was also recruited to the membrane by MinD and, if MinE was present, oscillated with the same pattern as MinD (11, 22). Subsequent studies with MinE-GFP revealed that MinE was mostly present as a ring that also oscillated, with its movement coupled to that of MinD (8, 9).
An oscillatory cycle of the Min proteins can be summarized as follows. MinD forms a polar cap at one pole with MinE mostly present as a ring at the rim of the polar cap near the middle of the cell. As the MinD cap and accompanying MinE ring recede towards the pole, a new polar MinD cap is formed at the opposite pole. Once this new polar cap is formed, the MinE ring, released from the first pole, reassembles at the rim of this new cap near the middle of the cell and the process is repeated. If MinC is present, it oscillates with the same pattern as MinD. The oscillation is rapid, with a complete cycle taking approximately 50 s (11, 22, 23).
Observation of the oscillatory behavior indicates that the Min proteins are at the membrane in the polar cap but in the cytoplasm during the transition phase. How do the Min proteins bind to the membrane and how is the process regulated? In vitro studies demonstrated that the MinD ATPase, known to be necessary for spatial regulation of cell division by the min system (4), was stimulated 10-fold by MinE in the presence of phospholipid vesicles (12). Importantly, examination of several MinE mutants revealed that the magnitude of the stimulation by MinE correlated inversely with the period of the oscillation. The requirement of phospholipid vesicles for ATPase stimulation led to the discovery that MinD binds directly to phospholipid vesicles in the presence of ATP and is released by MinE-induced ATP hydrolysis (10, 18). When MinC is present, it is recruited to the vesicles by MinD and also released by MinE (15, 18). Furthermore, the binding of MinD to the membrane appears to be largely due to hydrophobic interactions as the association is resistant to high ionic strength (13). These results demonstrated that the reversible membrane association of the Min proteins is dependent upon the MinD ATPase cycle.
Direct interaction of MinD with phospholipid vesicles was not predicted from the structure, which indicated a typical cytoplasmic protein (3). However, deletion analysis revealed that the C-terminal ∼10 amino acids, which are disordered in the crystal structure, are essential for membrane binding (13, 26). Deletion of as little as three amino acids reduces binding, although deletion of 10 further reduces binding. Interestingly, the C-terminal 11 amino acids of MinD are relatively conserved and have the potential to form an amphipathic helix. This led to a model in which membrane binding involves the insertion of the hydrophobic face of the amphipathic helix into the bilayer (13, 26). Consistent with this model, altering the phasing of the hydrophobic residues within the putative helix (26) or replacing hydrophobic residues with charged residues (13, 26) eliminates membrane binding. Furthermore, in this model the role of ATP is to regulate the formation or availability of this helix.
MinD binds to phospholipid vesicles and dimerizes (15) but undergoes further assembly on the vesicle surface to form polymers that tubulate the vesicles (10). Authors of another study also observed MinD assembly in the presence of phospholipid vesicles; however, the filaments did not appear to be on the surfaces of the vesicles (24). This difference is somewhat surprising considering MinD's well-characterized affinity for phospholipid vesicles. Tubulation of vesicles is also observed with eukaryotic cell proteins involved in vesicle formation, such as dynamin, endophilin, and amphiphysin (6, 25). These proteins are thought to tubulate vesicles as a result of insertion of an amphipathic helix into the vesicle bilayer and subsequent oligomerization. Another eukaryotic vesicle-promoting protein, epsin, also causes tubulation of vesicles in vitro; however, the mechanism appears distinct as epsin does not oligomerize and the tubes have smaller diameters (7). MinD's mechanism of tubulation appears to be very similar to that of the dynamin group (10).
To test the above model for MinD binding to the membrane, we have generated a series of mutants, including several with single tryptophan substitutions within the C-terminal region. Results from analysis of all these substitutions, both in vivo and in vitro, are consistent with the notion that the C terminus forms an amphipathic helix that mediates membrane binding. Furthermore, tryptophanyl fluorescence is a good reporter of the local environment and has been used previously to monitor insertion of a peripheral membrane protein into the lipid bilayer (2). We have therefore employed tryptophan fluorescence spectroscopy to determine whether the hydrophobic residues within the C-terminal amphipathic helix are embedded within the membrane bilayer.
MATERIALS AND METHODS
Strains and plasmids.
The E. coli K-12 strain JS964 (MC1061 malP::lacIq Δmin::kan) was used in this study (21). The plasmid pZH110 (minC) contains the minC gene with its own promoter cloned into the medium copy vector pGB2 and was described before (11). Plasmid pZH106 (gfp-minD) has been described previously and contains the gfp-minD fusion downstream of an arabinose-inducible promoter in the vector pBAD18 (11). Point mutations that produced mutants K260A K261A, K260E, K261E, L264E L267E, K265A R266A, F263E, F263W, L264E, and L264W were introduced into minD on pZH106 by using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, Calif.). Mutations generating mutants L267E, L267W, F268D, F268W, and L264W F268D were produced by a PCR procedure similar to that used for the construction of wild-type pZH106 but with the 3′ primers incorporating the desired mutations. Plasmid pZH115 (minD) contains the minD gene cloned downstream of the tac promoter in the expression vector pJF118EH (12). Similar plasmids were constructed to overexpress the various MinD mutants by cloning the PCR fragments corresponding to F263W, L264W, and L264W F268D into pJF118EH, resulting in plasmids pHJZ106-263, pHJZ106-264, and pHJZ106-WD. The PCR fragments corresponding to L267W and F268W were inserted into the multicloning site downstream of PBAD between the EcoRI and HindIII sites on the vector pBAD18, giving rise to the overexpression plasmids pHJZ106-267 and pHJZ106-268, respectively. All mutations were verified by DNA sequencing.
Fluorescence microscopy.
The effects of point mutations in gfp-minD on membrane localization of MinD were determined as described previously (11). Briefly, plasmids pZH106 and derivatives were introduced into strain JS964 (Δmin) by selecting for Ampr. Cultures were grown at 37°C until the optical density at 600 nm reached 0.05. At this point, 0.001% arabinose was added, and samples were examined 2 h later. Samples were fixed with 2% glutaraldehyde, placed on microscope slides, and photographed with a Nikon fluorescence microscope equipped with a MagnaFire charge-coupled device camera (Optronics). Images were imported into Adobe Photoshop for assembly.
Phenotypic analysis of minD mutants.
Strain JS964 (Δmin) was transformed with plasmids containing gfp-minD mutations and with pZH110 (minC), and Ampr and Spcr colonies were selected on Luria-Bertani plates containing ampicillin, spectinomycin, and 0.2% glucose (13). Colonies appeared after overnight incubation at 37°C. The cell morphology was examined microscopically, and the colonies were restreaked onto plates with ampicillin and spectinomycin. Arabinose was included at 0.0001% and 0.001% as indicated. The effect of minD mutations on cell morphology was determined after overnight incubation. Cells were examined by a Nikon phase-contrast microscope.
Protein purification.
MinE was purified from strain JS964 (Δmin) containing pJPB216 (minE), as previously described (12). The MinD mutants F263W and L264W and the double mutant L264W F268D were overexpressed by using plasmids pHJZ106-263, pHJZ106-264, and pHJZ106-WD, respectively. Isopropyl-β-d-thiogalactopyranoside (IPTG; 1 mM) was added to induce the overexpression. The MinD mutants L267W and F268W were overproduced by using plasmids pHJZ106-267 and pHJZ106-268, respectively. Arabinose (0.1%) was added to induce the overproduction. The purification procedure was the same as that for the wild-type MinD (12), and the mutants behaved the same as the wild-type protein during the purification. The protein concentrations were determined with the protein assay (Bio-Rad Laboratories, Hercules, Calif.), and the mutants were judged to be >95% pure by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Phospholipid vesicles.
E. coli phospholipids were purchased from Avanti Polar Lipids (Alabaster, Ala.). Multilamellar large vesicles were prepared as follows. The E. coli phospholipids were suspended on ice in buffer A (25 mM Tris-HCl [pH 7.5]-40% glycerol) with vigorous vortexing, followed by a rapid 50-fold dilution in buffer B (25 mM Tris-HCl [pH 7.5]-50 mM KCl). After a 30-min incubation on ice, phospholipid vesicles were collected by centrifugation for 20 min at 36,500 rpm in a Beckman 50.2 Ti rotor. The vesicles were suspended at 2 mg/ml and incubated at 65°C for 2 h with occasional vortexing. Small unilamellar vesicles (SUVs) used in the ATPase assay for MinD assembly and tryptophanyl fluorescence measurements were prepared by sonication as described previously (12). To prepare SUVs with a lipid-embedded quencher, synthetic 1-palmitoyl-2-stearoyl (n-doxyl)-sn-glycero-3-phosphocholine (doxyl-PC) with the spin label at the 7 position of the sn-2 acyl chain (Avanti Polar Lipids) was mixed with E. coli phospholipids at a ratio of 1:9, respectively, and processed as described above.
MinD ATPase and phospholipid vesicle binding assay.
The MinD ATPase assay was performed as described previously (12). MinD and MinD mutants (9 μM) and SUVs (400 μg/ml) were mixed with 1 mM [γ-32P]ATP in an ATPase buffer (25 mM Tris-HCl [pH 7.5]-50 mM KCl-5 mM MgCl2). MinE (9 μM) was added to the reaction mixtures as indicated. Reaction mixtures were incubated at 30°C, and samples were taken at 20- and 40-min time points. The amount of released radioactive Pi was determined, and the specific activities were calculated from the results of an average of three experiments. Binding of MinD mutants to phospholipid vesicles was determined by a sedimentation assay. The MinD mutant (4 μM), multilamellar large vesicles (400 μg/ml), and nucleotide (1 mM ADP or ATP) were mixed at room temperature in 50 μl of ATPase buffer. In experiments to determine whether MinE could release MinD from the vesicles, MinE (4 μM) was then added. The reaction mixtures were incubated at room temperature for 20 min. For reaction mixtures with MinE added, the samples were incubated at 30°C. Samples were centrifuged at 6,500 × g at room temperature in a tabletop centrifuge for 2 min. The supernatants were carefully removed, and the pellets were resuspended in 50 μl of SDS-sample buffer. Aliquots (20 μl) of the samples were electrophoresed on SDS-12.5% PAGE and stained with Coomassie brilliant blue.
Electron microscopy analysis of the interaction of MinD with phospholipid vesicles.
Preparation of samples for electron microscopy was described previously (10). In brief, MinD (7.5 μM), SUVs (40 μg/ml), and nucleotide (1 mM ATP) were mixed at room temperature in 25 μl of ATPase buffer. The reaction mixture was incubated at 30°C for 5 to 10 min. Then 10 μl of the sample was adsorbed onto glow discharge carbon-coated grids and negatively stained. The specimens were examined with a JEOL 1200EX microscope.
Protein fluorescence measurements.
Tryptophanyl fluorescence measurements were made with a Cary Eclipse fluorescence spectrophotometer utilizing Cary Eclipse software (Varian Instruments, Walnut Creek, Calif.). MinD mutants (4 μM) and nucleotide (1 mM ATP or ADP) were mixed at room temperature in 100 μl of ATPase buffer. SUVs (100 μg/ml) or SUVs containing the spin label were added as indicated. Samples were loaded into 100-μl cuvettes and incubated at 30°C in the cell holder. Samples were excited at 290 nm with a slit width of 5 nm. Emission spectra were collected from 300 to 400 nm. Spectra were corrected for the blank which contained buffer or buffer and vesicles. Data were assembled and plotted by Sigma Plot.
RESULTS
Effects of mutations in the amphipathic helix.
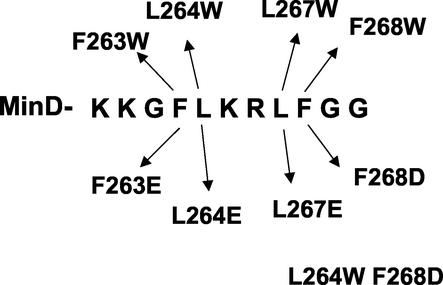
The binding of MinD to the membrane can be assessed in vivo through examination of a GFP-MinD fusion or in vitro through the use of purified MinD and a sedimentation assay with phospholipid vesicles. To assess the importance of the chemical nature of the amino acid residues in the C-terminal region to membrane binding, mutations were introduced into a gfp-minD fusion contained on an expression vector and first screened in vivo. Analysis of MinDs from various bacteria indicates that the E. coli MinD sequence, KKGFLKRLFGG, serves as a consensus sequence for this family. The most highly conserved features of the C-terminal region include four large hydrophobic residues separated by two basic residues and preceded by two basic residues and a glycine. The mutations were designed to assess the importance of the highly conserved basic and hydrophobic residues (Fig. 1), and the results are summarized in Fig. 2 and Table 1.
FIG. 1.
Sequence of the amphipathic helix and illustration of the amino acid substitutions examined in this study. The sequence of the last 11 amino acids of the MinD protein is depicted as well as the amino acid substitutions that were generated and examined in this study.
FIG. 2.
In vivo analysis of the effect of amino acid substitutions within the amphipathic helix on membrane localization. Mutations were introduced into the gfp-minD fusion carried on pZH106 (inducible with arabinose). The effect of the substitutions on localization of MinD was examined in strain JS964 (Δmin). Following induction with 0.001% arabinose for 2 h, cells were fixed and examined by fluorescence microscopy. One cell for each mutant is shown.
TABLE 1.
Effect of minD mutations on MinD localization and MinC activation
| MinD in GFP-MinD fusion | Localization | Ability to activate MinCa [in strain JS964(pGB2minC) (Δmin)] on plates with:
|
|||
|---|---|---|---|---|---|
| Glucose | 0% Ara | 0.0001% Ara | 0.001% Ara | ||
| K260A K261A | Membrane | + | + | + | + |
| K260E | Membrane | + | + | + | + |
| K261E | Membrane | + | + | + | + |
| L264E L267E | Cytoplasm | − | − | − | +/− |
| K265A R266A | Membrane | + | + | + | + |
| F263E | Cytoplasm | − | − | +/− | + |
| F263W | Membrane | + | + | + | + |
| L264E | Cytoplasm | − | + | + | + |
| L264W | Membrane | + | + | + | + |
| L267E | Cytoplasm | − | + | + | + |
| L267W | Membrane | + | + | + | + |
| F268D | Cytoplasm | − | − | − | − |
| F268W | Membrane | + | + | + | + |
| L264W F268D | Cytoplasm | − | +/− | +/− | + |
| Wild type | Membrane | + | + | + | + |
| K16Q | Cytoplasm | − | − | − | − |
Activation was indicated by cell morphology. +, filamentation; +/−, slight filamentation; −, minicell phenotype with no filamentation. Ara, arabinose.
Replacing the two leucines with glutamate eliminated the halo appearance of the corresponding GFP-MinD fusion, indicating a lack of membrane binding. In addition, replacing any one of the hydrophobic residues with a negatively charged residue eliminated the halo appearance of the GFP-MinD fusion, indicating a loss of membrane binding. In contrast, changing the pair of lysines preceding the hydrophobic residues to alanines did not eliminate the halo, indicating that the mutant protein still bound to the membrane. Likewise, changing the two basic residues that are flanked by the hydrophobic residues to alanines did not eliminate membrane binding. From this analysis, it appears that each of the hydrophobic residues is necessary for membrane binding whereas the basic residues appear to be dispensable. We have not determined whether all of the basic residues can be dispensed with simultaneously.
The results described above demonstrated the importance of the hydrophobic residues. To determine whether the specific amino acids were important, we changed each of the hydrophobic residues to tryptophan. If the mutants behaved similarly to the wild type they could be used in vitro for fluorescence spectroscopy to determine whether the residues were embedded in the membrane bilayer. Analysis of the fusions of GFP with each of the four tryptophan mutants indicated that the proteins were on the membrane (Fig. 2; Table 1). This result suggests that as long as the hydrophobic character of the amino acids was retained the proteins bound to the membrane (Fig. 2; Table 1). As a control we also introduced a negatively charged amino acid into one of the tryptophan-containing mutants to produce L264W F268D. This double mutant did not bind to the membrane in vivo (Fig. 2; Table 1).
Effects of substitutions in the amphipathic helix on MinC activation.
MinD recruits MinC to the membrane in an ATP-dependent manner (15, 18). The resulting membrane-associated complex has increased affinity for some septal component and is a potent inhibitor of cell division (17). In contrast, MinD mutants that have the C-terminal region deleted (3 or 10 amino acids removed) bind MinC normally but are much less efficient in activating MinC since they bind the membrane poorly and therefore lack the ability to confer high affinity for a septal component on the MinCD complex (13). To test the ability of the substitution mutants to activate MinC, strain JS964 (Δmin) containing pGB2minC was transformed with the various GFP-MinD fusions. Transformants were plated in the presence of glucose and restreaked onto plates containing no sugar or various concentrations of arabinose to induce the GFP-MinD fusions. This test is useful because one can compare MinD mutants over a range of MinD concentrations to determine to what extent their activity has been diminished. It should be noted that even on glucose plates (fully repressed conditions) the basal expression of GFP-MinD is sufficient to activate MinC and cause extensive filamentation. Based upon the in vivo localization data, the mutant proteins that contain hydrophobic to charged substitutions would be expected to be less active at inhibiting division whereas those that contain tryptophan substitutions should still display wild type-like activity. Also, the mutants in which basic residues were changed to alanines should still activate MinC.
Transformants with the wild-type GFP-MinD fusion gave only a few colonies on glucose plates, and the cells were extremely filamentous (Table 1). This result is similar to that previously reported (13) and demonstrates that even the fully repressed level of the GFP-MinD fusion is sufficient to inhibit division. Similar results were obtained with both of the mutants in which either of the two pairs of basic residues were changed to alanines (K260A K261A and K265A R266A) (Table 1). Also, changing either lysine 260 or 261 to glutamate did not affect the ability of MinD to activate MinC (Table 1). However, changing any one of the four large hydrophobic residues to a negatively charged amino acid resulted in transformants that were not filamentous on glucose, indicating that the corresponding mutant MinD was attenuated in its ability to activate MinC. By restreaking these transformants onto plates with various arabinose concentrations, it was apparent that replacing the phenylalanines attenuated MinD more than replacing the leucines. However, replacing both leucines resulted in a dramatically attenuated MinD (Table 1). From these results we conclude that substituting a negatively charged amino acid for any one of these hydrophobic residues results in a cytoplasmic MinD. Consistent with this conclusion, the mutant MinDs were less efficient in activating MinC. In contrast, each of the four single tryptophan mutants appeared as potent as the wild-type MinD in activating MinC and causing filamentation (Table 1). As expected, the double mutant L264W F268D was deficient in membrane binding and activation of MinC.
In vitro analysis of the MinD mutants.
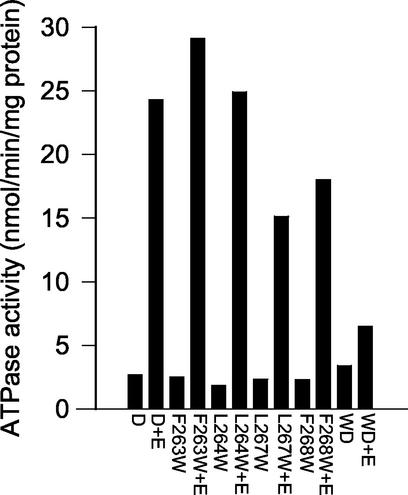
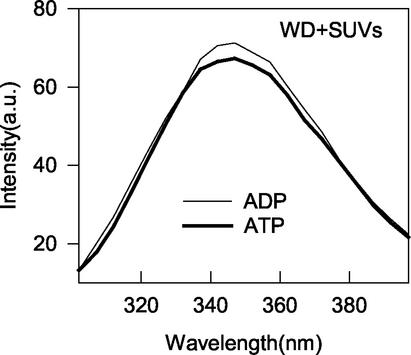
Each of the MinD tryptophan substitution mutants was cloned into an expression vector and expressed as an unfused protein. The mutant proteins were then overexpressed and purified. Each behaved similarly to the wild-type protein during purification. We then assessed these mutant proteins for the known in vitro activities of MinD, which include ATP-dependent membrane binding, ATPase activity, and tubulation of vesicles (10). Each of the mutant proteins expressed a basal ATPase activity in the presence of phospholipid vesicles that was comparable to the wild-type basal activity (Fig. 3). Addition of MinE to F263W and L264W resulted in an approximately 10- to 12-fold increase in the ATPase activity, whereas addition of MinE to L267W and F268W resulted in a six- to eightfold stimulation. The double mutant L264W F268D had a slightly higher basal activity than the wild type but was stimulated less than twofold by MinE. Thus, L264W and F263W behaved very similarly to wild-type MinD, and the behavior of L267W and F268W was near that of the wild type. In contrast, the poor stimulation of the ATPase activity of the double mutant is consistent with its deficiency in membrane binding in vivo.
FIG. 3.
ATPase activity of mutants containing tryptophan. The purified MinD mutant proteins were analyzed for ATPase activity along with the wild type. The proteins were incubated at 9 μM in the presence of phospholipid vesicles (400 μg/ml) and with or without MinE (9 μM). D, MinD; E, MinE; WD, double mutant L264W F268D.
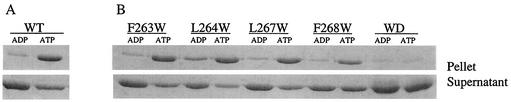
To confirm that these single tryptophan-containing mutants bound to phospholipid vesicles, we used the sedimentation assay. As shown in Fig. 4, these mutant proteins exhibited ATP-dependent binding to the vesicles. The extent of binding by F263W and L264W was quantitatively comparable to that by the wild-type protein. In contrast, the extent of binding by the other two mutants, L267W and F268W, was somewhat less. The addition of MinE resulted in a decrease in the amount of the MinD mutant proteins bound to the membrane, even for the two that bound less well (data not shown). This MinE-induced release of the protein is consistent with the ability of MinE to stimulate the ATPase activity of these mutant proteins. The double mutant did not bind significantly to the vesicles (Fig. 4).
FIG. 4.
Binding of MinD mutants to the membrane. Binding of the MinD mutant proteins to the membrane was determined by a sedimentation assay. Either MinD (4 μM) (A) or the mutant proteins (B) were mixed with phospholipid vesicles (400 μg/ml) and ADP or ATP (1 mM). After a 20-min incubation at room temperature, the samples were centrifuged and the pellets were solubilized in SDS-sample buffer. Equivalent aliquots of the pellets and supernatants were analyzed by SDS-PAGE. WT, wild type; WD, double mutant L264W F268D.
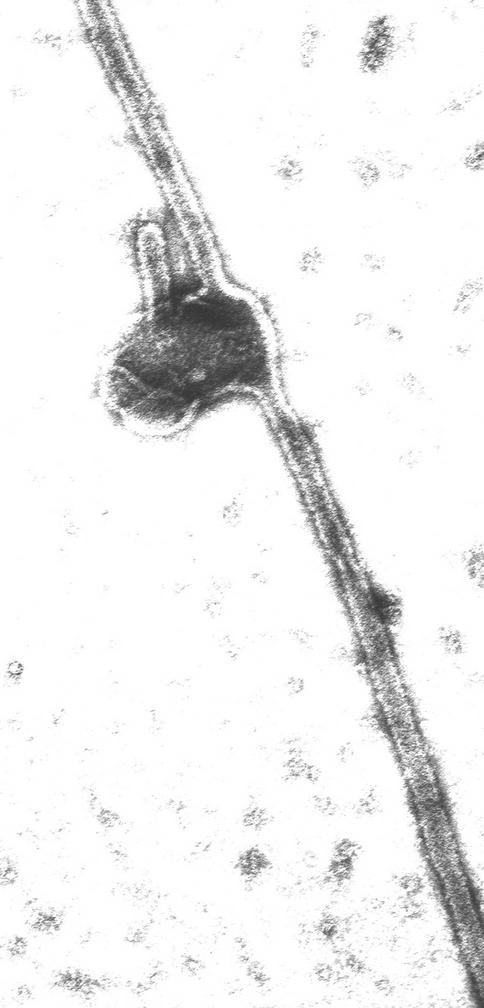
These in vitro results combined with the in vivo results indicate that the single mutants containing tryptophan interact with a phospholipid bilayer. However, as previously reported, MinD is also able to tubulate vesicles. This activity towards vesicles is observed with proteins that display two properties: (i) they bind to the membrane by virtue of an amphipathic helix, and (ii) they oligomerize. To determine whether these activities were retained by the tryptophan-containing mutants, three of the mutant proteins, L264W, F263W, and L264W F268D, were incubated with ATP and vesicles and examined by electron microscopy following negative staining. Both single mutants were able to tubulate vesicles in a manner similar to the that of the wild-type protein; however, the double mutant was not (Fig. 5 [only L264W is shown]). Thus, these single mutants containing tryptophan behave similarly in vivo and in vitro to the wild-type protein and were therefore used for fluorescence spectroscopy.
FIG. 5.
Tubulation of vesicles by L264W. The MinD mutant L264W (7.5 μM) was mixed with phospholipid vesicles (40 μg/ml) in ATPase buffer. After a 5-min incubation at 30°C, an aliquot was analyzed by electron microscopy following negative staining.
Examination of the trp-containing mutants by fluorescence spectroscopy.
The interaction of MinD with the phospholipid vesicles was further explored by exploiting the tryptophan-containing mutants. The fluorescence of tryptophan is sensitive to the local environment and exhibits a blueshift with altered intensity upon encountering a more hydrophobic environment (2). The wild-type protein, which lacks tryptophan, displayed little fluorescence when an excitation wavelength of 290 nm was used (data not shown). In contrast, the mutants all exhibited marked emission spectra in the absence of nucleotides, indicating the presence of tryptophan (data not shown).
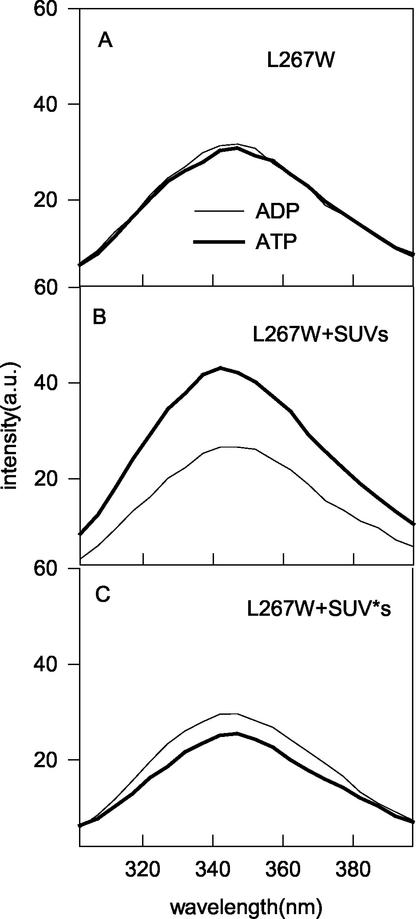
The intensity of the emission spectrum of L264W in the absence of phospholipid vesicles was slightly higher with ADP than with ATP (Fig. 6A). The addition of phospholipid vesicles did not affect the emission spectrum if ADP was present; however, the sample containing ATP showed a marked increase in the fluorescence intensity that was blueshifted 12 nm (Fig. 6B). This result indicated that the environment of the tryptophan had become more hydrophobic, which may be due to a conformational change in the protein that led to an altered environment or due to insertion into the bilayer.
FIG. 6.
Effect of phospholipid vesicles and nucleotides on the fluorescence spectra of MinD mutant L264W. MinD mutant L264W (4 μM) was incubated with ADP or ATP in the absence (A) or presence (B) of phospholipid vesicles (100 μg/ml) and examined by fluorescence spectroscopy at 30°C. The excitation wavelength was 290 nm. (C) Effect of incorporating a spin label into the phospholipid vesicles on the emission spectrum of MinD mutant L264W. MinD mutant L264W was incubated in the presence of phospholipid vesicles containing 10% doxyl-PC with the spin label at the 7 position of the sn-2 acyl chain. The asterisk indicates the presence of the spin label. a.u., arbitrary units.
To determine whether the change in the emission spectrum was due to the insertion of the tryptophan residue into the bilayer, we used phospholipid vesicles containing an embedded nitroxide spin label to see if the label could quench the fluorescence of the tryptophan. The addition of this spin label did not affect MinD binding (data not shown). This spin label (doxyl-PC labeled at the 7 position of the sn-2 acyl chain) quenches the fluorescence by collisional interaction with the fluorophore, which can only occur if the tryptophan is inserted into the interior of the bilayer (2). This is therefore a direct test of whether or not the tryptophan is immersed in the hydrophobic interior of the bilayer. The emission spectrum of L264W in the presence of ADP and these vesicles was identical to that in the presence of vesicles without the spin label (Fig. 6C). However, the emission spectrum of L264W in the presence of ATP and these vesicles was altered compared to that in the presence of vesicles without the spin label. The vesicle-induced blueshift was still present; however, the intensity was significantly reduced, similar to that observed with ADP. This result indicates that the tryptophan must be inserted into the interior of the bilayer, where the fluorescence is quenched by contact with the spin label.
As an additional control for the fluorescence spectroscopy, we examined the emission spectrum of the double mutant L264W F268D. As shown above, this mutant does not bind to the membrane due to the introduction of the negatively charged aspartate residue. As shown in Fig. 7, the spectrum of this mutant does not display a blueshift or change in intensity upon addition of ATP and phospholipid vesicles; the spectrum is superimposable upon that obtained with ADP. This result further demonstrates that membrane binding is required for observation of a blueshift and an increase in the intensity of the spectrum of the L264W mutant.
FIG. 7.
Effect of substituting a negatively charged amino acid for F268 on the fluorescence spectra of MinD mutant L264W. The double MinD mutant L264W F268D (WD; 4 μM) was incubated with phospholipid vesicles (100 μg/ml) and ADP or ATP (1 mM), and the emission spectra were obtained. a.u., arbitrary units.
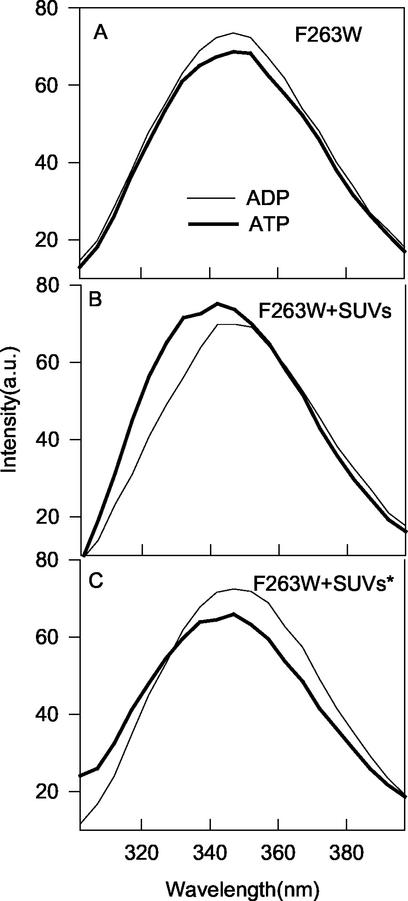
The fluorescence spectrum of F263W was qualitatively similar to that of L264W (Fig. 8). The above assays showed that F263W bound well to phospholipid vesicles in the presence of ATP and that its ATPase activity was stimulated about 10-fold by MinE. It was also released from vesicles by MinE. In the absence of vesicles, the intensity of F263W was slightly higher with ADP than with ATP (Fig. 8A). In the presence of vesicles, the fluorescence intensity of F263W was higher with ATP than with ADP and displayed an ATP-dependent blueshift of 7 nm (Fig. 8B). As with L264W, the increase in intensity was quenched by the use of phospholipid vesicles containing the nitroxide spin label (Fig. 8C). Thus, we conclude that the tryptophan residue in this mutant also inserts into the membrane.
FIG. 8.
Effect of phospholipid vesicles and nucleotides on the fluorescence spectra of MinD mutant F263W. MinD mutant F263W (4 μM) was incubated with ADP or ATP (1 mM) in the absence (A) or presence (B) of phospholipid vesicles (100 μg/ml) and examined by fluorescence spectroscopy at 30°C. (C) Effect of incorporating a spin label into the phospholipid vesicles on the emission spectra of MinD F263W. MinD F263W was incubated in the presence of phospholipid vesicles containing 10% doxyl-PC with the spin label at the 7 position of the sn-2 acyl chain. The asterisk indicates the presence of the spin label. a.u., arbitrary units.
The other two tryptophan mutants did not bind to the vesicles as well as the first two; however, they did display some binding that was reduced by MinE. In addition, their ATPase activities were stimulated quite well by MinE. Therefore, these mutant proteins were also analyzed by fluorescence microscopy. In the absence of vesicles, the emission spectra of L267W with ADP and ATP were comparable (Fig. 9A). In the presence of vesicles, the fluorescence intensity of the sample with ATP was greater than that of the sample with ADP (Fig. 9B). The blueshift was less than that observed with the first two mutants examined; however, the increased intensity was effectively quenched by incorporation of the nitroxide spin label into the vesicles (Fig. 9C). We conclude that the L267W tryptophan residue is also embedded in the bilayer. The emission spectra obtained for F268W in the presence of vesicles were not significantly different in the presence of ATP or ADP and therefore the tryptophan residue of this mutant may not embed in the bilayer (data not shown). Nonetheless, three of the four tryptophan substitution mutants showed blueshifts and increased intensity that was quenched by a membrane-embedded quencher, indicating that the tryptophans were penetrating into the lipid bilayer.
FIG. 9.
Effect of phospholipid vesicles and nucleotides on the fluorescence spectra of MinD mutant L267W. MinD mutant L267W (4 μM) was incubated with ADP or ATP in the absence (A) or presence (B) of phospholipid vesicles (100 μg/ml) and examined by fluorescence spectroscopy at 30°C. The excitation wavelength was 290 nm. (C) Effect of incorporating a spin label into the phospholipid vesicles on the emission spectra of MinD mutant L267W. MinD mutant L267W was incubated in the presence of phospholipid vesicles containing 10% doxyl-PC with the spin label at the 7 position of the sn-2 acyl chain. The asterisk indicates the presence of the spin label. a.u., arbitrary units.
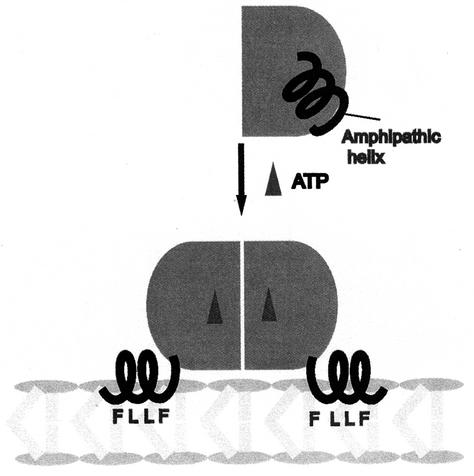
FIG. 10.
Model for MinD binding to the membrane. In the presence of ADP, MinD is a monomer and the C-terminal amphipathic helix is unavailable for interaction with the membrane. In this model, the helix is formed but sequestered by interaction with the rest of the protein. Upon ATP binding, the amphipathic helix is available for interaction with the membrane. Upon association with the membrane, the hydrophobic residues are inserted into the interior of the bilayer, rendering the association resistant to high ionic strength.
DISCUSSION
MinD binds to phospholipid vesicles in an ATP-dependent manner, recruits MinE, and is released following MinE-stimulated ATP hydrolysis (10). The binding of MinD to phospholipid vesicles requires a short conserved C-terminal tail of 10 amino acids which has the potential to form an amphipathic helix (13, 26). The binding is resistant to high ionic strength, indicating that the interaction is mostly hydrophobic (13). The goal of this study was to determine whether the amphipathic helix at the C terminus of MinD was directly involved in membrane binding. The results obtained demonstrate that three of the large hydrophobic residues (when replaced by tryptophan) within the amphipathic helix are inserted into the hydrophobic interior of the phospholipid bilayer. We therefore conclude that the binding of MinD to the membrane is due to insertion of the hydrophobic surface of this amphipathic helix into the bilayer (Fig. 10).
In this study we used tryptophan as a reporter to monitor the chemical environment of the hydrophobic residues within the amphipathic helix. Although this approach involves the substitution of tryptophan for the resident leucine or phenylalanine residues, it did not appear to significantly affect MinD function. All four of the tryptophan mutants bound to the membrane in vivo and were able to activate MinC as well as the wild-type MinD. Two of the four mutants behaved like the wild type in vitro, whereas two appeared slightly diminished in activity. The two mutant proteins with substitutions at positions 263 and 264 bound to phospholipid vesicles and had their ATPase stimulated by MinE, similar to wild-type MinD. Thus, tryptophan substitutions at these positions should be excellent reporters of the environment of the wild-type residues. The mutants with substitutions at positions 267 and 268 did not bind to the membrane as well as the wild type did and their ATPase activity was somewhat less stimulated, however, indicating that tryptophan at these positions may slightly impact MinD activity. However, the mutants behaved similarly to MinD in vivo and are therefore also likely to be good reporters.
This study provides strong evidence that several of the hydrophobic residues within the amphipathic helix are embedded in the hydrophobic interior of the bilayer. Both L264W and F263W showed significant blueshifts and increases in intensity that were dependent upon ATP and phospholipid vesicles. Likewise, L267W displayed an increase in intensity, although it displayed less of a blueshift than the other two mutants. Importantly, the increases in intensities of these three mutant proteins were quenched by the addition of a nitroxide spin label embedded in the bilayer through coupling to the 7 position of a fatty acid chain at the sn-2 position. Since this type of quencher is collisional, only tryptophan residues that insert into the bilayer would come into contact with the spin label so that the fluorescence is quenched (2). Thus, this study provides strong evidence that this region of the amphipathic helix is inserted into the phospholipid bilayer.
In addition to using the vesicles with the spin label, we examined a double mutant, L264W F268D, as a control in the fluorescence experiments. It did not bind to the membrane in vivo or activate MinC. It did not display significant vesicle binding or stimulation by MinE in vitro. Importantly, its emission spectrum was not affected by the addition of ATP and phospholipid vesicles. This result is consistent with the notion that the blueshift and intensity changes of the L264W single mutant are due to insertion into the bilayer.
In contrast to the results with the three tryptophan mutants described above, the F268W mutant did not show a significant change in the emission spectrum in the presence of ATP and phospholipid vesicles. It localized to the membrane and activated MinC in vivo but interacted less well with a bilayer in vitro than did wild-type MinD. Thus, tryptophan inserted at the distal end of the amphipathic helix does not appear to embed into the hydrophobic interior of the bilayer to the same extent as tryptophan substitutions at the other hydrophobic positions. Why tryptophan at this position and at position 267 appears to be less well tolerated than that at the beginning of the amphipathic helix is not clear. It is quite possible that the wild-type phenylalanine residue at position 268 is inserted into the bilayer.
We also examined the role of the positively charged amino acids by replacing them with alanines. Two mutants were made in which the two lysines at the beginning of the helix or the lysine-arginine pair located between the hydrophobic residues were changed to alanines. These mutants were still localized to the membrane in vivo and activated MinC as well as wild-type MinD.
How might the insertion of the amphipathic helix in the membrane be regulated? Since membrane binding by MinD is ATP dependent, it has been proposed that the binding of ATP by MinD affects the conformation of the C-terminal region such that it is available for interaction with the bilayer (13, 26). This effect of ATP could be mediated by dimerization of MinD, which is also ATP dependent (15).
Many proteins interact with the membrane through amphipathic helices (16). The prevailing notion is that amphipathic helices are disordered and become structured upon interaction with the membrane. Several have been studied in some detail and are interesting for comparison. Recently, the structure of epsin has been determined in the presence and absence of a ligand which promotes membrane binding by ordering an N-terminal amphipathic helix (7). In the absence of the ligand the structure of the N-terminal region is disordered; however, upon binding of the ligand the N-terminal region forms an amphipathic helix, held in place by the ligand. On the other hand, several Arf family members have also been studied in some detail (19). Arf6 and Arf1 have been crystallized in the presence of GDP and GTP. In the presence of GDP, the N terminus, which carries an amphipathic helix similar to that of MinD, is folded back on the protein such that it is unavailable for interaction with the membrane. However, GTP binding induces a conformational change that expels the N-terminal amphipathic helix such that it is free to interact with the membrane. Thus, there appear to be two possible mechanisms for regulating an amphipathic helix. In the first case, the potential helix is induced to form by a ligand, whereas in the second the helix is sequestered and made available by ligand binding. We suspect that MinD is more like the Arf example and that the helix is sequestered and released upon ATP binding.
Acknowledgments
This work was supported by grant GM 29764 from the National Institutes of Health.
REFERENCES
- 1.Bi, E., and J. Lutkenhaus. 1993. Cell division inhibitors SulA and MinCD prevent formation of the FtsZ ring. J. Bacteriol. 175:1118-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chapman, E. R., and A. F. Davis. 1998. Direct interaction of a Ca2+-binding loop of synaptotagmin with lipid bilayers. J. Biol. Chem. 273:13995-14001. [DOI] [PubMed] [Google Scholar]
- 3.Cordell, S. C., and J. Lowe. 2001. Crystal structure of the bacterial cell division regulator MinD. FEBS Lett. 492:160-165. [DOI] [PubMed] [Google Scholar]
- 4.de Boer, P. A., R. E. Crossley, A. R. Hand, and L. I. Rothfield. 1991. The MinD protein is a membrane ATPase required for the correct placement of the Escherichia coli division site. EMBO J. 10:4371-4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Boer, P. A., R. E. Crossley, and L. I. Rothfield. 1989. A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell 56:641-649. [DOI] [PubMed] [Google Scholar]
- 6.Farsad, K., N. Ringstad, K. Takei, S. R. Floyd, K. Rose, and P. De Camilli. 2001. Generation of high curvature membranes mediated by direct endophilin bilayer interactions. J. Cell Biol. 155:193-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ford, M. G., I. G. Mills, B. J. Peter, Y. Vallis, G. J. Praefcke, P. R. Evans, and H. T. McMahon. 2002. Curvature of clathrin-coated pits driven by epsin. Nature 419:361-366. [DOI] [PubMed] [Google Scholar]
- 8.Fu, X., Y. L. Shih, Y. Zhang, and L. I. Rothfield. 2001. The MinE ring required for proper placement of the division site is a mobile structure that changes its cellular location during the Escherichia coli division cycle. Proc. Natl. Acad. Sci. USA 98:980-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hale, C. A., H. Meinhardt, and P. A. de Boer. 2001. Dynamic localization cycle of the cell division regulator MinE in Escherichia coli. EMBO J. 20:1563-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu, Z., E. P. Gogol, and J. Lutkenhaus. 2002. Dynamic assembly of MinD on phospholipid vesicles regulated by ATP and MinD. Proc. Natl. Acad. Sci. USA 99:6671-6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu, Z., and J. Lutkenhaus. 1999. Topological regulation of cell division in Escherichia coli involves rapid pole to pole oscillation of the division inhibitor MinC under the control of MinD and MinE. Mol. Microbiol. 34:82-90. [DOI] [PubMed] [Google Scholar]
- 12.Hu, Z., and J. Lutkenhaus. 2001. Topological regulation of cell division in E. coli. Spatiotemporal oscillation of MinD requires stimulation of its ATPase by MinE and phospholipid. Mol. Cell 7:1337-1343. [DOI] [PubMed] [Google Scholar]
- 13.Hu, Z., and J. Lutkenhaus. 2003. A conserved sequence at the C-terminus of MinD is required for binding to the membrane and targeting MinC to the septum. Mol. Microbiol. 47:345-355. [DOI] [PubMed] [Google Scholar]
- 14.Hu, Z., A. Mukherjee, S. Pichoff, and J. Lutkenhaus. 1999. The MinC component of the division site selection system in Escherichia coli interacts with FtsZ to prevent polymerization. Proc. Natl. Acad. Sci. USA 96:14819-14824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu, Z., C. Saez, and J. Lutkenhaus. 2003. Recruitment of MinC, an inhibitor of Z-ring formation, to the membrane in Escherichia coli: role of MinD and MinE. J. Bacteriol. 185:196-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson, J. E., and R. B. Cornell. 1999. Amphitropic proteins: regulation by reversible membrane interactions. Mol. Membr. Biol. 16:217-235. [DOI] [PubMed] [Google Scholar]
- 17.Johnson, J. E., L. L. Lackner, and P. A. de Boer. 2002. Targeting of DMinC/MinD and DMinC/DicB complexes to septal rings in Escherichia coli suggests a multistep mechanism for MinC-mediated destruction of nascent FtsZ rings. J. Bacteriol. 184:2951-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lackner, L. L., D. M. Raskin, and P. A. de Boer. 2003. ATP-dependent interactions between Escherichia coli Min proteins and the phospholipid membrane in vitro. J. Bacteriol. 185:735-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pasqualato, S., L. Renault, and J. Cherfils. 2002. Arf, Arl, Arp and Sar proteins: a family of GTP-binding proteins with a structural device for ′front-back' communication. EMBO Rep. 3:1035-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pichoff, S., and J. Lutkenhaus. 2001. Escherichia coli division inhibitor MinCD blocks septation by preventing Z-ring formation. J. Bacteriol. 183:6630-6635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pichoff, S., B. Vollrath, C. Touriol, and J. P. Bouche. 1995. Deletion analysis of gene minE which encodes the topological specificity factor of cell division in Escherichia coli. Mol. Microbiol. 18:321-329. [DOI] [PubMed] [Google Scholar]
- 22.Raskin, D. M., and P. A. de Boer. 1999. MinDE-dependent pole-to-pole oscillation of division inhibitor MinC in Escherichia coli. J. Bacteriol. 181:6419-6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raskin, D. M., and P. A. de Boer. 1999. Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc. Natl. Acad. Sci. USA 96:4971-4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suefuji, K., R. Valluzzi, and D. RayChaudhuri. 2002. Dynamic assembly of MinD into filament bundles modulated by ATP, phospholipids, and MinE. Proc. Natl. Acad. Sci. USA 99:16776-16781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sweitzer, S. M., and J. E. Hinshaw. 1998. Dynamin undergoes a GTP-dependent conformational change causing vesiculation. Cell 93:1021-1029. [DOI] [PubMed] [Google Scholar]
- 26.Szeto, T. H., S. L. Rowland, L. I. Rothfield, and G. F. King. 2002. Membrane localization of MinD is mediated by a C-terminal motif that is conserved across eubacteria, archaea, and chloroplasts. Proc. Natl. Acad. Sci. USA 99:15693-15698. [DOI] [PMC free article] [PubMed] [Google Scholar]