Abstract
The filamentous cyanobacterium Anabaena (Nostoc) sp. strain PCC 7120 maintains a genome that is divided into a 6.4-Mb chromosome, three large plasmids of more that 100 kb, two medium-sized plasmids of 55 and 40 kb, and a 5.5-kb plasmid. Plasmid copy number can be dynamic in some cyanobacterial species, and the genes that regulate this process have not been characterized. Here we show that mutations in an open reading frame, all1076, reduce the numbers of copies per chromosome of several plasmids. In a mutant strain, plasmids pCC7120δ and pCC7120ζ are both reduced to less than 50% of their wild-type levels. The exogenous pDU1-based plasmid pAM1691 is reduced to less than 25% of its wild-type level, and the plasmid is rapidly lost. The peptide encoded by all1076 shows similarity to members of the GntR family of transcriptional regulators. Phylogenetic analysis reveals a new domain topology within the GntR family. PlmA homologs, all coming from cyanobacterial species, form a new subfamily that is distinct from the previously identified subfamilies. The all1076 locus, named plmA, regulates plasmid maintenance functions in Anabaena sp. strain PCC 7120.
Certain species of filamentous cyanobacteria can use oxygenic photosynthesis while simultaneously performing oxygen-labile nitrogen fixation (reviewed in reference 32). The ability to fix both carbon and nitrogen produces an exceptional degree of autonomy within the biosphere. Despite that autonomy, several of these species interact with a variety of symbiotic partners (1). These flexible organisms persist in a range of environments that include extremes of temperature, salinity, aridity, and pH (35). Ancestral organisms arose 2.5 billion to 3.6 billion years ago (reviewed in reference 38; see also reference 39), although the earliest dates have been challenged (6). Phylogenetic analysis also reveals deep roots for the cyanobacteria, which cluster with gram-positive bacteria in 16S rRNA analysis despite having an outer membrane (11). Cells from these organisms can undergo ordinary vegetative growth or differentiate into specialized cell types. These cell types include nitrogen-fixing heterocysts, spore-like akinetes, and cells that comprise motile hormogonia (32). Heterocysts are terminally differentiated in some species of filamentous cyanobacteria (21), providing an instance of cellular specialization in a multicellular organism akin to the development of tissues. A relatively advanced suite of genetic tools exists for the manipulation of the heterocystous strain Anabaena (Nostoc) sp. strain PCC 7120, making it the most commonly used organism for studying heterocyst development.
The complex properties of nitrogen-fixing strains might argue for a complex genome. The genome sequence of Anabaena sp. strain PCC 7120 contains roughly the same number of genes as the eukaryote Saccharomyces cerevisiae (27) and is comprised of a single chromosome and six plasmids. Nostoc punctiforme, another filamentous cyanobacterium with multiple developmental fates and symbiotic interactions, has a genome that is about one-third larger than that of Anabaena sp. strain PCC 7120 (33). Unlike Escherichia coli, cyanobacteria are thought to carry several genome equivalents of DNA in each cell. An estimate of 24 genome equivalents per cell in Calothrix sp. strain PCC 7601 has been published previously (46). The number of genome equivalents per cell can be calculated for two other strains, with the caveat that the data were obtained in different laboratories. Synechococcus elongatus PCC 6301 (Anacystis nidulans) has a 2.7-Mb chromosome (26) and contains 3.0 × 10−15 g of DNA per cell (13), which corresponds to 10 genome equivalents per cell. Similarly, the Anabaena variabilis genome has been estimated to be 5.7 Mb (24) and to contain 3.6 × 10−14 g of DNA per cell (13), which corresponds to approximately 6 genome equivalents per cell. In cyanobacteria, the amount of DNA per cell has been shown to differ in response to culture age, cell type, or other conditions (31, 43). The mechanisms that regulate this variation have not been characterized.
It has long been thought that the genome of Anabaena sp. strain PCC 7120 encodes a diffusible inhibitor of heterocyst development, which would provide a mechanism to place heterocysts at ordered intervals along each filament (48). Our laboratory has described such a signal, namely, a peptide named PatS. Strains that overexpress patS make no heterocysts. Strains deficient for patS form multiple contiguous heterocysts (52). The fact that only a subset of cells become heterocysts in a patS deletion strain indicates that there must be other signaling mechanisms, possibly including the diffusion of nitrogen fixation products (53) and a pathway requiring the hetN gene (9, 29).
This report describes a screening for suppressors of the patS overexpression phenotype. When plasmid-carried patS is overexpressed from a glnA promoter, suppressors might arise from genes required for plasmid maintenance, genes that regulate the glnA promoter, or genes encoding elements of the patS signaling pathway. We present an analysis of one such suppressor, plmA, having the first of those roles.
MATERIALS AND METHODS
Strains and culture conditions.
The strains, plasmids, and real-time PCR primers used in this study are described in Table 1; further details of the constructions are available from the authors. Anabaena sp. strain PCC 7120 and its derivatives were grown in BG-11 medium (which contains sodium nitrate) or BG-110 medium (which lacks sodium nitrate) at 30°C as previously described (20). For Anabaena sp. strain PCC 7120 cultures, antibiotics were used at the following final concentrations: chloramphenicol (Cm), 10 μg/ml; erythromycin (Em), 10 μg/ml; neomycin (Nm), 25 μg/ml; spectinomycin (Sp), 2 μg/ml; and streptomycin (Sm), 2 μg/ml. Concentrations were halved when antibiotics were used in combination, during the initial isolation of Anabaena exconjugants, or for strains that grew poorly. Blue-white screening of E. coli strains was performed on LB (Lennox L broth) plates with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (40 μg/ml).
TABLE 1.
Strains and plasmids used in this study
| Strain, plasmid, or primer | Characteristics | Source and/or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| AM1181 | HB101 with pRL1087b (carries transposon Tn5-1087) and pRL1045 (provides DNA methyltransferases) | This study |
| AM1460 | HB101 with conjugal plasmid pRK2013 | 30 |
| Anabaena sp. strain | ||
| PCC 7120 strains | ||
| PCC 7120 | Wild-type Anabaena (Nostoc) sp. strain PCC 7120 | R. Haselkorn |
| AMC450 | PCC7120(pAM1691) (carries PglnA-patS) | 52 |
| AMC451 | patS replaced with Sp/Sm cassette | 52 |
| AMC455 | PCC 7120(pAM1714) (carries PpetE-patS) | 52 |
| AMC484 | PCC 7120(pAM1951) (carries PpatS-gfp) | 52 |
| AMC486 | PCC 7120(pAM1954) (carries PrbcL-gfp) | 52 |
| AMC720 | patS overexpression suppressor isolated in Tn5-1087 mutagenesis | This study |
| AMC787 | patS overexpression suppressor isolated in Tn5-1087 mutagenesis | This study |
| AMC1050 | plmA single-recombination knockout constructed with nonreplicating plasmid pAM2563 | This study |
| AMC1051 | AMC1050(pAM1691), plmA background with patS overexpression plasmid; this isolate grows better than AMC1084 and may have a secondary mutation | This study |
| AMC1080 | PCC 7120(pAM1690) (carries PrbcL-patS plasmid) | This study |
| AMC1082 | AMC1050(pAM1690) (plmA background carrying PrbcL-patS) | This study |
| AMC1084 | AMC1050(pAM1691) (plmA background carrying PglnA-patS plasmid) | This study |
| AMC1086 | AMC1050(pAM1714) (plmA background carrying PpetE-patS) | This study |
| AMC1108 | PCC 7120(pAM2842) (carries PplmA-gfp) | This study |
| AMC1109 | PCC 7120(pAM2850) [carries PplmA(rev)-gfp], a control construct in which the promoter for plmA points away from gfp | This study |
| AMC1115 | AMC1050(pAM2904) library isolate pdl1, which relieves the senescence phenotype | This study |
| AMC1116 | AMC1050(pAM2905) library isolate pdl5, which relieves the senescence phenotype | This study |
| Plasmids | ||
| pAM123 | pRL444 digested with BamHI to remove luxAB and then recircularized by T4 ligase treatment; Kmr | J. Brusca, J. Goldena |
| pAM504 | An EcoRI-XbaI fragment from the pUC18 multiple cloning site was end filled and inserted into the BamHI site (also end filled) of pAM123; Kmr | 47 |
| pAM542 | An approximately 400-bp BamHI/SalI fragment, carrying PrbcL from pAM522, was cloned into BamHI/SalI-digested pAM504, with the promoter pointing toward the BamHI site; Kmr | T. S. Ramasubramanian, J. Goldena |
| pAM743 | A 270-bp SalI fragment bearing the glnA promoter was digested from pAM658 and inserted into the SalI site of pAM504, with the promoter pointing toward the multiple cloning site; Kmr | L. Whorff, J. Goldena |
| pAM1689 | A PCR fragment carrying patS, amplified with AMO-807 and AMO-808, was DraI/TaqI digested and cloned into ClaI/EcoRV-digested pBluescript II SK+; Apr | H. S. Yoon, J. Goldena |
| pAM1690 | A 103-bp BamHI-KpnI fragment from pAM1689, carrying patS, was cloned into similarly digested pAM542 to produce PrbcL-patS on a shuttle vector; Kmr | H. S. Yoon, J. Goldena |
| pAM1691 | A 103-bp BamHI-KpnI fragment from pAM1689, carrying patS, was cloned into similarly digested pAM743 to produce PglnA-patS on a shuttle vector; Kmr | 52 |
| pAM1693 | A control construct, essentially the same as pAM1689 except the pBluescript SK+ was digested with ClaI/HincII, reversing the orientation of the patS insert; Apr | H. S. Yoon, J. Goldena |
| pAM1694 | A BamHI-KpnI fragment from pAM1693, carrying patS, cloned into similarly digested pAM542 to produce a PrbcL-patS(reversed) control on a shuttle vector; Kmr | H. S. Yoon, J. Goldena |
| pAM1695 | A BamHI-KpnI fragment from pAM1693, carrying patS, cloned into similarly digested pAM1248 (pAM743 containing lacZ on a BamHI-KpnI fragment), to produce a PglnA-patS(reversed) control on a shuttle vector; Kmr | 52 |
| pAM1697 | The PCR fragment described for pAM1689 was DraI/SacI digested and cloned into pPet1 to make PpetE-patS; Apr | 8, 52 |
| pAM1698 | The PCR fragment described for pAM1689 was SmaI/XbaI digested and cloned into pPet1 to make a PpetE-patS(reversed) fusion; Apr | 8; H. S. Yoon, J. Goldena |
| pAM1714 | A ScaI/SacI fragment carrying PpetE-patS from pAM1697 was cloned into SmaI/SacI-digested pAM504; Kmr | 52 |
| pAM1716 | A ScaI/SacI (partial) digest of pAM1698 released PpetE-patS (reversed orientation), which was cloned into SmaI/SacI of pAM504 Kmr | 52 |
| pAM1951 | PpatS-gfp in a shuttle vector pAM505; Kmr | 52 |
| pAM1954 | PrbcL-gfp in a shuttle vector pAM505; Kmr | 52 |
| pAM2563 | An internal fragment from plmA was amplified with primers AMO-449 and AMO-450, digested with BglII and PstI, and cloned into similarly digested pRL277; Spr Smr | This study/PICK> |
| pAM2586 | The unique NdeI site in lacZα of pUC18 was removed by NdeI digestion, end filling, and recircularization; isolate pAM2585 had lost the NdeI site but (unexpectedly) retained lacZ activity; this isolate was used as template for reverse PCR to replace the lacZ promoter sequences with unique XhoI/NdeI sites such that the NdeI site overlapped the lacZ start codon; Apr | This study |
| pAM2600 | Accession no. AY263154; an XhoI-PpetE-NdeI cassette was amplified from the chromosome of PCC 7120 with AMO-471 and AMO-472 and cloned into pAM2586 to make pAM2588. Reverse PCR using pAM2588 as template and oligonucleotides AMO-473 and AMO-474 produced a product carrying SapI-6 His (stop)-ClaI and a 22-bp direct repeat at both ends; in E. coli, the repeats permitted recombination to circularize the molecule; essentially, this is a pUC18 derivative with an insert of XhoI-PpetE-NdeI-lacZα-SapI(Cys)-6 His (stop)-ClaI; Apr | This study |
| pAM2770 | Accession no. AY265466; pAM123 was digested with NdeI, end filled, and recircularized to make pAM2625; pAM2625 was SalI digested, end filled, BamHI digested, and ligated with an EcoRV/BclI-digested PCR product carrying EcoRV-XhoI-cat-ClaI-BclI to make pAM2742; the cat gene was then replaced with the XhoI-ClaI cassette from pAM2600 to make pAM2758; finally, about 2.2 kb of DNA was removed by BsrGI digestion and recircularization; Kmr | This study |
| pAM2834 | Reverse PCR was used to create a silent mutation; removing an NdeI site internal to gfp on pKEN2-GFPmut2; the original CATATG was changed to CACATG (His→His); Apr | This study |
| pAM2842 | A 376-bp fragment carrying the region upstream of plmA was amplified with AMO-581 and AMO-611 and cloned as an XhoI-NdeI fragment into pAM2770, replacing the PpetE sequences (making pAM2839); the modified gfp (gfpΔNdeI) on pAM2834 was amplified with flanking NdeI and ClaI sites and cloned into pAM2839 (replacing lacZ) to create a PplmA-gfp fusion; Kmr | This study |
| pAM2850 | Similar to pAM2842, except that flanking restriction sites on the amplified plmA upstream region were reversed, producing a fusion in which the putative plmA promoter points away from gfp; Kmr | This study |
| pAM2904 | An isolate from an overexpression library (the construction of which is described in reference 30) which relieves the senescence phenotype of a plmA mutant strain; Kmr | This study |
| pAM2905 | Like pAM2904, an isolate from an overexpression library that relieves the senescence phenotype of a plmA mutant strain; Kmr | This study |
| pAM2980 | Contains a pCC7120ɛ fragment amplified with AMO-709 and AMO-710 and inserted into SmaI-digested pWB19-12; Apr | This study |
| pDU1 | A naturally occurring plasmid isolated from Nostoc sp. strain PCC 7524 used to provide a cyanobacterial replication origin for shuttle vectors | 49 |
| pKEN2-GFPmut2 | High fluorescence gfp mutant on high-copy-number plasmid; Apr | 12 |
| pPet1 | Anabaena sp. strain PCC 7120 petE promoter in pUC19 | 8 |
| pRK2013 | RK2-based plasmid with an added ColE1 origin of replication; Kmr | 19 |
| pRL277 | oriT and oriV from pMB1, aadA (Spr Smr), sacB (for sucrose counter selection); does not replicate in Anabaena sp. strain PCC 7120 | 5 |
| pRL443 | Conjugal plasmid derived from RP-4; Kms Apr Tcr | 16 |
| pRL444 | A conjugal shuttle plasmid bearing both a pMB1 oriV and the Nostoc pDU1 ori; contains luxAB and aphA-2; Kmr | 15 |
| pRL623 | Helper plasmid, encoding three DNA methyltransferases; Cmr | 16 |
| pRL1045 | Helper plasmid encoding two DNA methyltransferases; does not contain mob gene; Kmr | 16 |
| pRL1087b | Nonreplicating plasmid carrying oriT and Tn5-1087 transposon; Cmr Emr | 18 |
| pUC18 | Cloning vector with lacZα-MCS and oriV from pMB1; Apr | 51 |
| pWB19-12 | Contains both hetR and aphA-2; used to make standard curve for real time PCR assay of pAM1691/chromosome ratio; Kmr | 7 |
| Oligonucleotide primers | ||
| AMO-449 | Amplifies a plmA internal fragment with flanking BglII/PstI sites (BglII site, underlined); GCGCAGATCTAAGTCTATCGTCAGTTAGAGG | This study |
| AMO-450 | Amplifies a plmA internal fragment with flanking BglII/PstI sites (PstI site underlined); GCAACTGCAGGCAGATAATCACTTCGG | This study |
| AMO-471 | Amplifies PpetE with flanking XhoI (underlined)/NdeI sites; CCAACCCTCGAGCACAGGACTCAGAACACAG | This study |
| AMO-472 | Amplifies PpetE with flanking XhoI/NdeI (underlined) sites; CCAACCCATATGGTTCTCCTAACCTGTAGTTTT | This study |
| AMO-473 | Reverse PCR primer adding a 22-bp repeat (double underlined), stop codon, and a ClaI site (underlined) to one end of a product generated from pAM2588; CTGCCATCATCACCATCACCACTAAATCGATGCCGACACC CGCCAACAC | This study |
| AMO-474 | Reverse PCR primer adding a 22-bp repeat (double underlined) and a SapI site (underlined) to one end of a product generated from pAM2588 opposite the end produced by AMO-473; GTGGTGATGGTGATGATGGCAGGAAGAGCGGCTGGCTTA ACTATGCG | This study |
| AMO-645 | Amplifies hetR fragment for real-time PCR; TAAGTCCGCTCTTGGTCGTCTG | This study |
| AMO-646 | Amplifies hetR fragment for real-time PCR; TAAGTCCGCTCTTGGTCGTCTG | This study |
| AMO-679 | Amplifies nptA fragment from pAM1691 for real-time PCR; AGCTGTGCTCGACGTTGTCA | This study |
| AMO-680 | Amplifies nptA fragment from pAM1691 for real-time PCR; GCAGGAGCAAGGTGAGATGA | This study |
| AMO-701 | Amplifies alr7243 fragment from pCC7120α for real-time PCR; TGAAAAGTGGCTACCGCTCAAC | This study |
| AMO-702 | Amplifies alr7243 fragment from pCC7120α for real-time PCR; ATCTCCTTTCCCATCCTTGGC | This study |
| AMO-703 | Amplifies all7629 fragment from pCC7120β for real-time PCR; TCCAGAACAACACGCCGAA | This study |
| AMO-704 | Amplifies all7629 fragment from pCC7120β for real-time PCR; TGCGACCAACTGCATTGCT | This study |
| AMO-705 | Amplifies all8089 fragment from pCC7120γ for real-time PCR; CATTGAGCAAGCAGCAGGAA | This study |
| AMO-706 | Amplifies all8089 fragment from pCC7120γ for real-time PCR; GCTTGCAAACCCTTTTCGG | This study |
| AMO-707 | Amplifies all8519 fragment from pCC7120δ for real-time PCR; TCGAAAGGCGTTACCCCAA | This study |
| AMO-708 | Amplifies all8519 fragment from pCC7120δ for real-time PCR; AGTGCGTTTCATCAGTGCTGC | This study |
| AMO-709 | Amplifies all9018 fragment from pCC7120ɛ for real-time PCR; TCGTATTGCCGCCGTAACA | This study |
| AMO-710 | Amplifies all9018 fragment from pCC7120ɛ for real-time PCR; CCAACTAGCTCCCGAATCACAA | This study |
| AMO-711 | Amplifies asl9502 fragment from pCC7120ζ for real-time PCR; ACCAGTTGGATGAAGTAGCCAA | This study |
| AMO-712 | Amplifies asl9502 fragment from pCC7120ζ for real-time PCR; GGCTATGTTCTGCTGTTCACCT | This study |
| AMO-807 | Amplifies patS with DraI flanking sequence CTGTTTAAAAGTAATTCACCG | This study |
| AMO-808 | Amplifies patS with overlapping XbaI/SacI flanking sequence (underlined); GCTCTAGAGCTCTCTACATGATAACGAC | This study |
Unpublished data.
DNA manipulation.
Standard protocols were used for cloning, E. coli transformation, PCR, Southern blotting, and Northern blotting (3). Anabaena chromosomal preparations were performed as described previously (20). Qiagen (Valencia, Calif.) plasmid preparation procedures and Concert-Kit (Life Technologies, Grand Island, N.Y.) DNA purification of PCR products was performed as recommended by the manufacturers. Big Dye sequencing (Applied Biosystems, Foster City, Calif.) was performed with one-quarter-volume reaction mixtures with 200 ng of template. Chromosomal sequencing was performed using 2× volume Big Dye sequencing reaction mixtures and 6 μg of chromosomal DNA as template. High-fidelity PCR was performed using Pwo polymerase or an Expand mixture (Roche Applied Science, Indianapolis, Ind.).
Plasmid segregation assay.
Anabaena strains were streaked on BG-11 agar plates, allowed to grow to small colonies (7 days), and then streaked for heavy growth on BG-11 plates without Nm selection. Plates were used instead of liquid culture, because the plmA mutant filaments fragmented and grew slowly in liquid. After 10 days of growth without selection, strains were suspended in 1 ml of BG-11 medium and sonicated for 10 s in a Branson 2200 Ultrasonic Cleaner bath to produce short filaments containing an average of 2.1 ± 1.4 cells and approximately 45% single cells. The sonicated filaments were diluted and plated both on plates with Nm selection (to determine plasmid-containing CFU numbers) and on plates without Nm selection (to determine total CFU). The plmA mutant grows slowly, raising a concern that the stress of sonication and plating might have killed these slowly dividing cells even when they retained the plasmid. To control for this, both wild-type and plmA mutant strains were grown for 10 days on Nm plates (forcing plasmid retention) before resuspending, sonicating, diluting, and plating.
Real-time PCR.
Real-time PCR was performed using an ABI 7700 apparatus (Applied Biosystems) and the Quantitech SYBR Green PCR kit (Qiagen). Each reaction produced short products with sizes in the range of 102 to 108 bp. Primers for the hetR gene were used to assay the concentration of Anabaena sp. strain PCC 7120 chromosomes. Primers for the Nm resistance gene, aphA-2, were used to assay the concentration of the patS overexpression plasmid (pAM1691). Using an arbitrarily chosen gene on each plasmid, the genome sequence was used to design primers for each of the endogenous plasmids. Standard curves were generated for each plasmid and the chromosome. Plasmid pWB19-12 carries both hetR and aphA-2, so known concentrations of this one molecule generated both standard curves in the assay comparing pAM1691 concentration to chromosome concentration. Similarly, plasmid pAM2980 carries hetR and a PCR-amplified pCC7120ɛ sequence. Known concentrations of this molecule generated both standard curves for the assay of pCC7120ɛ.
For each of the remaining plasmids, the real-time PCR primers were used to produce a solution of amplified product. Known concentrations of this product were used to generate a standard curve for plasmid concentration. DNA samples used as standards were resolved in an agarose gel and quantified using a Kodak 1D system (Kodak, Rochester, N.Y.). Generally, a 25-μl real-time PCR mixture produced a linear response to template of 10−17 to 10−22 moles/reaction (although the assay occasionally remained linear up to 10−16 moles/reaction). A total of seven experiments were performed (one experiment per plasmid). Each experiment included a standard curve for assaying the amount of chromosome (in duplicate), a standard curve for assaying the amount of one particular plasmid (in duplicate), an assay of the amount of chromosome prepared from strains PCC 7120, AMC1084, and AMC1051 (in triplicate), and an assay of the amount of plasmid in those same DNA preparations (in triplicate).
Transposon mutagenesis and screening.
The transposon-bearing strain AM1181 and conjugal strain AM1460 were grown overnight on LB agar plates with antibiotic. A loopful of each strain was resuspended and diluted into 5 ml of LB (plus antibiotic). The two cultures were grown for 5 h at 37°C. The cells were pelleted by centrifugation, washed twice in LB, combined in a 15-ml conical tube, pelleted again, and (after the supernatant was decanted) resuspended in about 50 μl of the remaining supernatant. The combined culture was incubated at 37°C for 1 h to permit the conjugal plasmid to enter the transposon-bearing strain. A 10-ml sample of Anabaena sp. strain AMC450, which overexpresses patS, was added to the E. coli, and the cells were pelleted at 1,700 × g for 7 min, decanted, and resuspended in the remaining 300 to 500 μl of supernatant. BG-11 agar plates were spread with 40 μl of the resuspended mixture and then incubated overnight at 30°C with 1% CO2 at 30 to 80 μM photons m−2 s−1. The next day, Nm was added beneath the agar pads to produce a final concentration of 12.5 μg/ml and plates were incubated (as described above) until small colonies appeared. Colony lifts and rec-85 filters (Whatman, Clifton, N.J.) were used to transfer colonies to BG-110 agar. These plates were incubated for 5 to 12 weeks and scored for the appearance of green colonies or green papillae, small green extrusions from a colony that is otherwise yellow-brown. Total DNA was recovered from the mutants, digested with either ClaI or PvuI, and then treated with ligase. The circularized DNA was transformed into E. coli, permitting recovery of the replicon carried in the transposon and the chromosomal sequences on either side of the transposon.
Targeted inactivation.
The plmA gene was targeted for inactivation by cloning a PCR-amplified internal fragment (with flanking BglII and PstI sites) into suicide plasmid pRL277 to make pAM2563. The new construct was transferred by conjugation into Anabaena sp. strain PCC 7120. Single-recombination mutants were identified by Sp and Sm selection. The gene disruption was confirmed by PCR and Southern blot analysis. The patS overexpression plasmid pAM1691 was transferred by conjugation into the new mutant strain to complete the reconstruction.
Construction of PpetE-lacZα-6His plasmid.
A shuttle plasmid permitting blue-white screening, the use of a copper-inducible promoter (PpetE [8]), and fusion to a 6-His tag was constructed in three phases. First, a pUC18 derivative was modified in four steps to produce pAM2600, containing the following elements: XhoI, PpetE, NdeI, lacZα, SapI (cys), 6-His (stop), and ClaI. Here PpetE is the Anabaena sp. strain PCC 7120 petE (plastocyanin) promoter (8), SapI (cys) is a SapI cognate site in which the degenerate 3-bp overhang carries a cysteine codon, and 6-His (stop) is a string of six histidine codons and a stop codon. All four of the listed restriction sites are unique in pAM2600. This plasmid produces a blue colony color in a DH10B background after 2 days at 37°C on LB Ap X-Gal plates. The NdeI site overlaps the start codon of lacZα. The SapI site can be used to make a translational fusion between a cloned gene and the 6-His tag. Second, shuttle plasmid pRL444 was modified in three steps to remove the luxAB genes, eliminate the NdeI site, remove the multiple cloning site, and introduce a cat gene flanked by unique XhoI and ClaI sites. The final product is called pAM2742. Third, the XhoI-ClaI cassette from pAM2600 was moved into pAM2742, replacing the cat gene. Then a 2.1-kb BsrGI fragment carrying nonessential sequences was removed. The final product, pAM2770, is a blue-white cloning plasmid exploiting PpetE expression of inserted sequences. The SapI site introduced by the cassette is not unique in pAM2770.
Construction of PplmA-gfp reporter and microscopy.
The region upstream of plmA was fused to gfp to test for promoter activity. This region extends from the plmA start codon to the start codon of the divergently transcribed upstream gene (alr1077) and was amplified by PCR. The amplified product was used to replace the PpetE promoter on pAM2770, resulting in a PplmA-lacZα transcriptional fusion. The lacZα fragment was then replaced with gfp from pKEN2-GFPmut2 to produce pAM2842 (carrying PplmA-gfp). As a control, we inverted the PplmA region to make PplmA(reversed)-gfp on plasmid pAM2850.
Photomicrographs were taken with an IX70 microscope with Nomarski differential interference contrast (DIC) optics (Olympus, Melville, N.Y.) and a Proscan automation system for automatic switching between light sources (Prior Scientific, Rockland, Mass.). A Piston green fluorescent protein (GFP) filter cube (set ID 41025; Chroma Technology Corp., Brattleboro, Vt.) was used for fluorescence images. Images were captured with a cooled ORCA charge-coupled device camera (Hamamatsu, Bridgewater, N.J.). Composite images of Nomarski DIC and GFP images were made using SimplePCI software (C-imaging Inc., Cranberry Township, Pa.). Contrast in the composite images was improved by inverting the Nomarski DIC images so that cells appear dark gray.
Bioinformatics.
Genome sequences were obtained from the Anabaena sp. strain PCC 7120 genome database (http://www.kazusa.or.jp/cyano/Anabaena/index.html). Similarity searches were performed using BLAST (2). General sequence analysis was performed using Biology Workbench (44) (http://workbench.sdsc.edu), and Pfam (4) (http://pfam.wustl.edu/) was used for motif searches. Selection of plmA homologs, multiple protein sequence alignments, secondary structure predictions, and phylogenetic tree constructions were performed as described previously (36).
RESULTS
Transposon mutagenesis was used to produce suppressor strains from a derivative of Anabaena sp. strain PCC 7120 that overexpresses patS. Strain AMC450 carries patS on plasmid pAM1691. This strain fails to produce heterocysts on BG-110 agar medium, which lacks a source of combined nitrogen. Colonies do form, but they rapidly turn yellow-brown. A total of 50,000 transposon-mutagenized AMC450 colonies were screened. Of these, 62 were identified as suppressors. These mutants formed green colonies or papillae on BG-110 agar, remained green when transferred to fresh BG-110 agar, and exhibited heterocysts when examined microscopically. Mutants AMC720 and AMC787 each contained a Tn5-1087 transposon inserted in open reading frame all1076 which was named plmA (for plasmid maintenance). The patS overexpression plasmid was recovered from AMC787 and reintroduced into Anabaena sp. strain PCC 7120, where it was still able to confer a heterocyst suppression phenotype (data not shown).
A new subfamily of GntR-like transcriptional regulators.
A Pfam search revealed that PlmA is similar to peptides of the GntR family of transcriptional regulators. The peptides in this family share a region of homology within the DNA-binding domain found near the N terminus. A recent analysis indicates that the GntR family of proteins clusters into five subfamilies on the basis of heterologies in the C-terminal sequences (the effector-binding-oligomerization domain) (36). When aligned with these homologous sequences, PlmA also shared highest homology with the DNA-binding domain of the family. However, PlmA did not fit into any of the existing subfamilies. Instead, a search of various databases uncovered seven cyanobacterial sequences that cluster with PlmA in a new subfamily. An unrooted tree that highlights the clustering of the cyanobacterial sequences relative to the five previously identified subfamilies is shown in Fig. 1. The genes used to construct the tree are described in Table 2. The GntR family contains six subfamilies, MocR, YtrA, FadR, AraR, HutC, and PlmA. We found that each of the subfamilies could be discerned from alignments of the DNA-binding domains alone (in an alignment employing 20 sequences; data not shown). Using just this DNA-binding alignment, the PlmA subfamily shared highest similarity with the YtrA and MocR subfamilies (data not shown). We infer that the PlmA subfamily arose from an ancestral sequence shared by one of these subfamilies, diverging through a process of replacement in the effector-binding-oligomerization domain.
FIG. 1.
PlmA clusters with a new subfamily within the GntR family. Full-length sequences were aligned using MULTIALIN and then manually adjusted using each protein's predicted secondary structure to guide the alignment. Distances between aligned proteins were computed with a PRODIST program (using maximum likelihood estimates on the Dayhoff PAM matrix). A FITCH program was used with the Fitch-Margoliash algorithm to estimate phylogenies from the distances. The trees were drawn using TREEVIEW.
TABLE 2.
GntR proteins used in phylogenetic analysis
| ORF | Organism | Length (bp) | Accession no. |
|---|---|---|---|
| FadR | E. coli | 238 | P09371 |
| HutC | Pseudomonas putida | 248 | P22773 |
| MocR | Rhizobium meliloti | 493 | P49309 |
| YtrA | Bacillus subtilis | 130 | O34712 |
| AraR | Bacillus subtilis | 364 | P96711 |
| PlmA (all1076) | Anabaena sp. strain PCC 7120 | 328 | Q8YXY0 |
| sll1961 | Synechocystis sp. strain PCC 6803 | 388 | P73804 |
| tll2117 | T. elongatus BP-1 | 367 | BAC09669 |
| NZ_AAAX01000001 (Pmit_p_0101) | Prochlorococcus marinus sp. strain MIT 9313 | 329 | ZP_00112619 |
| NZ_AAAW01000001 (Pmar_p_1604) | Prochlorococcus marinus subsp. pastoris strain CCMP1378 | 323 | ZP_00105573 |
| NZ_AAAU01000098 (Tery_p_4269) | Trichodesmium erythraeum IMS101 | 327 | ZP_00074937 |
| NZ_AABD01000001 (Synwh_p_89) | Synechococcus sp. strain WH 8102 | 440 | ZP_00114559 |
| NZ_AABC01000142 (Npun_p_2326) | N. punctiforme | 328 | ZP_00107916 |
Characterization of plmA.
A plmA null mutant was constructed via targeted inactivation. Plasmid pAM2563 carries an internal fragment from plmA and does not replicate in Anabaena sp. strain PCC 7120. It was used to inactivate plmA via single recombination, producing strain AMC1050. To test the constructed plmA mutant strain, we introduced the patS overexpression plasmid pAM1691 into AMC1050, creating strain AMC1084. An otherwise wild-type strain failed to produce heterocysts when patS was overexpressed, but the reconstructed plmA mutant strain, AMC1084, suppresses this phenotype. Strain AMC1084, like the original transposon plmA mutants, produced colonies that were smaller and more yellow than those of the wild type (data not shown). In summary, the newly constructed mutant had the same phenotype as the original transposon mutants.
Many heterocyst development genes are upregulated in heterocysts or proheterocysts (5, 25, 50, 52). We fused the presumed promoter sequence for plmA to gfp (encoding GFP) to determine whether plmA expression is limited to a specific cell type. The promoter sequence includes the entire 362-bp intergenic sequence between plmA and all1077, the adjacent and divergently expressed open reading frame. For controls, we also examined the fluorescence of strains in which gfp had been fused to a developmentally regulated promoter (PpatS [52]) or to a vegetative-cell promoter (PrbcL) from the gene encoding ribulose bisphosphate carboxylase (17). All three constructs were transferred by conjugation into wild-type Anabaena sp. strain PCC 7120.
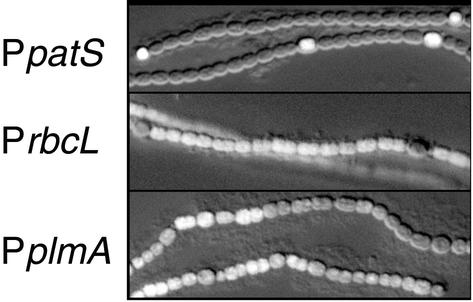
Figure 2 shows a composite image for each of these strains which combines an inverted Nomarski DIC image and a GFP fluorescence image. The strains were photographed 24 h after nitrogen step-down. Expression of gfp from the patS promoter (Fig. 2, top panel) produced a pattern of fluorescence in regularly spaced single cells that had the morphology of heterocysts or proheterocysts. Expression of gfp from the rbcL promoter (middle panel) produced a pattern of fluorescence from vegetative cells. Some heterocysts showed a slight fluorescence, possibly because GFP persists for some time in newly developed heterocysts. Unlike that of the two control constructs, expression of gfp from the plmA promoter (bottom panel) did not produce cell type-specific fluorescence. Instead, expression was markedly patchy. Stretches of cells had bright florescence, while adjoining stretches were dark. Fluorescence was neither limited to nor excluded from either vegetative cells or heterocysts.
FIG. 2.
The activity of the plmA promoter is not cell type specific. Each composite photograph shows an inverted Nomarski image overlaid with a GFP fluorescence image from an Anabaena sp. strain PCC 7120 strain carrying a promoter-gfp construct. (Inverting the Nomarski image improved the contrast.) PpatS, patS promoter driving gfp in strain AMC484; PrbcL, rbcL promoter driving gfp in strain AMC486; PplmA, plmA promoter driving gfp in strain AMC1108.
If plmA was essential for heterocyst development, then plmA mutant strain AMC1050 would be expected to show a heterocyst defect. However, the plmA mutant filaments exhibited wild-type patterns of heterocysts. No unusual morphology was detected when individual heterocysts were examined using light microscopy (data not shown). Neither the expression pattern from the plmA promoter nor the phenotype of the plmA mutant supports a role for plmA in heterocyst development.
Plasmid maintenance.
It seemed plausible that plmA had a role in the stable maintenance of the patS overexpression plasmid. Unequal segregation between daughter cells might have led to patches of cells with low levels of patS expression, which would permit heterocyst development. Similarly, a decrease in copy number could have reduced the level of aphA-2 expression (Nmr), reducing the mutant's growth rate under Nm selection.
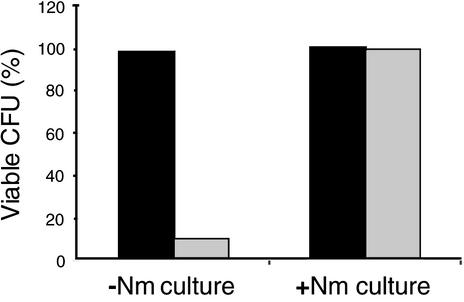
A partitioning defect should have produced cells in which the pAM1691 copy number had fallen below the levels needed for producing Nm resistance. Wild-type and plmA mutant (AMC1084) strains harboring pAM1691 were grown without selection, sonicated to shorten the filaments, and then plated with antibiotic selection to test for plasmid loss (Fig. 3). The plmA mutant strain retained Nm resistance in only 10% of the CFU (in the form of single cells and short filaments averaging 2.1 cells in length). The wild-type strain retained Nm resistance in 100% of the CFU.
FIG. 3.
The plmA mutant strain segregates cells that lack shuttle vector plasmids. plmA+ strain AMC450 (black columns) and plmA mutant strain AMC1084 (gray columns) carry pAM1691, which provides Nm resistance. Strains were grown for 10 days without Nm selection (−Nm culture) to permit plasmid segregation. Additionally, both strains were grown with Nm selection (+Nm culture) as controls. Dilutions of both strains were then plated both with and without Nm selection. The y axis indicates the ratio of CFU arising on plates with Nm selection to CFU arising on plates without Nm selection (expressed as a percentage).
As in earlier experiments, the plmA strain grew poorly. If this mutation compromised the strain's ability to adapt to our experimental conditions, then stresses from sonication and plating might have killed plmA cells even if they retained pAM1691 at levels sufficient for Nm resistance. To control for this, the wild-type and plmA mutant strains were grown in cultures with Nm selection so that only cells with a suitably high plasmid copy number remained viable. Both strains were washed, sonicated, and plated as described above. Both strains retained the ability to survive plating and form colonies on Nm in nearly 100% of the CFU (Fig. 3; see columns labeled +Nm). This shows that a plmA mutant strain carrying sufficient pAM1691 to confer Nm resistance survived plating as well as did a wild-type strain. Therefore, the decrease in levels of Nm-resistant CFU arising during growth without selection can be attributed to decreases in the pAM1691 copy number or defective segregation. The CFU in this experiment included short filaments, which may have contained a mixture of cells with high levels of pAM1691 content and cells with low levels of pAM1691 content. Thus, the data reported here probably understate the tendency for plmA mutant cells to become Nm sensitive.
Plasmid partitioning defects are sometimes associated with a decrease in plasmid copy number. Real-time PCR was used to examine the relative copy number (the number of plasmids per chromosome) for exogenous plasmid pAM1691 and the six endogenous plasmids. Three strains were used in this assay. AMC450 is wild type for plmA. AMC1051 carries a targeted inactivation in plmA and was grown in subcultures for months before being used in the assay. AMC1084 was constructed in exactly the same fashion as AMC1051, but all AMC1084 isolates produced colonies that were smaller and lighter than AMC1051 colonies. It is possible that AMC1051 acquired a second-site mutation that partially relieves the slow-growth phenotype associated with the plmA insertional inactivation. All three strains carry plasmid pAM1691.
The results from the real-time PCR analyses are shown in Fig. 4. Plasmids pCC7120α, pCC7120β, and pCC7120γ are relatively large plasmids of 408, 187, and 102 kb, respectively (27). The assay showed that these large plasmids are under stringent copy number control, as the number of copies per chromosome in AMC450 was close to 1. Anabaena sp. strain PCC 7120 is presumed to have several chromosomes per cell, which makes it possible for the cell to have a plasmid-to-chromosome ratio of less than 1. We did not determine the number of chromosomes per cell in our experiments. The plmA mutation did not significantly influence the relative copy numbers of the large plasmids or of the 40-kb plasmid pCC7120ɛ. However, the plmA mutation did have a significant effect on the relative copy numbers of pCC7120δ, pCC7120ζ, and pAM1691. In a wild-type background, pAM1691 accumulated to 17 copies per chromosome. In both plmA mutant strains, the concentrations decreased to less than four copies per chromosome. Endogenous plasmids pCC7120δ and pCC7120ζ also exhibited reduced plasmid copy numbers (down to 50 and 35% of the wild-type number, respectively).
FIG. 4.
The plmA mutation reduces copy numbers of endogenous plasmids pCC7120δ and pCC7120ζ and of shuttle vector pAM1691. The relative numbers of plasmids per chromosome (y axis) were determined for each of the six endogenous plasmids (pCC7120α to pCC7120ζ) and for the pDU1-based plasmid pAM1691. Total DNA was extracted from a strain with wild-type plmA (AMC450, black columns), a slow-growing plmA mutant strain (AMC1084, gray columns), and a relatively fast-growing plmA mutant strain (AMC1051, hatched columns). Each strain was independently cultured in BG-11 medium with Nm, extracted, and assayed in triplicate. Error bars show standard deviations (n = 3).
Senescence.
It proved to be difficult to maintain pDU1-based plasmids (such as pAM1691) in plmA mutants. Conjugation experiments using either wild-type or plmA mutant recipients produced roughly equal numbers of exconjugant colonies. However, when the exconjugants of plmA mutant strains were restreaked they showed poor growth. Wild-type controls produced a dense, dark-green patch in the initial streak, while the mutant strain produced much thinner, lighter patches. Typically, the next transfer of plmA+ strains still produced dense growth while plmA mutants produced little more than a string of overlapping, small, and yellow colonies. It was common for a third serial transfer to produce no growth at all for the plmA mutant strains. This progressive loss of viability was termed senescence and was observed on Nm plates with or without a source of combined nitrogen.
We screened for bypass mutations that would relieve the senescence phenotype as a means of identifying genes that operate in the same pathway as plmA. A library carrying random Anabaena sp. strain PCC 7120 fragments was introduced into plmA mutant AMC1050, and exconjugants were plated on BG-11 medium. Senescence is most clearly seen after restreaking exconjugants. To avoid individually transferring thousands of colonies, however, we collected the entire lawn of exconjugants by suspension in liquid medium and then plated dilutions of the filaments. On a plate with approximately 1,000 to 2,000 colonies, most colonies were small and yellow (senescent), which was expected because the library was constructed with a pDU1-based vector. On a typical plate, however, as many as 100 colonies developed that were larger and greener than those of the background. Two isolates were identified that conferred improved viability after serially repeated restreaking. Plasmids from these isolates were recovered and transferred by conjugation back into the plmA strain. The new exconjugants showed approximately wild-type levels of growth after repeated restreaking, which confirmed that the senescence phenotype was suppressed in these two library clones. Figure 5 shows maps of the senescence-suppressing fragments cloned in these plasmids. The fragments originated from endogenous plasmids pCC7120ζ and pCC7120γ. Each fragment carries a gene that is similar to genes having ascribed roles in controlling plasmid copy numbers. Plasmid pCC7120ζ carries open reading frame asl9502 (Fig. 5A), which has homology to copG from Streptococcus agalactiae. Plasmid pCC7120γ carries open reading frame asl8050 (Fig. 5B), which has homology to copB from Klebsiella pneumoniae. The identification of two plasmid fragments as senescence suppressors is consistent with a role for plmA in plasmid maintenance.
FIG. 5.
Two fragments from Anabaena sp. strain PCC 7120 endogenous plasmids suppress the plmA-dependent senescence phenotype of a shuttle vector. A library of chromosomal fragments was used to screen for cloned fragments that suppressed the senescence phenotype in a plmA mutant strain carrying the library shuttle vector. Two isolates were recovered. (A) Map of the insert identified in plasmid pAM2904, containing a fragment derived from pCC7120ζ. (B) Map of the insert identified in plasmid pAM2905, containing a fragment derived from pCC7120γ. Partial open reading frames are shown as hatched arrows, genes with no assigned function are shown as open arrows, and genes with a functional assignment determined on the basis of sequence similarity are shown as black arrows. Arrows labeled “Not Annotated” represent open reading frames not annotated by the genome site at the Kazusa Institute.
The glnA promoter is not essential for plmA activity.
The observed reduction in plasmid copy numbers does not preclude the hypothesis that plmA acts as an activator of the glnA promoter. Two additional plasmids were constructed to determine whether suppression of the patS overexpression phenotype was dependent on the use of the glnA promoter. The patS gene is expressed from the petE promoter in pAM1714 and from the rbcL promoter in pAM1690. In control constructs, the patS gene was fused to the same promoters but in an inverted orientation. These plasmids were introduced into wild-type and plmA mutant strains, and the exconjugants were scored on medium without a source of combined nitrogen.
As originally observed, expression of patS from the glnA promoter in a wild-type background (strain AMC450) produced yellow-brown colonies without heterocysts, and the plmA mutation (strain AMC1084) suppressed both phenotypes. Expression of patS from the rbcL promoter produced identical results. The wild-type control (strain AMC1080) produced yellow-brown colonies and no heterocysts, whereas a plmA mutant strain (AMC1082) suppressed both phenotypes. Expression of patS from the petE promoter (activated by 400 nM copper in the medium) only partially suppressed heterocysts in the wild-type control (strain AMC455). Colonies were nearly wild type in size and color, but microscopic inspection revealed that heterocyst frequency was markedly reduced. This decreased frequency of heterocysts was suppressed by the plmA mutation (strain AMC1086). In sum, the suppressor phenotype of plmA did not depend on the heterologous promoter used to express patS.
DISCUSSION
A plmA mutation was identified as a suppressor of patS overexpression when patS was carried on plasmid pAM1691. The mutant also produced smaller and lighter-colored colonies than the wild type and an elevated rate of plasmid loss resulting in decreasing viability under antibiotic selection (senescence). These characteristics suggested that the plmA mutation produced a defect in plasmid maintenance. Therefore, we assayed the relative copy numbers of the Anabaena sp. strain PCC 7120 endogenous plasmids and pAM1691 in both plmA mutant and wild-type backgrounds. Significant reductions in copy number were observed for plasmids pCC7120δ and pCC7120ζ as well as for the pDU1-based shuttle plasmid, pAM1691. Sequence analysis suggests that plmA carries a regulator of transcription. We conclude that the protein product of plmA plays a role, possibly indirect, in regulating plasmid maintenance.
This report presents an analysis of relative copy numbers for each of the endogenous plasmids and a pDU1 replicon in a growing culture of Anabaena sp. strain PCC 7120. The three large plasmids were present at a ratio of about 1:1 with the chromosome. The intermediate-size plasmids pCC7120δ and pCC7120ɛ were present at about a 2:1 ratio with the chromosome. The small plasmid pCC7120ζ was present at a ratio of 6:1. Exogenous plasmid pAM1691 was present at a ratio of 17:1. Anabaena sp. strain PCC 7120 is thought to carry 10 to 20 copies of the chromosome per cell (28). This leads to an estimate of 170 to 340 copies per cell for pAM1691.
In earlier work, plasmid pJL3, also a pDU1 replicon, was estimated to have a relative copy number of 1 (28). Both pJL3 and pAM1691 are derived from shuttle plasmid pRL25, differing chiefly in the inserts (consisting of either patS or cat, a gene conferring Cm resistance). It is not clear why such similar plasmids appear to have such different copy numbers. It is possible that assay methods, the influence of the different inserts, or differences in growth conditions had an effect.
It has been previously shown that a substantial fraction of the DNAs recovered from Anabaena sp. strain PCC 7120 had an high relative copy number; 5.8% of the genome renatured at a rate indicative of a relative copy number of 40 (24). It was suggested that the rapidly renaturing DNA fraction might stem from plasmids or from insertion sequences. None of the endogenous plasmids had such a high relative copy number. It is possible that a major component of the rapidly renaturing portion of the genome was derived from insertion sequences; 145 presumptive transposases have been identified in the genome (27).
Regulated changes in plasmid copy number have previously been described for a marine Synechococcus sp. (45) and for Agmenellum quadruplicatum (37). It is not known whether Anabaena sp. strain PCC 7120 can similarly regulate plasmid content in response to growth and environmental conditions. However, mutations in plmA alter the relative copy numbers for several plasmids and do so in a manner that may explain the mutant's three phenotypes. First, in an otherwise wild-type background, plmA mutants grow slowly. If essential genes are carried on pCC7120δ or pCC7120ζ, then the reduction in their relative copy numbers to less than 50% of that of the wild type could retard growth. Second, the plmA mutant permits heterocysts to develop even when patS is being overexpressed from a plasmid. This may stem directly from the global reduction in the plasmid's relative copy number to 25% of that of the wild type. However, the segregation of Nm-sensitive cells after growth without selection suggests that the plasmid segregates unequally between daughter cells. Heterocysts would tend to form in those segments of the filament in which the plasmid was at an especially low copy number. An alternative model, in which plmA affects expression from the patS promoter, is unlikely, since the plmA mutation repressed the effects of patS overexpression from glnA, rbcL, and petE promoters. Finally, the slow-growth phenotype escalates markedly when a plasmid based on a pDU1 replicon is transferred by conjugation into a plmA mutant strain and subjected to antibiotic selection. Such exconjugants become senescent; that is, they lose viability with each new replating. Presumably, the partial growth defect is compounded by decreased Nm resistance provided by the shuttle vector plasmid.
Phylogenetic analysis placed PlmA in the GntR family of transcriptional regulators. This family was recently divided into the FadR, HutC, MocR, YtrA, and AraR subfamilies on the basis of the heterogeneity of their effector-binding-oligomerization domains (22, 36). These five subfamilies contain genes from both gram-positive and gram-negative bacteria. In contrast, PlmA clusters with members of a new subfamily that is composed exclusively of genes from cyanobacterial species. The effector-binding-oligomerization domain that identifies the new subfamily may respond to a cue that is most commonly found in cyanobacteria such as circadian rhythm signals or stresses due to oxygenic photosynthesis. PlmA affects plasmid maintenance in Anabaena sp. strain PCC 7120, but there are no identified plasmids in the two Prochlorococcus species, in Thermosynechococcus elongatus BP-1, or Synechococcus sp. strain WH 8102, all of which contain genes similar to plmA. Therefore, it is unclear whether all of the members of the cyanobacterial PlmA subfamily are involved in plasmid maintenance.
Within the larger GntR family, however, there are examples of proteins that are known to affect plasmid maintenance. These genes are found in Streptomyces species and fall into the HutC subfamily. One example is the KorSA peptide, which is encoded on the integrative element pSAM2 and autoregulates its own expression as well as the expression of another plasmid-carried peptide, Pra (42). Pra is an activator of pSAM2 replication, integration, and excision (40, 41). When KorSA is inactivated, the element loses its ability to integrate into the chromosome.
The screening for genes that suppressed senescence provided additional evidence that plmA has a role in plasmid maintenance. Library shuttle vector clones that carried a fragment from pCC7120γ or a fragment from pCC7120ζ each produced viable exconjugants. It is possible that the cloned fragments carry an origin of replication from the endogenous plasmids, which would mean that the library clone was not dependent on the pDU1 origin of replication. However, it is striking that both fragments carry genes with homology to regulators of plasmid copy number. The pCC7120ζ fragment carries asl9502, encoding a protein similar to members of the CopG family. The copG gene was identified on a streptococcal plasmid, pMV158. In a regulatory process similar to that used by KorSA (see above), CopG represses its own expression as well as the expression of repB, which encodes a nickase required for the initiation of replication (reviewed in reference 14). The pCC7120γ fragment carries asl8050, which encodes a protein with 73% sequence similarity to CopB from K. pneumoniae plasmid pGSH500. The copy number function of the Klebsiella gene was inferred through homology with peptides from the incFII family (34). The rescue of plmA by two separate plasmid sequences (especially plasmid sequences with presumptive copy number functions) is consistent with the hypothesis that plmA regulates plasmid maintenance functions.
The screening for senescence suppressors did not identify wild-type plmA itself. A plasmid carrying plmA and its downstream neighbor (all1075) complemented the heterocyst suppression phenotype of plmA (data not shown). However, the poor-growth phenotype was not complemented by this construct (or by plmA or all1075 alone). The poor-growth phenotype associated with plmA may be sensitive to the locus's copy number or its location within the genome.
This report demonstrates the influence of plmA on a cell's ability to maintain its relative plasmid content, but it is not clear how the influence is effected. The effect could be indirect, as plasmid maintenance in other organisms has been shown to be influenced by markedly nonspecific mechanisms. For example, the pcnB gene from E. coli encodes a poly(A) polymerase (10) but was identified by its effect on plasmid copy numbers. Loss of pcnB globally alters RNA transcript stability. The copy number of the pUC18 plasmid is affected by two transcripts. RNAII acts as a primer for replication. RNAI is an antisense transcript. When annealed with RNAII, RNAI effectively sequesters the primer and reduces pUC18 copy numbers. The pcnB mutation happens to preferentially stabilize RNAI, leading to decreased copy numbers (23). The mechanism by which plmA influences plasmid maintenance (direct or indirect) remains to be determined.
Acknowledgments
We thank Ho-Sung Yoon for providing several previously undescribed plasmids and Jayna Ditty for her critical reading of the manuscript. We thank Mark Zoran for help with microscopy performed in the Cell Physiology and Molecular Imaging Core Facility (NINDS grant PO1 NS-39546).
This work was supported by National Institutes of Health grant GM36890, Department of Energy grant DE-FG03-98ER020309, and Texas Advanced Research Program grant 010366-0010-1999.
REFERENCES
- 1.Adams, D. G. 2000. Symbiotic interactions, p. 523-561. In B. A. Whitton and M. Potts (ed.), The ecology of cyanobacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. Greene Publishing Associates and Wiley-Interscience, New York, N.Y.
- 4.Bateman, A., E. Birney, L. Cerruti, R. Durbin, L. Etwiller, S. R. Eddy, S. Griffiths-Jones, K. L. Howe, M. Marshall, and E. L. Sonnhammer. 2002. The Pfam protein families database. Nucleic Acids Res. 30:276-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Black, T. A., Y. Cai, and C. P. Wolk. 1993. Spatial expression and autoregulation of hetR, a gene involved in the control of heterocyst development in Anabaena. Mol. Microbiol. 9:77-84. [DOI] [PubMed] [Google Scholar]
- 6.Brasier, M. D., O. R. Green, A. P. Jephcoat, A. K. Kleppe, M. J. Van Kranendonk, J. F. Lindsay, A. Steele, and N. V. Grassineau. 2002. Questioning the evidence for Earth's oldest fossils. Nature 416:76-81. [DOI] [PubMed] [Google Scholar]
- 7.Buikema, W. J., and R. Haselkorn. 1991. Characterization of a gene controlling heterocyst differentiation in the cyanobacterium Anabaena 7120. Genes Dev. 5:321-330. [DOI] [PubMed] [Google Scholar]
- 8.Buikema, W. J., and R. Haselkorn. 2001. Expression of the Anabaena hetR gene from a copper-regulated promoter leads to heterocyst differentiation under repressing conditions. Proc. Natl. Acad. Sci. USA 98:2729-2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Callahan, S. M., and W. J. Buikema. 2001. The role of HetN in maintenance of the heterocyst pattern in Anabaena sp. PCC 7120. Mol. Microbiol. 40:941-950. [DOI] [PubMed] [Google Scholar]
- 10.Cao, G. J., and N. Sarkar. 1992. Identification of the gene for an Escherichia coli poly(A) polymerase. Proc. Natl. Acad. Sci. USA 89:10380-10384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castenholz, R. W. 2001. Phylum BX. Cyanobacteria, p. 473-487. In D. R. Boone and R. W. Castenholz (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 1. Springer, New York, N.Y.
- 12.Cormack, B. P., R. H. Valdivia, and S. Falkow. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173:33-38. [DOI] [PubMed] [Google Scholar]
- 13.Craig, I. W., C. K. Leach, and N. G. Carr. 1969. Studies with deoxyribonucleic acid from blue-green algae. Arch. Mikrobiol. 65:218-227. [DOI] [PubMed] [Google Scholar]
- 14.Del Solar, G., A. M. Hernandez-Arriaga, F. X. Gomis-Ruth, M. Coll, and M. Espinosa. 2002. A genetically economical family of plasmid-encoded transcriptional repressors involved in control of plasmid copy number. J. Bacteriol. 184:4943-4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elhai, J. 1993. Strong and regulated promoters in the cyanobacterium Anabaena PCC 7120. FEMS Microbiol. Lett. 114:179-184. [DOI] [PubMed] [Google Scholar]
- 16.Elhai, J., A. Vepritskiy, A. M. Muro-Pastor, E. Flores, and C. P. Wolk. 1997. Reduction of conjugal transfer efficiency by three restriction activities of Anabaena sp. strain PCC 7120. J. Bacteriol. 179:1998-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elhai, J., and C. P. Wolk. 1990. Developmental regulation and spatial pattern of expression of the structural genes for nitrogenase in the cyanobacterium Anabaena. EMBO J. 9:3379-3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ernst, A., T. Black, Y. Cai, J. M. Panoff, D. N. Tiwari, and C. P. Wolk. 1992. Synthesis of nitrogenase in mutants of the cyanobacterium Anabaena sp. strain PCC 7120 affected in heterocyst development or metabolism. J. Bacteriol. 174:6025-6032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Golden, J. W., L. L. Whorff, and D. R. Wiest. 1991. Independent regulation of nifHDK operon transcription and DNA rearrangement during heterocyst differentiation in the cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 173:7098-7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haselkorn, R. 1992. Developmentally regulated gene rearrangements in prokaryotes. Annu. Rev. Genet. 26:113-130. [DOI] [PubMed] [Google Scholar]
- 22.Haydon, D. J., and J. R. Guest. 1991. A new family of bacterial regulatory proteins. FEMS Microbiol. Lett. 63:291-295. [DOI] [PubMed] [Google Scholar]
- 23.He, L., F. Soderbom, E. G. Wagner, U. Binnie, N. Binns, and M. Masters. 1993. PcnB is required for the rapid degradation of RNAI, the antisense RNA that controls the copy number of ColE1-related plasmids. Mol. Microbiol. 9:1131-1142. [DOI] [PubMed] [Google Scholar]
- 24.Herdman, M., M. Janvier, R. Rippka, and R. Y. Stanier. 1979. Genome size of cyanobacteria. J. Gen. Microbiol. 111:73-85. [Google Scholar]
- 25.Jones, K. M., and R. Haselkorn. 2002. Newly identified cytochrome c oxidase operon in the nitrogen-fixing cyanobacterium Anabaena sp. strain PCC 7120 specifically induced in heterocysts. J. Bacteriol. 184:2491-2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaneko, T., T. Matsubayashi, M. Sugita, and M. Sugiura. 1996. Physical and gene maps of the unicellular cyanobacterium Synechococcus sp. strain PCC6301 genome. Plant Mol. Biol. 31:193-201. [DOI] [PubMed] [Google Scholar]
- 27.Kaneko, T., Y. Nakamura, C. P. Wolk, T. Kuritz, S. Sasamoto, A. Watanabe, M. Iriguchi, A. Ishikawa, K. Kawashima, T. Kimura, Y. Kishida, M. Kohara, M. Matsumoto, A. Matsuno, A. Muraki, N. Nakazaki, S. Shimpo, M. Sugimoto, M. Takazawa, M. Yamada, M. Yasuda, and S. Tabata. 2001. Complete genomic sequence of the filamentous nitrogen-fixing cyanobacterium Anabaena sp. strain PCC 7120. DNA Res. 8:205-213, 227-253. [DOI] [PubMed]
- 28.Lang, J. D., and R. Haselkorn. 1991. A vector for analysis of promoters in the cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 173:2729-2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, B., X. Huang, and J. Zhao. 2002. Expression of hetN during heterocyst differentiation and its inhibition of hetR up-regulation in the cyanobacterium Anabaena sp. PCC 7120. FEBS Lett. 517:87-91. [DOI] [PubMed] [Google Scholar]
- 30.Liu, D., and J. W. Golden. 2002. hetL overexpression stimulates heterocyst formation in Anabaena sp. strain PCC 7120. J. Bacteriol. 184:6873-6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mann, N., and N. G. Carr. 1974. Control of macromolecular composition and cell division in the blue-green algae Anacystis nidulans. J. Gen. Microbiol. 83:399-405. [DOI] [PubMed] [Google Scholar]
- 32.Meeks, J. C., and J. Elhai. 2002. Regulation of cellular differentiation in filamentous cyanobacteria in free-living and plant-associated symbiotic growth states. Microbiol. Mol. Biol. Rev. 66:94-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meeks, J. C., J. Elhai, T. Thiel, M. Potts, F. Larimer, J. Lamerdin, P. Predki, and R. Atlas. 2001. An overview of the genome of Nostoc punctiforme, a multicellular, symbiotic cyanobacterium. Photosyn. Res. 70:85-106. [DOI] [PubMed] [Google Scholar]
- 34.Osborn, A. M., F. M. da Silva Tatley, L. M. Steyn, R. W. Pickup, and J. R. Saunders. 2000. Mosaic plasmids and mosaic replicons: evolutionary lessons from the analysis of genetic diversity in IncFII-related replicons. Microbiology 146:2267-2275. [DOI] [PubMed] [Google Scholar]
- 35.Paerl, H. W., J. L. Pinckney, and T. F. Steppe. 2000. Cyanobacterial-bacterial mat consortia: examining the functional unit of microbial survival and growth in extreme environments. Environ. Microbiol. 2:11-26. [DOI] [PubMed] [Google Scholar]
- 36.Rigali, S., A. Derouaux, F. Giannotta, and J. Dusart. 2002. Subdivision of the helix-turn-helix GntR family of bacterial regulators in the FadR, HutC, MocR, and YtrA subfamilies. J. Biol. Chem. 277:12507-12515. [DOI] [PubMed] [Google Scholar]
- 37.Roberts, T. M., and K. E. Koths. 1976. The blue-green alga Agmenellum quadruplicatum contains covalently closed DNA circles. Cell 9:551-557. [DOI] [PubMed] [Google Scholar]
- 38.Schopf, J. W. 2000. The paleobiologic record of cyanobacterial evolution, p. 105-129. In Y. V. Brun and L. J. Shimkets (ed.), Prokaryotic development. American Society for Microbiology, Washington, D.C.
- 39.Schopf, J. W., A. B. Kudryavtsev, D. G. Agresti, T. J. Wdowiak, and A. D. Czaja. 2002. Laser-Raman imagery of Earth's earliest fossils. Nature 416:73-76. [DOI] [PubMed] [Google Scholar]
- 40.Sezonov, G., A. M. Duchene, A. Friedmann, M. Guerineau, and J. L. Pernodet. 1998. Replicase, excisionase, and integrase genes of the Streptomyces element pSAM2 constitute an operon positively regulated by the pra gene. J. Bacteriol. 180:3056-3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sezonov, G., J. Hagege, J. L. Pernodet, A. Friedmann, and M. Guerineau. 1995. Characterization of pra, a gene for replication control in pSAM2, the integrating element of Streptomyces ambofaciens. Mol. Microbiol. 17:533-544. [DOI] [PubMed] [Google Scholar]
- 42.Sezonov, G., C. Possoz, A. Friedmann, J. L. Pernodet, and M. Guerineau. 2000. KorSA from the Streptomyces integrative element pSAM2 is a central transcriptional repressor: target genes and binding sites. J. Bacteriol. 182:1243-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simon, R. D. 1977. Macromolecular composition of spores from the filamentous cyanobacterium Anabaena cylindrica. J. Bacteriol. 129:1154-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Subramaniam, S. 1998. The Biology Workbench—a seamless database and analysis environment for the biologist. Proteins 32:1-2. [PubMed] [Google Scholar]
- 45.Takeyama, H., H. Nakayama, and T. Matsunaga. 2000. Salinity-regulated replication of the endogenous plasmid pSY10 from the marine cyanobacterium Synechococcus sp. Appl. Biochem. Biotechnol. 84-86:447-453. [DOI] [PubMed] [Google Scholar]
- 46.Tandeau de Marsac, N. 1994. Differentiation of hormogonia and relationships with other biological processes, p. 825-842. In D. A. Bryant (ed.), The molecular biology of cyanobacteria, vol. 1. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 47.Wei, T.-F., T. S. Ramasubramanian, and J. W. Golden. 1994. Anabaena sp. strain PCC 7120 ntcA gene required for growth on nitrate and heterocyst development. J. Bacteriol. 176:4473-4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wolk, C. P. 1967. Physiological basis of the pattern of vegetative growth of a blue-green alga. Proc. Natl. Acad. Sci. USA 57:1246-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wolk, C. P., A. Vonshak, P. Kehoe, and J. Elhai. 1984. Construction of shuttle vectors capable of conjugative transfer from Escherichia coli to nitrogen-fixing filamentous cyanobacteria. Proc. Natl. Acad. Sci. USA 81:1561-1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu, X., and C. P. Wolk. 2001. Role for hetC in the transition to a nondividing state during heterocyst differentiation in Anabaena sp. J. Bacteriol. 183:393-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 52.Yoon, H. S., and J. W. Golden. 1998. Heterocyst pattern formation controlled by a diffusible peptide. Science 282:935-938. [DOI] [PubMed] [Google Scholar]
- 53.Yoon, H. S., and J. W. Golden. 2001. PatS and products of nitrogen fixation control heterocyst pattern. J. Bacteriol. 183:2605-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]