Abstract
The proteome of Haemophilus influenzae strain Rd KW20 was analyzed by liquid chromatography (LC) coupled with ion trap tandem mass spectrometry (MS/MS). This approach does not require a gel electrophoresis step and provides a rapidly developed snapshot of the proteome. In order to gain insight into the central metabolism of H. influenzae, cells were grown microaerobically and anaerobically in a rich medium and soluble and membrane proteins of strain Rd KW20 were proteolyzed with trypsin and directly examined by LC-MS/MS. Several different experimental and computational approaches were utilized to optimize the proteome coverage and to ensure statistically valid protein identification. Approximately 25% of all predicted proteins (open reading frames) of H. influenzae strain Rd KW20 were identified with high confidence, as their component peptides were unambiguously assigned to tandem mass spectra. Approximately 80% of the predicted ribosomal proteins were identified with high confidence, compared to the 33% of the predicted ribosomal proteins detected by previous two-dimensional gel electrophoresis studies. The results obtained in this study are generally consistent with those obtained from computational genome analysis, two-dimensional gel electrophoresis, and whole-genome transposon mutagenesis studies. At least 15 genes originally annotated as conserved hypothetical were found to encode expressed proteins. Two more proteins, previously annotated as predicted coding regions, were detected with high confidence; these proteins also have close homologs in related bacteria. The direct proteomics approach to studying protein expression in vivo reported here is a powerful method that is applicable to proteome analysis of any (micro)organism.
The genomes of dozens of microorganisms have been sequenced already, and many more sequencing projects are currently under way. A period of extensive postgenome analysis with a variety of high-throughput methods should incorporate the information gained from these genome sequences and should yield a better understanding of microbial biology (9). In a popular approach DNA microarrays are used to generate data on gene expression and to monitor the entire cellular genetic orchestra as a dynamic system (26, 49, 58). However, a number of experimental and computational studies have suggested that to adequately describe and model cellular metabolism, information on gene expression should be complemented by protein expression data (12, 13, 19, 21). There are numerous cases in which cellular regulation occurs either posttranscriptionally or through posttranslational modifications of proteins. These situations can be addressed by a global postgenomic high-throughput approach commonly referred to as proteome analysis. Proteome analysis is aimed at studying all proteins expressed in a certain organism under certain conditions (1, 5, 7, 8, 18, 27, 34, 35, 37, 38, 53, 60, 61, 63-67). One of the central issues surrounding proteome analysis is accurately identifying proteins in complex mixtures extracted from the cells.
Haemophilus influenzae strain Rd KW20, a nontypeable strain, was the first free-living organism to have a completely sequenced genome (15). Consequently, it has become a commonly used model organism for whole-genome annotation, computational analysis, and cross-genome comparisons (15, 29, 56). H. influenzae strain Rd KW20 is an experimentally tractable, genetically manipulatable organism that grows well on defined media, and it has a single natural host, humans. It is an important pathogen that causes meningitis, otitis media, sinusitis, and chronic bronchitis, as well as life-threatening invasive infections (3, 5-7, 32, 41, 44, 46, 59). Nontypeable H. influenzae is the most common bacterium isolated from the lower respiratory tract of patients with chronic obstructive pulmonary disease (chronic bronchitis) (5, 42, 47, 51, 52, 55), which is the fourth-most-common cause of death in the United States (51). Serotype b disease is a serious problem in much of the developing world, where conjugate vaccines are not readily available (5, 46).
Due to its relatively small genome size and its phylogenetic proximity to Escherichia coli, H. influenzae is an extremely convenient model organism for proteomic studies (5, 7, 12, 13, 19, 32, 34, 37). It was the first organism for which a genome-scale model of metabolic fluxes was constructed (10, 45, 50), and whole-genome transposon mutagenesis analysis also has been implemented (2, 23). In the present study, H. influenzae strain Rd KW20 (1,830 kb; 1,705 predicted open reading frames [56, 57]) was used as a test microorganism to evaluate the performance of a direct proteomics approach to proteome analysis, with the ultimate aim of determining the in vivo properties of the protein set expressed by the bacterium under certain conditions. In this method a relatively inexpensive ion trap mass spectrometer is used to analyze unlabeled trypsinized protein mixtures obtained directly from cell preparations disrupted with a French press, which ensures minimal perturbation of the protein content. In this paper the results which we obtained are compared with the predictions made by computational analysis and the experimental results obtained by a variety of other approaches.
MATERIALS AND METHODS
Media and growth conditions.
The proteome of a classic facultative anaerobe, H. influenzae strain Rd KW20, which belongs to the gamma subdivision of the Proteobacteria (25), was analyzed after growth under two different conditions, anaerobic and microaerobic. As this organism is more frequently isolated from clinical material after anaerobic incubation, these conditions are expected to be close to the natural state of H. influenzae in its human host (A. L. Smith, unpublished data), as well as similar to each other. Thus, we expected to find a relatively high number of common proteins for these two conditions, which was an important prediction to examine.
H. influenzae strain Rd KW20 cells (initial inocula, 1.2 × 105 to 4.6 × 105 CFU/ml) were grown at 37°C in brain heart infusion (BHI) broth (Difco) containing 10 mg of β-NAD per liter, 10 mg of equine hemin hydrochloride per liter, and 10 mg of l-histidine per liter (all obtained from Sigma Chemical Co., St. Louis, Mo.). A sterile loop was used to transfer an inoculum from a fresh BHI agar plate into 50 ml of supplemented BHI (sBHI) broth in a 250-ml Erlenmeyer flask. For microaerobic growth, the cultures were incubated on a rotary platform at 200 rpm. The partial pressure of oxygen in the cell suspensions remained steady in the range from 128 to 145 Torr in early the logarithmic phase and in the stationary phase and decreased to an average of 53 Torr (n = 3) in the mid-logarithmic phase (A650, 0.64). Thus, the conditions were microaerobic during late logarithmic growth.
The cultures were also incubated anaerobically in the same flasks fitted with butyl rubber stoppers, in which the air in the headspace was purged with nitrogen in a Difco anaerobic chamber. Nitrogen was bubbled through the sBHI broth prior to inoculation. Anaerobiosis was indicated by a disposable GasPak anaerobic indicator (Becton Dickenson, Franklin Lakes, N.J.) and was verified by growth of spores of the obligate anaerobe Clostridium butyricum on chocolate agar plates placed in the incubation chamber. H. influenzae cells were grown overnight under both conditions and harvested by centrifugation. Pseudomonas aeruginosa strain PAO1 (inoculum size, 2.0 × 105 to 5.2 × 105 CFU/ml; n = 3) failed to grow in sBHI under the anaerobic conditions. The incubation times varied from 19.0 to 23.5 h, after which the bacterial densities ranged from 1.9 × 109 to 6.5 × 109 CFU/ml (for both microaerobic and anaerobic growth). The initial pH of the sBHI broth was 7.34 ± 0.12 (n = 9), while at the conclusion of incubation under anaerobic conditions the pH was 4.69 ± 0.59 (n = 6) and at the conclusion of incubation under microaerobic conditions the pH was 5.97 ± 0.16 (n = 4).
Protein preparation.
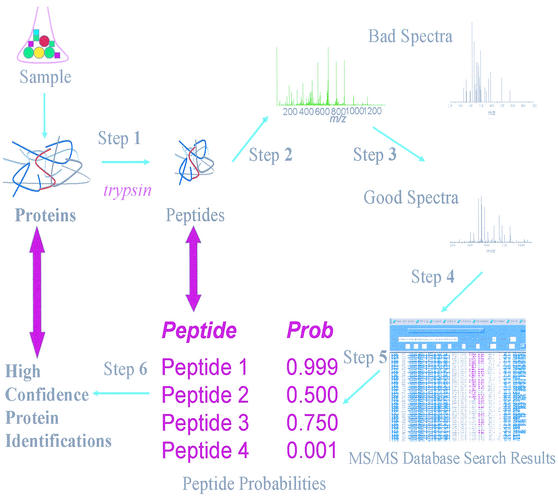
Cells were resuspended in phosphate-buffered saline and were disrupted by passage through a French pressure cell (SLM Instrument, Urbana, Ill.) at 15,000 lb/in2. Soluble and membrane fractions of H. influenzae strain Rd KW20 cells were separated by ultracentrifugation at 140,000 × g for 4 h at 4°C in a Ti80 rotor. The pellet was used as the membrane fraction, and the supernatant was used as the soluble fraction. Both soluble and membrane samples were boiled for 3 min, precipitated with cold acetone, and resuspended in phosphate-buffered saline containing 0.05% sodium dodecyl sulfate. Protein concentrations were determined with a Bio-Rad protein assay kit (Bio-Rad, Munich, Germany) and were adjusted to 2 μg/μl. One microgram of porcine modified trypsin (Promega, Madison, Wis.) per 100 μg of H. influenzae strain Rd KW20 proteins was added to each sample, and this was followed by incubation at 37°C overnight. Protein mixtures were dried and resuspended in 0.4% acetic acid to a concentration of 2 μg of protein per μl (Fig. 1, step 1).
FIG.1.
Proteomics approach implemented in this study. For details see Materials and Methods.
Experimental design.
Soluble and membrane peptide mixtures were injected onto a capillary column (10 cm by 100 μm) with an autosampler (FAMOS; Dionex, Sunnyvale, Calif.) connected online via an electrospray ionization source (Brechbuhler, Spring, Tex.) to an ion trap mass spectrometer (LCQ DECA XP; ThermoFinnigan, San Jose, Calif.). Peptides were eluted with a linear gradient of acetonitrile (10 to 70%) for 1 h against 0.4% acetic acid in water.
The complex peptide mixture was analyzed by liquid chromatography (LC) with an electrospray ionization source-ion trap mass spectrometer. We used a standard top-down data-dependent ion selection approach for tandem mass spectrometry (MS/MS), in which the base peak ion was selected for collision-induced dissociation and this was followed by 3 min of dynamic exclusion to prevent reselection of previously selected ions. To optimize proteome coverage and improve protein identification in complex mixtures of total cell tryptic digests, multiple narrowly overlapping m/z window ranges (so-called gas phase fractionation) were employed (30, 54, 67). Three sets of experimental conditions were utilized in this study; each set of experiments was carried out in duplicate, and the sets differed in the number of m/z ranges and thus the number of total experiments used for the data-dependent dynamic exclusion ion selection for collision-induced dissociation. The numbers of LC-MS/MS analyses conducted for the different m/z ranges were as follows: (i) one analysis for m/z 400 to 2000; (ii) three analyses for m/z 400 to 800, m/z 700 to 1200, and m/z 1100 to 2000; and (iii) 16 analyses for m/z 400 to 510, m/z 490 to 610, m/z 590 to 710, m/z 690 to 810, m/z 790 to 910, m/z 890 to 1010, m/z 990 to 1110, m/z 1090 to 1210, m/z 1190 to 1310, m/z 1290 to 1410, m/z 1390 to 1510, m/z 1490 to 1610, m/z 1590 to 1710, m/z 1690 to 1810, m/z 1790 to 1910, and m/z 1890 to 2000.
An amount of peptide equivalent to 2 μg was injected for each LC-MS/MS analysis (Fig. 1, step 2). The spectra resulting from the LC-MS/MS analyses were tested by determining their spectral quality scores (Fig. 1, step 3). If the spectral quality scores were equal to or greater than a certain threshold (a normalized value of 2.0 within a range from 0.0 to 4.0 was used), the so-called good spectra (http://www.biatech.org/publications/) were then searched with the SEQUEST program (11) (Fig. 1, step 4). By using different parameters (Table 1) (http://www.biatech.org/publications/), SEQUEST searches were performed against the H. influenzae strain Rd KW20 protein database (57). Then the observed peptide and protein identifications were reevaluated by using peptide (31) and protein (A. I. Nesvizhskii, A. Keller, E. Kolker, and R. Aebersold, submitted for publication) statistical models (Fig. 1, steps 5 and 6, respectively). Proteins with confidence levels of at least 90% were manually validated, and the confirmed identification results were listed as high-confidence results (Table 1).
TABLE 1.
H. influenzae strain Rd KW20 candidate protein assignments as determined by SEQUEST and high-confidence protein assignments as determined by statistical models
| Method | No. of identified proteins
|
No. of tryptic termini | |||
|---|---|---|---|---|---|
| Total | Common | Microaerobic conditions | Anaerobic conditions | ||
| SEQUEST_1a | 1,295 | 584 | 471 | 240 | 0, 1, 2 |
| SEQUEST_1 (≥2 distinct peptides) | 808 | 258 | 397 | 189 | |
| SEQUEST_2a | 725 | 245 | 300 | 180 | 1, 2 |
| SEQUEST_2 (≥2 distinct peptides) | 304 | 126 | 116 | 62 | |
| High confidence | 414 | 221 | 138 | 55 | 0, 1, 2 |
| High confidence (≥2 distinct peptides) | 331 | 180 | 112 | 39 | |
The SEQUEST default parameters were 1.5/2.0/2.5/0.1/50/1.
Control protein mixture.
Control mixtures were prepared by using 18 selected pure proteins (purchased from Sigma and Prozyme, San Leandro, Calif.) having different physicochemical properties (Table 2). The initial concentrations ranged from 2 to 1,000 nM to mimic the concentrations in MS/MS experiments performed with complex protein samples (30). A previously characterized preparation of microaerophilically grown H. influenzae strain Rd KW20 membrane proteins was spiked with the purified digest of the control mixture. A 1:1 volumetric combination showed that the H. influenzae peptides eluted with approximately 10% of the intensity of the peptides from the control mixture. Thus, a 9:1 volumetric combination was used with the resulting protein concentrations indicated in Table 2. LC-MS/MS analyses for five m/z ranges were conducted with replicates by using the spiked sample. This experiment was aimed at assessing the sensitivity (reliable limit of detection) of protein identification by our direct proteomics approach.
TABLE 2.
Protein identifications for experimental control mixture
| No. | Protein | Accession no.a | Mol wt | Concn (nM) | Identifi- cationb |
|---|---|---|---|---|---|
| 1 | Bovine beta-casein | P02666 | 25,107 | 100 | + |
| 2 | Bovine carbonic anhydrase | P00921 | 28,980 | 100 | + |
| 3 | Bovine serum albumin | P02769 | 69,293 | 40 | + |
| 4 | Bovine beta-lactoglobulin | P02754 | 19,883 | 20 | + |
| 5 | E. coli alkaline phosphatase | P00634 | 49,438 | 20 | + |
| 6 | Bovine serotransferrin | Q29443 | 77,753 | 10 | + |
| 7 | Rabbit GAPDHc | P46406 | 35,688 | 2 | + |
| 8 | Bovine catalase | P00432 | 57,585 | 2 | + |
| 9 | Rabbit glycogen phosphorylase | P00489 | 97,158 | 1 | + |
| 10 | Bovine cytochrome c | P00006 | 11,572 | 40 | − |
| 11 | E. coli beta-galactosidase | P00722 | 116,351 | 0.4 | − |
| 12 | Bovine alpha-lactalbumin | P00711 | 16,246 | 10 | − |
| 13 | Bacillus lichenformis alpha-amylase | Q04977 | 66,924 | 4 | − |
| 14 | Horse myoglobin | P02188 | 16,951 | 4 | − |
| 15 | Saccharomyces cerevisiae mannose-6-phosphate isomerase | P29952 | 48,057 | 1 | − |
| 16 | Chicken ovalbumin | P01012 | 42,750 | 0.4 | − |
| 17 | Bovine gamma-actin | ATBOG | 41,661 | 0.2 | − |
| 18 | Rabbit myosin | P02562 | 241,852 | 0.2 | − |
Most protein accession numbers are from the SWISS-PROT database (http://www.expasy.org/sprot); the only exception is the accession number of bovine gamma-actin, which is from the PIR database (http://pir.georgetown.edu).
Proteins 1 through 9 were identified with high confidence (see Table 1) and were subsequently confirmed manually.
GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Statistical models.
Fast, automated, and reliable assessment of the quality of thousands of tandem mass spectra is recognized as one of the major elements required for successful implementation of high-throughput proteomics approaches. Data obtained from digested mixtures of 18 selected proteins (30) analyzed by LC-MS/MS as described above were monitored by using the spectral quality assignments (http://www.biatech.org/publications/) (Fig. 1, step 3). If the spectral quality scores were equal to or greater than the threshold value, 2.0, then the spectra were subjected to a SEQUEST (11) database search (Fig. 1, step 4). The SEQUEST program compares experimentally observed tandem mass spectra to theoretical spectra of all possible peptide fragments from the sequence database of choice, which in this study was the H. influenzae strain Rd KW20 annotated protein database (57).
Statistical models for peptide and protein identification were used to analyze the results obtained from the SEQUEST search. The peptide model estimates the probability that peptide sequences are correctly assigned to spectra by the database search. On the basis of the SEQUEST scores and the number of allowed tryptic termini, these probabilities have been shown to be accurate and to have high power for discriminating between correct and incorrect assignments (31) (Fig. 1, step 5). The number of tryptic termini has been routinely used to assess whether assignments are correct (30, 38, 64, 67). This number, which may be 0, 1, or 2, measures how many of the peptide termini, based on the amino acid sequence, are consistent with cleavage by trypsin at the amino-terminal side of arginine and lysine.
The protein model (Nesvizhskii et al., submitted) estimates the likelihood of the presence of proteins in a sample as determined from the probabilities of the corresponding peptides assigned to tandem mass spectra (31) (Fig. 1, step 6). Combining these statistical approaches allowed us to compile a set of protein identifications with confidence levels of at least 90%. These identifications were then manually examined, which reduced the error rates even further (Table 1) (http://www.biatech.org/publications/). The resulting set of high-confidence identifications permitted reliable assessment of the protein contents of a sample.
RESULTS
Sensitivity.
An estimate of the sensitivity (or reliable limit of detection) of the direct proteomics approach was obtained by utilizing a controlled mixture of proteins. Table 2 summarizes the information for the various proteins and the confidence levels at which their presence in the samples analyzed could be established (see Materials and Methods). These experiments revealed the detection levels for proteins present at concentrations as low as 1 nM (Table 2). However, only one of the two proteins present in the experimental mixture at a concentration of 40 nM was identified with high confidence. Therefore, we conservatively estimated that the minimal level of a protein for reliable detection must be approximately 100 nM. Assuming that the efficiency for proteolytic activity is 90%, this value represents 18,000 amol of digested protein (and therefore peptide) in a 200-μl reaction mixture. Thus, the sensitivity level was estimated to be approximately 90 amol of reliably detected peptide per μl of injected sample. This is a fairly conservative and preliminary estimate of the sensitivity of our direct proteomics approach. Extrapolation of this estimate depends on multiple parameters, including the type of mass spectrometer utilized; the number of replicate experiments investigated; the data-dependent dynamic exclusion ion selection variables employed; the amount and complexity of the samples analyzed; the statistical models used to estimate the probability that a given peptide or protein is present; and the set of parameters for SEQUEST database searches, quality assignments, and peptide and protein probabilities employed in the study. It should be recognized that this is not an exhaustive list. Nevertheless, the estimate of the reliable limit of detection given above provided a baseline for our direct proteomics approach and should be directly applicable to studying proteomes of diverse (micro)organisms.
Protein identification.
Proteins from the soluble and membrane fractions were analyzed in duplicate experiments that differed in the number of m/z ranges used. The spectra resulting from these LC-MS/MS experiments were examined, as shown in Fig. 1 and described in Materials and Methods. Then their spectral qualities were evaluated, and the good spectra were subjected to a SEQUEST search against the protein database (57). The resulting peptide and protein identifications were reevaluated by using the statistical models described above (31; Nesvizhskii et al., submitted). Finally, protein identifications with a confidence level of at least 90% were manually examined and, if they were confirmed, were considered to be high-confidence identifications (see Materials and Methods). Multiple identifications of distinct peptides corresponding to the same protein were obtained for a majority of the proteins identified with high confidence (Table 1) (http://www.biatech.org/publications/).
Table 1 shows the numbers of the H. influenzae strain Rd KW20 proteins assigned by using different SEQUEST (11) thresholds, as well as the numbers of high-confidence identifications. A total of 436 proteins were identified in the soluble and membrane fractions of the microaerobically and anaerobically grown H. influenzae cells when spectral quality assignments (quality assignment threshold, 2.0) were coupled with the peptide and protein probability (confidence level, at least 90%) estimates. These identifications were then manually validated, which reduced the number of identifications to 414 high-confidence protein identifications, 138 of which were found exclusively in microaerobic samples and 55 of which were found exclusively in anaerobic samples (Table 1). As expected (see Materials and Methods), a high number of high-confidence proteins (221 proteins; more than 53% of the proteins) were identified under both conditions (Table 1). The 414 proteins accounted for approximately 25% of all assigned H. influenzae strain Rd KW20 open reading frames (predicted proteins) identified by whole-genome analysis (57).
Alternatively, the SEQUEST thresholds coupled with nonspecific trypsin digestion yielded 1,295 candidate proteins (Table 1), which accounted for more than 75% of the H. influenzae strain Rd KW20 theoretical proteome. When one or two tryptic termini were required (see Materials and Methods), the total number of candidate protein identifications was reduced by about 40%, to 725 proteins (more than 42% of the theoretical proteome). Our method resulted in much higher specificity than conventional approaches (38, 64, 67), whose false-positive rates require laborious manual verification.
Ribosomal proteins.
In addition to our observation that a large number of proteins were expressed under both growth conditions, ribosomal proteins (which constituted one of the most abundantly expressed protein types in cells) were also expected to be detected in high numbers. The success of the direct proteomics approach can therefore be estimated from a comparison of sets of ribosomal proteins identified in this study and obtained by conventional gel electrophoresis coupled with mass spectrometry. A recent compilation of multiple protein identifications obtained in numerous experiments by using two-dimensional gel electrophoresis (2DE) to separate and visualize proteins followed by identification by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MS) showed that overall, the 2DE-MS approach detected 18 ribosomal proteins (34) or only one-third of the 54 known ribosomal proteins. In contrast, with our approach we were able to identify 43 ribosomal proteins with high confidence (approximately 80% of the total number of ribosomal proteins [Table 3 ]).
TABLE 3.
Ribosomal proteins of H. influenzae strain Rd KW20 identified in this study
| Protein | Genome identification no. | Size (amino acids) | Microaerobic conditionsa
|
Anaerobic conditionsa
|
Detection by 2DEc | ||||
|---|---|---|---|---|---|---|---|---|---|
| No. of peptides | No. of unique peptides | Identificationb | No. of peptides | No. of unique peptides | Identificationb | ||||
| RpS1 | HI1220 | 550 | 14 | 5 | + | 17 | 10 | + | − |
| RpS2 | HI0913 | 252 | 19 | 4 | + | 12 | 3 | + | <0.1 |
| RpS3 | HI0783 | 236 | 19 | 5 | + | 13 | 7 | + | <0.1 |
| RpS4 | HI0801 | 207 | 4 | 1 | + | 10 | 4 | + | + |
| RpS5 | HI0795 | 167 | 11 | 2 | + | 4 | 2 | + | + |
| RpS6 | HI0547 | 131 | 10 | 4 | + | 2 | 2 | + | − |
| RpS7 | HI0580 | 157 | 33 | 4 | + | 11 | 4 | + | − |
| RpS8 | HI0792 | 131 | − | 3 | 2 | + | + | ||
| RpS9 | HI1442 | 131 | − | − | + | ||||
| RpS10 | HI0776 | 119 | 9 | 1 | + | 16 | 13 | + | + |
| RpS11 | HI0800 | 130 | − | − | − | ||||
| RpS12 | HI0581 | 125 | − | − | − | ||||
| RpS13 | HI0799 | 123 | 8 | 2 | + | − | − | ||
| RpS14 | HI0791 | 102 | − | 2 | 1 | + | − | ||
| RpS15 | HI1468 | 90 | − | − | − | ||||
| RpS16 | HI0204 | 83 | 5 | 2 | + | 2 | 1 | + | − |
| RpS17 | HI0786 | 86 | 3 | 2 | + | 7 | 1 | + | − |
| RpS18 | HI0545 | 76 | − | 2 | 2 | + | − | ||
| RpS19 | HI0781 | 92 | 22 | 3 | + | 11 | 4 | + | − |
| RpS20 | HI0965 | 90 | 13 | 2 | + | 1 | 1 | ± | − |
| RpS21 | HI0531 | 72 | − | − | − | ||||
| RpL1 | HI0516 | 230 | 45 | 3 | + | 32 | 8 | + | − |
| RpL2 | HI0780 | 274 | 10 | 1 | + | 14 | 5 | + | <0.1 |
| RpL3 | HI0777 | 209 | 27 | 4 | + | 11 | 4 | + | + |
| RpL4 | HI0778 | 201 | 2 | 2 | + | 19 | 5 | + | + |
| RpL5 | HI0790 | 180 | 10 | 4 | + | 12 | 6 | + | + |
| RpL6 | HI0793 | 178 | 8 | 3 | + | 22 | 6 | + | + |
| RpL7/1 | HI0641 | 124 | 316 | 19 | + | 96 | 16 | + | − |
| RpL9 | HI0544 | 150 | 16 | 4 | + | 22 | 5 | + | − |
| RpL10 | HI0640 | 164 | 41 | 3 | + | 11 | 4 | + | − |
| RpL11 | HI0517 | 143 | 8 | 2 | + | 21 | 7 | + | − |
| RpL13 | HI1443 | 143 | 10 | 2 | + | 13 | 3 | + | + |
| RpL14 | HI0788 | 124 | − | 7 | 3 | + | − | ||
| RpL15 | HI0797 | 145 | 3 | 2 | + | 21 | 4 | + | − |
| RpL16 | HI0784 | 137 | 11 | 2 | + | 1 | 1 | + | − |
| RpL17 | HI0803 | 129 | 21 | 6 | + | 7 | 3 | + | + |
| RpL18 | HI0794 | 118 | 8 | 2 | + | 1 | 1 | + | − |
| RpL19 | HI0201 | 117 | − | 3 | 2 | + | − | ||
| RpL20 | HI1320 | 118 | 6 | 2 | + | − | − | ||
| RpL21 | HI0880 | 104 | 17 | 3 | + | 6 | 1 | + | + |
| RpL22 | HI0782 | 110 | 22 | 3 | + | 11 | 4 | + | − |
| RpL23 | HI0779 | 100 | 5 | 2 | + | 1 | 1 | ± | + |
| RpL24 | HI0789 | 104 | 24 | 1 | + | 43 | 3 | + | + |
| RpL25 | HI1630 | 96 | 15 | 3 | + | 3 | 1 | + | + |
| RpL27 | HI0879 | 86 | − | − | − | ||||
| RpL28 | HI0951 | 79 | 1 | 1 | +/− | 5 | 1 | + | − |
| RpL29 | HI0785 | 64 | 70 | 3 | + | 14 | 5 | + | − |
| RpL30 | HI0796 | 60 | − | − | − | ||||
| RpL31 | HI0758 | 71 | 3 | 2 | + | − | − | ||
| RpL32 | HI0158 | 57 | − | − | 4 | 2 | + | − | |
| RpL33 | HI0950 | 57 | − | − | − | − | |||
| RpL34 | HI0998 | 45 | − | − | − | − | |||
| RpL35 | HI1319m | 66 | − | − | − | − | |||
| RpL36 | HI0798.1 | 38 | − | − | − | − | |||
Cells were grown under microaerobic or anaerobic conditions as described in Materials and Methods.
+, protein identified with high confidence (see Table 1); ±, candidate protein; −, protein not detected.
Data from reference 34. <0.1, protein that was found on the gel at a low level and comprises no more than 0.1% of the total cell protein.
It also should be noted that the classical 2DE-MS approach (34) was unable to identify ribosomal proteins S1 and L1, two of the largest ribosomal proteins, while several distinct high-probability peptides for these two proteins were observed multiple times in our study. Only one ribosomal protein that has been detected previously by the 2DE-MS approach, S9, was not detected in our experiments. Other ribosomal proteins that we were unable to detect were short proteins (no more than 90 amino acid residues), with two exceptions (Table 3). These data indicate that the direct proteomics approach, based on the LC-MS/MS method coupled with a database search, quality assignments, and statistical models, offers a good alternative to the classical 2DE-MS approach.
Protein synthesis machinery.
Besides ribosomal proteins, many other components of the translational machinery were typically detected with high confidence. These include all aminoacyl-tRNA synthetases (except the asparaginyl- and cysteinyl-tRNA synthetases, HI1302 and HI0078), translation elongation factors EF-G, EF-P, EF-Ts, and EF-Tu, methionyl-tRNA formyltransferase, and some other proteins. All three translation initiation factors, InfA, InfB, and InfC, were determined to be candidate proteins (unless indicated otherwise, all the data below are available at the BIATECH website [http://www.biatech.org/publications/]), although only InfB was consistently detected with high confidence. Several additional translation components were detected, albeit not with high confidence, which might be indicative of lower abundance. These include, for example, peptide chain release factors PrfA (HI1561) and PrfC (HI1735) and N-formylmethionyl-tRNA deformylase (HI0622). In contrast, peptidyl-tRNA hydrolase (HI0394) and several other proteins were not detected in this study. Of all the tRNA- and rRNA-modifying enzymes, only tRNA-guanine transglycosylase (HI0244) was detected with high confidence.
DNA replication, repair, and transcription.
Although DNA replication comprises a crucial part of cell growth, DNA polymerase and other DNA-interacting enzymes appeared to be less abundant than the components of protein synthesis machinery. DNA primase, DNA polymerase I, DNA polymerase III subunits, two subunits of DNA gyrase, DNA topoisomerases I and III, and several DNA repair proteins were determined to be candidate proteins in our experiments (http://www.biatech.org/publications/).
In contrast, transcriptional proteins were well represented in H. influenzae cells. All the subunits of the DNA-dependent RNA polymerase (RpoA, RpoB, RpoC, and RpoZ), including the main sigma subunit (RpoD, HI0533), were identified with high confidence. Additional sigma factors, such as RpoE and RpoH, were clearly less abundant, and RpoH (HI0269) was not even detected. This observation is consistent with the roles of these proteins in stress response, which apparently does not occur in cells grown in near-optimal conditions in rich media. The amounts of transcriptional factors varied. While transcription antitermination protein NusG and transcription termination factor Rho were both identified with high confidence, transcription elongation factor GreA was identified as a candidate protein, and elongation factor GreB was not detected. In contrast, a large number of peptides derived from the DNA-binding protein HU-alpha (HupA, HI0430) were identified, indicating that there was an abundance of this protein in the cell. Additionally, another DNA-binding protein, Hns, was also identified with high confidence.
Cell division proteins.
Given the reliable limit of detection reported above and the relatively low cellular concentrations of cell division proteins, it was not surprising that most of these proteins were not detected in the present work (http://www.biatech.org/publications/). The notable exceptions are the FtsY protein, which was confidently identified in the microaerobically grown cells, and the FtsH protein, which was confidently identified in the anaerobically grown cells. It is not clear at this time whether these findings reflect actual differential expression of these two proteins or represent spurious hits. In any case, given the paucity of data on the mechanisms of cell division in H. influenzae, which lacks MinC, MinD, and MinE proteins (16), evaluations of relative protein expression by the direct proteomics approach should result in better understanding in this area.
Membrane proteins.
The H. influenzae strain Rd KW20 genome contains genes encoding a wide variety of respiratory enzymes, including Na+-translocating NADH:ubiquinone oxidoreducatase (NqrABCDEF, HI0164 to HI0171 and HI1683 to HI1688), periplasmic nitrate reductase (NrfBCFGH, HI0342 to HI0348), nitrite reductase (NrfABCD, HI1066 to HI1069), dimethyl sulfoxide reductase (DmsABC, HI1045 to HI1047), and cytochrome d ubiquinol oxidase (CydAB, HI1075 and HI1076). Identification of these enzymes posed a significant challenge in that some of their subunits were identified with high confidence, while others were not. For example, while the alpha (HI0164) and gamma (HI0167) subunits of Na+-translocating NADH:ubiquinone oxidoreducatase were detected with high confidence in both microaerobically and anaerobically grown cells, the beta (HI0171) and delta (HI0168) subunits were detected with high confidence only in anerobically grown cells, and the two remaining subunits, NqrB (HI0166) and NqrE (HI0170), either were not detected or were determined to be candidate proteins. While these results paralleled the original order of discovery of the Nqr subunits (first the alpha, beta, and gamma subunits were discovered, followed by three other subunits [6, 22]), they indicate that there is a potential problem with recovery and identification of integral membrane proteins. Remarkably, we detected no expression of the predicted second nqr operon (HI1683 to HI1688 [20]).
The most interesting results regarding multiple-subunit membrane enzymes were obtained with the FoF1-type H+-ATPase (ATP synthase) that consists of a cytoplasmic F1 sector with α6β6γδɛ stoichiometry and a membrane Fo sector with an approximate ab2c10-12 stoichiometry (14). Subunits α (HI0481), β (HI0479), and b (HI0483) were detected with high confidence in both microaerobically and anaerobically grown cells, subunit γ was detected with high confidence only in microaerobically grown cells, and the remaining subunits either were determined to be candidate proteins or were not detected. These data again emphasize that detection depends not just on the relative abundance of each polypeptide but can be affected by the size of the protein, its hydrophobicity, the number of trypsin cleavage sites, and other parameters.
Conserved hypothetical proteins.
In a recent whole-genome transposon mutagenesis study of H. influenzae, 478 genes were identified as genes that are essential for microaerobic growth; 259 of these genes were originally annotated as hypothetical or putative genes (2). In the present study, 47 of 414 proteins detected with high confidence also fell into this category of hypothetical or putative genes. As discussed previously, short of a systematic mistake in sequencing or gene calling, a protein that is conserved across diverse phylogenetic lineages should not be considered hypothetical (16). Furthermore, conserved proteins that are encoded in relatively small genomes of diverse parasitic bacteria are likely to be essential for growth of the bacteria (17). Indeed, 15 conserved hypothetical genes detected under microaerobic conditions (Table 4) also belong on the list of essential genes (2). Thus, the results of the present study support the mutagenesis data and indicate that these 15 hypothetical genes indeed encode expressed proteins. In fact, additional BLAST searches (4) and manual reannotation revealed predicted or experimentally determined functions for a majority of these hypothetical proteins (Table 4). Some of these updated protein functions have recently been incorporated into the database (57).
TABLE 4.
Expression of essential conserved hypothetical proteins of H. influenzae strain Rd KW20a
| No. | Genome identifi- cation no. | SWISS-PROT symbol | Current annotationb | Expression conditionsc |
|---|---|---|---|---|
| 1 | HI0056 | YGDQ_HAEIN | Integral membrane protein, homolog of tellurium resistance protein TerC | MAE |
| 2 | HI0119 | ZNUA_HAEIN | Periplasmic Zn-binding component of an ABC-type Mn/Zn transport system | Both |
| 3 | HI0131 | Y131_HAEIN | Periplasmic Fe-binding component of an ABC-type iron transport system | Both |
| 4 | HI0148 | YJHT_HAEIN | Uncharacterized protein containing a kelch-type beta-propeller domain | Both |
| 5 | HI0172 | APBE_HAEIN | Lipoprotein involved in thiamine biosynthesis | Both |
| 6 | HI0405 | PDXY_HAEIN | Pyridoxal kinase | Both |
| 7 | HI0458 | SURA_HAEIN | Periplasmic peptidyl-prolyl cis-trans isomerase | Both |
| 8 | HI0467 | YICC_HAEIN | Uncharacterized protein, induced by stress and in the stationary phase | MAE |
| 9 | HI1001 | 60IM_HAEIN | Preprotein translocase subunit YidC/Oxalp/A1b3 | MAE |
| 10 | HI1021 | THIQ_HAEIN | ATP-binding component of ABC-type thiamine transport system | MAE |
| 11 | HI1245 | MAO2_HAEIN | Fusion of NADP-dependent malate dehydrogenase (malic enzyme) and phosphotransacetylase | Both |
| 12 | HI1349 | YD49_HAEIN | Stress-induced DNA-binding protein Dps | Both |
| 13 | HI1556 | YF56_HAEIN | 2-Hydroxyacid dehydrogenase | Both |
| 14 | HI1603 | YG03 HAEIN | Putative regulator of phosphate transport | MAE |
| 15 | HI1682 | SOHB_HAEIN | Periplasmic serine protease SohB, C1pP-like family | MAE |
Whether a gene product is essential was based on the data in reference 2.
Annotations from the latest GenBank entry or from the COG database (57).
Growth conditions under which the protein was identified with high confidence in this study. MAE, microaerobic conditions; Both, both microaerobic and anaerobic conditions.
The remaining conserved hypothetical proteins that were detected with high confidence, while apparently not essential for microaerobic growth, are expressed at substantial levels and can be expected to perform some important functions. In certain cases, keys to their functions already exist. This is exemplified by the HI1426 protein, which is expressed under both microaerobic and anaerobic growth conditions, is closely related to the universal stress protein UspA (HI0815), and, like E. coli UspA, must be a nucleotide-binding protein. The HI0409 protein, which is expressed exclusively under microaerobic conditions, is a member of a large family of membrane proteins related to metalloendopeptidases and might play a role in protein maturation and/or secretion (57).
In other cases, there appears to be no discernible function(s) for certain conserved proteins, whose wide phylogenetic distribution suggests their importance for cell biology. A good example is the HI0442 protein (YbaB, COG0718), which is almost universally conserved in bacteria and whose gene is usually sandwiched between the dnaX and recR genes, likely forming operons and suggesting that there is a functional association. Although the structure of this protein has been resolved recently (36), its role in DNA replication or repair, if any, remains obscure. In our experiments, two distinct peptides derived from this protein were detected in H. influenzae cells with high confidence under both microaerobic and anaerobic growth conditions, when expression of many DNA repair genes (uvrA, uvrC, uvrD, mutH, mutL, radA, radC, recN, and recO) was absent or barely detectable. These observations suggest that YbaB plays a role in normal DNA replication rather than in DNA repair.
For several more proteins, such as HI0065 (YjeE, COG0802), HI0315 (YebC, COG0217), HI0656 (YciO, COG0009), and HI0719 (YjgF, COG0251), the exact functions remain unknown even though the crystal structures have been resolved in structural genomics studies (28, 62) and the predicted biochemical functions have been listed in the database (57). Our observation that these proteins are detected either under both growth conditions (HI0719) or predominantly during microaerobic growth (HI0065, HI0315, HI0656) might eventually help in pinpointing their functions.
Putative proteins.
In addition to encoding conserved hypothetical proteins, the H. influenzae genome includes a certain number of putative or hypothetical open reading frames that do not have detectable homologs in other organisms and therefore cannot be automatically assumed to encode real proteins. These open reading frames do not belong to any clusters of orthologous genes (33, 57), and most of them are appropriately annotated in the current genome database as H. influenzae predicted coding regions. Remarkably, of 112 proteins, only 2 were identified with high confidence in our samples, several more were determined to be candidate proteins, and expression of the rest was never detected. The data for the two expressed proteins, HI0246 and HI1624, show that although their functions are not known, these proteins have close homologs in Pasteurella multocida and Pseudomonas putida, respectively (57). Interestingly, the genes encoding most of the other putative proteins determined to be candidate proteins also turned out to have homologs in other sequenced genomes. These data illustrate the value of our direct proteomics and comparative genomics approaches for better genome annotation and assessment of protein functions.
Another area in which our direct proteomics approach is likely to be very useful in genome annotation is in distinguishing between the functions of close paralogs. Interestingly, the H. influenzae strain Rd KW20 genome contains three paralogs (HI0052, HI0146, and HI1028) of the E. coli gene yiaO that encodes a periplasmic C4-dicarboxylate-binding component of the tripartite ATP-independent periplasmic transporter. This is a rare case in which the H. influenzae genome has more paralogs than the 2.5-fold-larger E. coli genome, and the individual functions of the three paralogs are obscure. In our experiments, one of the three paralogs, HI0146, was identified with high confidence in both microaerobically and anaerobically grown cells. Another paralog, HI0052, was barely detectable, and only in microaerobically grown cells; the third paralog, HI1028, was never detected. These data suggest that although the three proteins might perform the same function, they are likely differentially regulated and expressed in different amounts and under different conditions.
While comparison of these genome-scale data must be interpreted carefully in view of differences in experimental methods and the types of data obtained, this example illustrates how integration of complementary high-throughput approaches dramatically refines our knowledge of genetic functions and protein identification.
DISCUSSION
2DE or not 2DE?
Currently, the most popular method employed to explore microbial proteomes is a combination of 2DE for separating and visualizing proteins and then MS for protein identification (7, 8, 12, 13, 19, 60, 61). The highest coverage for any proteome reported so far is a set of 502 proteins from H. influenzae (29% of all proteins predicted from the genome analysis), which was compiled from the results of multiple studies performed by several leading groups during a significant time period (34). This method requires several laborious and time-consuming steps, including individual spot extraction, digestion, and analysis. Classical 2DE also has severe limitations when it comes to identification of membrane proteins, proteins with extreme physicochemical properties, or low-abundance proteins (27, 35). Another unfortunate outcome of the classical 2DE-MS approach (34) is its inability to identify a majority of the ribosomal proteins, one of the most overabundant cellular protein types (see above). Finally, the majority of the 502 protein identifications lacked any independent experimental reevaluation and/or statistical estimation (34). This makes any further usage of the data rather difficult.
MS is becoming a method of choice for rapid identification of large numbers of proteins, their modified versions, and protein complexes (1, 18, 35, 38, 53, 55, 63-65, 67). For example, in studies of the yeast proteome (18, 38, 64, 65) and human proteins (1, 54), LC coupled with MS/MS has been used successfully. The development of statistical approaches for automated assignment of quality scores to experimental tandem mass spectra and for probability assignments of peptide and protein identifications has been described recently (31, 43; http://www.biatech.org/publications/; Nesvizhskii et al., submitted). The last three methods, along with analysis of control mixtures of peptides derived from digests of selected proteins (30), were used in this work.
As mentioned above, analysis of several membrane-bound protein complexes (e.g., NADH:ubiquinone oxidoreductase, fumarate reductase, nitrate reductase, nitrite reductase, dimethyl sulfoxide reductase, ATP synthase) showed that while certain subunits of these enzymes were identified with high confidence, other subunits of the same enzymes either were determined to be candidate proteins or were not detected. Thus, soluble subunits of enzymes were more frequently identified with high confidence than the corresponding membrane subunits were. These observations indicate that there are a number of experimental, technological, methodological, and biological sources of uncertainty in delineating the exact spectrum of proteins expressed by given cells under certain conditions.
Some possible experimental factors include small protein size (e.g., in most cases of undetected control and ribosomal proteins [Tables 2 and 3, respectively]), the physicochemical properties of a protein, and under- or overrepresentation of trypsin cleavage sites in a protein. Correct data-dependent dynamic exclusion ion spectra acquisition parameters, with which reduction of high-intensity peptide signals blocking detection of coeluting low-intensity signals can be achieved, is critical on the technological side. Some proteins were not detected because of the methodological approach used in this study. Proteins with an extensive membrane-spanning domain(s) may not have been well solubilized from the membrane fractions and therefore would have been underrepresented. Finally, some proteins may not have been identified due to their (relatively) low expression levels, which could have been below the reliable limit of detection of our direct proteomics approach. These limitations aside, our direct proteomics analysis is a powerful method for proteome analysis of any (micro)organism, as illustrated by this study of the H. influenzae strain Rd KW20 proteome.
From peptides to function.
The goals of this study were to assess the proteome of H. influenzae, to find correlations between protein expression data under two key growth conditions with significantly higher accuracy than that obtained previously, and to learn more about the cellular organization and behavior of this model microorganism. To do these things, several experimental and computational methods were utilized and integrated, as follows: multiple narrowly overlapping m/z windows as determined by LC-MS/MS were used to optimize proteome coverage (30, 54, 67); control mixtures of peptides were derived from digests of selected proteins having known concentrations (30); the sensitivity (reliable limit of detection) of our direct proteomics approach was estimated; and statistical analyses were used to estimate quality spectral assignments (http://www.biatech.org/publications/) and to assess peptide (31) and protein (Nesvizhskii et al., submitted) identifications. Finally, the recent compendia resulting from H. influenzae proteome analysis by the 2DE-MS approach (34) and from identification of essential genes by mutational analysis (2) allowed a genome-wide comparison with the observed results. The combination of diverse genome-wide analyses employed in this work provides a first glimpse of the integrated approaches that are needed to gain a full understanding of the genomes of H. influenzae and other microorganisms. In a recent review, Saier emphasized the fact that although E. coli is “perhaps the best-understood organism on earth,” our understanding of its biology is still extremely limited (48). A similar sentiment was expressed in the recently published book by Koonin and Galperin (33), who stated that our level of understanding other microorganisms, including H. influenzae, is even lower. At the current rate of experimental characterization of putative and hypothetical genes, completing the task for E. coli could take as long as 100 years (Galperin, unpublished data), and the results would make only a small dent in the set of over 110,000 predicted but unannotated proteins that are currently found in public databases.
Significant advancements are clearly needed to address this bottleneck, and the present study promises some improvement. This study resulted in detection of at least 15 proteins originally annotated as conserved hypothetical. These proteins have also been found to be essential by mutation analysis (2), so they certainly are expressed in vivo, at least under microaerobic conditions. As conserved hypothetical proteins, these proteins had clear homologs in the protein databases, which allowed assignment of (putative) functions for a majority of them through sequence similarity searches. An analysis of 30 more conserved hypothetical proteins is currently under way (E. Kolker et al., unpublished data).
Another intriguing result of functional assignment concerns the proteins that were originally annotated as putative, hypothetical, or encoded by H. influenzae predicted coding regions. Of 112 predicted coding regions, only two (HI0246 and HI1624) were identified with high confidence in this study, confirming that the proteins are expressed. For both of these proteins, sequence similarity searches successfully detected close homologs in related proteobacteria. Given the limitations of the direct proteomics approach, as discussed above, one could speculate that other proteins that were originally described as putative and which were detected here only with low confidence could still be expressed in H. influenzae cells in vivo. The case for these candidate proteins becomes even stronger when homologs of them are found encoded in other sequenced genomes. Verification of the expression and delineation of the cellular functions of previously putative proteins represents an important avenue for future research.
From peptides to metabolism.
This study of the H. influenzae proteome can serve as a first step towards a detailed analysis of this organism's metabolism. Even though it has been 8 years since the genome of H. influenzae strain Rd KW20 was sequenced, some critical questions about the metabolic capacities of this organism remain unanswered. For example, although H. influenzae is known to ferment glucose (24, 25, 39), its glucose transporter remains to be unidentified and properly annotated in the genome database (15, 57). The only protein annotated as a component of the glucose transport machinery, a homolog of the E. coli crr gene product, is apparently a part of the fructose-specific phosphoenolpyruvate-dependent phosphotransferase system (40). To address this and other open issues concerning the metabolism of H. influenzae, further comprehensive studies are necessary, in which genomic information for metabolic modeling (10, 45, 50) and mutational analyses (2, 23) should be used. Protein expression data, as reported in this work, should form the basis for such a multifaceted analysis of H. influenzae metabolism.
Acknowledgments
We greatly appreciate the critical comments of Brian Akerley, Alex Beliaev, Ludmila Chistoserdova, Krassen Dimitrov, Daniel Drell, James Fredrickson, Evelyne Kolker, Eugene Koonin, Mary Lidstrom, Robert Munson, Kenneth Nealson, Nathan Price, Anu Raghunathan, Monica Riley, Margaret Romine, Joseph St. Geme, Anna Stolyar, and Judy Wall.
This work was supported by National Institutes of Health grant AI44002 to A.L.S., by discretionary funds of L.H., and by Department of Energy Offices of Biological and Environmental Research and Advanced Scientific Computing Research Genomes to Life grant DE-FG08-01ER63218 to E.K.
REFERENCES
- 1.Aebersold, R., and D. R. Goodlett. 2001. Mass spectrometry in proteomics. Chem. Rev. 101:269-295. [DOI] [PubMed] [Google Scholar]
- 2.Akerley, B. J., E. J. Rubin, V. L. Novick, K. Amaya, N. Judson, and J. J. Mekalanos. 2002. A genome-scale analysis for identification of genes required for growth or survival of Haemophilus influenzae. Proc. Natl. Acad. Sci. USA 99:966-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alexander, H., and G. Leidy. 1953. Haemophilus influenzae Garf d dissociated to become Rd. J. Exp. Med. 97:17-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 5.Ball, P. 1996. Infective pathogenesis and outcomes in chronic bronchitis. Curr. Opin. Pulm. Med. 2:181-185. [DOI] [PubMed] [Google Scholar]
- 6.Beattie, P., K. Tan, R. M. Bourne, D. Leach, P. R. Rich, and F. B. Ward. 1994. Cloning and sequencing of four structural genes for the Na+-translocating NADH-ubiquinone oxidoreductase of Vibrio alginolyticus. FEBS Lett. 356:333-338. [DOI] [PubMed] [Google Scholar]
- 7.Cash, P., E. Argo, P. Langford, and S. J. Kroll. 1997. Development of a Haemophilus two-dimensional protein database. Electrophoresis 18:1472-1482. [DOI] [PubMed] [Google Scholar]
- 8.Cordwell, S. J., A. S. Nouwens, N. M. Verrills, J. C. McPherson, P. G. Hains, D. D. Van Dyk, and B. J. Walsh. 1999. The microbial proteome database—an automated laboratory catalogue for monitoring protein expression in bacteria. Electrophoresis 20:3580-3588. [DOI] [PubMed] [Google Scholar]
- 9.Drell, D. 2002. The Department of Energy Microbial Cell Project: a 180° paradigm shift for biology. OMICS J. Integr. Biol. 6:3-10. [DOI] [PubMed] [Google Scholar]
- 10.Edwards, J. S., and B. O. Palsson. 1999. Systems properties of the Haemophilus influenzae Rd metabolic genotype. J. Biol. Chem. 274:17410-17416. [DOI] [PubMed] [Google Scholar]
- 11.Eng, J. K., A. L. McCormack, and J. R. Yates III. 1994. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5:976-979. [DOI] [PubMed] [Google Scholar]
- 12.Evers, S., K. Di Padova, M. Meyer, M. Fountoulakis, W. Keck, and C. P. Gary. 1998. Strategies towards a better understanding of antibiotic action: folate pathway inhibition in Haemophilus influenzae as an example. Electrophoresis 19:1980-1988. [DOI] [PubMed] [Google Scholar]
- 13.Evers, S., K. Di Padova, M. Meyer, H. Langen, M. Fountoulakis, W. Keck, and C. P. Gary. 2001. Mechanism-related changes in the gene transcription and protein synthesis patterns of Haemophilus influenzae after treatment with transcriptional and translational inhibitors. Proteomics 4:522-544. [DOI] [PubMed] [Google Scholar]
- 14.Fillingame, R. H., and S. Divall. 1999. Proton ATPases in bacteria: comparison to Escherichia coli F1F0 as the prototype. Novartis Found. Symp. 221:218-229. [DOI] [PubMed] [Google Scholar]
- 15.Fleischmann, R. D., M. D. Adams, O. White, R. A. Clayton, E. F. Kirkness, A. R. Kerlavage, C. J. Bult, J.-F. Tomb, B. A. Dougherty, J. M. Merrick, K. McKenney, G. G. Sutton, W. FitzHugh, C. A. Fields, J. D. Gocayne, J. D. Scott, R. Shirley, L. I. Liu, A. Glodek, J. M. Kelley, J. F. Weidman, C. A. Phillips, T. Spriggs, E. Hedblom, M. D. Cotton, T. Utterback, M. C. Hanna, D. T. Nguyen, D. M. Saudek, R. C. Brandon, L. D. Fine, J. L. Fritchman, J. L. Fuhrmann, N. S. Geoghagen, C. L. Gnehm, L. A. McDonald, K. V. Small, C. M. Fraser, H. O. Smith, and J. C. Venter. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:496-512. [DOI] [PubMed] [Google Scholar]
- 16.Galperin, M. Y. 2001. Conserved “hypothetical” proteins: new hints and new puzzles. Comp. Funct. Genomics 2:14-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galperin, M. Y., and E. V. Koonin. 1999. Searching for drug targets in microbial genomes. Curr. Opin. Biotechnol. 10:571-578. [DOI] [PubMed] [Google Scholar]
- 18.Gavin, A. C., M. Bosche, R. Krause, P. Grandi, M. Marzioch, A. Bauer, J. Schultz, J. M. Rick, A. M. Michon, C. M. Cruciat, M. Remor, C. Hofert, M. Schelder, M. Brajenovic, H. Ruffner, A. Merino, K. Klein, M. Hudak, D. Dickson, T. Rudi, V. Gnau, A. Bauch, S. Bastuck, B. Huhse, C. Leutwein, M. A. Heurtier, R. R. Copley, A. Edelmann, E. Querfurth, V. Rybin, G. Drewes, M. Raida, T. Bouwmeester, P. Bork, B. Seraphin, B. Kuster, G. Neubauer, and G. Superti-Furga. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415:141-147. [DOI] [PubMed] [Google Scholar]
- 19.Gmuender, H., K. Kuratli, K. Di Padova, C. P. Gray, W. Keck, and S. Evers. 2001. Gene expression changes triggered by exposure of Haemophilus influenzae to novobiocin or ciprofloxacin: combined transcription and translation analysis. Genome Res. 11:28-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hase, C. C., N. D. Fedorova, M. Y. Galperin, and P. A. Dibrov. 2001. Sodium ion cycle in bacterial pathogens: evidence from cross-genome comparisons. Microbiol. Mol. Biol. Rev. 65:353-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hatzimanikatis, V., and K. H. Lee. 1999. Dynamical analysis of gene networks requires both mRNA and protein expression information. Metab. Eng. 1:275-281. [DOI] [PubMed] [Google Scholar]
- 22.Hayashi, M., K. Hirai, and T. Unemoto. 1995. Sequencing and the alignment of structural genes in the nqr operon encoding the Na+-translocating NADH-quinone reductase from Vibrio alginolyticus. FEBS Lett. 363:75-77. [DOI] [PubMed] [Google Scholar]
- 23.Herbert, M. A., S. Hayes, M. E. Deadman, C. M. Tang, D. W. Hood, and E. R. Moxon. 2002. Signature tagged mutagenesis of Haemophilus influenzae identifies genes required for in vivo survival. Microb. Pathog. 33:211-223. [DOI] [PubMed] [Google Scholar]
- 24.Hollander, R. 1976. Energy metabolism of some representatives of the Haemophilus group. Antonie Leeuwenhoek 42:429-444. [DOI] [PubMed] [Google Scholar]
- 25.Holt, J. G., N. Krieg, P. Sneath, and J. W. S. Staley. 1994. Bergey's manual of determinative bacteriology, p. 195, 277-278, and 286. Williams & Wilkins, Baltimore, Md.
- 26.Hughes, T. R., M. J. Marton, A. R. Jones, C. J. Roberts, R. Stoughton, C. D. Armour, H. A. Bennett, E. Coffey, H. Dai, Y. D. He, M. J. Kidd, A. M. King, M. R. Meyer, D. Slade, P. Y. Lum, S. B. Stepaniants, D. D. Shoemaker, D. Gachotte, K. Chakraburtty, J. Simon, M. Bard, and S. H. Friend. 2000. Functional discovery via a compendium of expression profiles. Cell 102:109-126. [DOI] [PubMed] [Google Scholar]
- 27.Jenkins, R. E., and S. R. Pennington. 2001. Arrays for protein expression profiling: towards a viable alternative to two-dimensional gel electrophoresis? Proteomics 1:13-29. [DOI] [PubMed] [Google Scholar]
- 28.Jia, J., V. V. Lunin, V. Sauve, L. W. Huang, A. Matte, and M. Cygler. 2002. Crystal structure of the YciO protein from Escherichia coli. Proteins 49:139-141. [DOI] [PubMed] [Google Scholar]
- 29.Karlin, S., J. Mrazek, and A. M. Campbell. 1996. Frequent oligonucleotides and peptides of the Haemophilus influenzae genome. Nucleic Acids Res. 24:4263-4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keller, A., S. Purvine, A. I. Nesvizhskii, S. Stolyar, D. R. Goodlett, and E. Kolker. 2002. Experimental protein mixture for validating tandem mass spectral analysis. OMICS J. Integr. Biol. 6:207-212. [DOI] [PubMed] [Google Scholar]
- 31.Keller, A., A. I. Nesvizhskii, E. Kolker, and R. Aebersold. 2002. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74:5383-5392. [DOI] [PubMed] [Google Scholar]
- 32.Kolker, E., S. Purvine, A. Picone, T. Cherny, B. J. Akerley, R. Munson, Jr., B. O. Palsson, D. A. Dainess, and A. L. Smith. 2002. H. influenzae consortium: integrative study of H. influenzae-human interactions. OMICS J. Integr. Biol. 6:341-348. [DOI] [PubMed] [Google Scholar]
- 33.Koonin, E. V., and M. Y. Galperin. 2002. Sequence-evolution-function. Computational approaches in comparative genomics. Kluwer Academic Publishers, Boston, Mass. [PubMed]
- 34.Langen, H., B. Takacs, S. Evers, P. Berndt, H. W. Lahm, B. Wipf, C. Gray, and M. Fountoulakis. 2000. Two-dimensional map of the proteome of Haemophilus influenzae. Electrophoresis 21:411-429. [DOI] [PubMed] [Google Scholar]
- 35.Liebler, D. C. 2002. Introduction to proteomics. Humana Press, Totowa, N.J.
- 36.Lim, K., A. Tempczyk, J. F. Parsons, N. Bonander, J. Toedt, Z. Kelman, A. Howard, E. Eisenstein, and O. Herzberg. 2003. Crystal structure of YbaB from Haemophilus influenzae (HI0442), a protein of unknown function coexpressed with the recombinational DNA repair protein RecR. Proteins 50:375-379. [DOI] [PubMed] [Google Scholar]
- 37.Link, A. J., L. G. Hays, E. B. Carmack, and J. R. Yates III. 1997. Identifying the major proteome components of Haemophilus influenzae type-strain NCTC 8143. Electrophoresis 18:1314-1334. [DOI] [PubMed] [Google Scholar]
- 38.Link, A. J., J. Eng, D. M. Schieltz, E. Carmack, G. J. Mize, D. R. Morris, B. M. Garvik, and J. R. Yates III. 1999. Direct analysis of protein complexes using mass spectrometry. Nat. Biotechnol. 17:676-682. [DOI] [PubMed] [Google Scholar]
- 39.Macfadyen, L. P., and R. J. Redfield. 1996. Life in mucus: sugar metabolism in Haemophilus influenzae. Res. Microbiol. 147:541-551. [DOI] [PubMed] [Google Scholar]
- 40.Macfadyen, L. P., I. R. Dorocicz, J. Reizer, M. H. Saier, Jr., and R. J. Redfield. 1996. Regulation of competence development and sugar utilization in Haemophilus influenzae Rd by a phosphoenolpyruvate:fructose phosphotransferase system. Mol. Microbiol. 21:941-952. [DOI] [PubMed] [Google Scholar]
- 41.Marrs, C. F., G. P. Krasan, K. W. McCrea, D. L. Clemans, and J. R. Gilsdorf. 2001. Haemophilus influenzae—human specific bacteria. Front. Biosci. 6:41-60. [DOI] [PubMed] [Google Scholar]
- 42.Miravitlles, M., C. Espinosa, E. Fernandez-Laso, J. A. Martos, J. A. Maldonado, and M. Gallego. 1999. Relationship between bacterial flora in sputum and functional impairment in patients with acute exacerbations of COPD. Study Group of Bacterial Infection in COPD. Chest 116:40-46. [DOI] [PubMed] [Google Scholar]
- 43.Moore, R. E., M. K. Young, and T. D. Lee. 2002. QSCORE: an algorithm for evaluating SEQUEST database search results. J. Am. Soc. Mass Spectrom. 13:378-386. [DOI] [PubMed] [Google Scholar]
- 44.Moxon, E. R., D. W. Hood, N. J. Saunders, E. K. Schweda, and J. C. Richards. 2002. Functional genomics of pathogenic bacteria. Philos. Trans. R. Soc. Lond. B Biol. Sci. 357:109-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Papin, J. A., N. D. Price, J. S. Edwards, and B. O. Palsson. 2002. The genome-scale extreme pathway structure in Haemophilus influenzae shows significant network redundancy. J. Theor. Biol. 215:67-82. [DOI] [PubMed] [Google Scholar]
- 46.Peltola, H. 2000. Worldwide Haemophilus influenzae type b disease at the beginning of the 21st century: global analysis. Clin. Microbiol. Rev. 13:302-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Read, R. C. 1999. Infection in acute exacerbations of chronic bronchitis: a clinical perspective. Respir. Med. 93:845-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saier, M. H., Jr. 2003. Answering fundamental questions in biology with bioinformatics. ASM News 69:175-181. [Google Scholar]
- 49.Schena, M., D. Shalon, R. W. Davis, and P. O. Brown. 1995. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270:467-470. [DOI] [PubMed] [Google Scholar]
- 50.Schilling, C. H., and B. O. Palsson. 2000. Assessment of the metabolic capabilities of Haemophilus influenzae Rd through a genome-scale pathway analysis. J. Theor. Biol. 203:249-283. [DOI] [PubMed] [Google Scholar]
- 51.Sethi, S., and T. F. Murphy. 2001. Bacterial infection in obstructive pulmonary disease in 2000: a state-of-the-art review. Clin. Microbiol. Rev. 14:336-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sethi, S., N. Evans, B. J. Grant, and T. F. Murphy. 2002. New strains of bacteria and exacerbations of chronic obstructive pulmonary disease. N. Engl. J. Med. 347:465-471. [DOI] [PubMed] [Google Scholar]
- 53.Smith, R. D., G. A. Anderson, M. S. Lipton, C. Masselon, L. Pasa-Tolic, Y. Shen, and H. R. Udseth. 2002. The use of accurate mass tags for high-throughput microbial proteomics. OMICS J. Integr. Biol. 6:61-90. [DOI] [PubMed] [Google Scholar]
- 54.Spahr, C. S., M. T. Davis, M. D. McGinley, J. H. Robinson, E. J. Bures, J. Beierle, J. Mort, P. L. Courchesne, K. Chen, R. C. Wahl, W. Yu, R. Luethy, and S. D. Patterson. 2001. Towards defining the urinary proteome using liquid chromatography-tandem mass spectrometry. I. Profiling an unfractionated tryptic digest. Proteomics 1:93-107. [DOI] [PubMed] [Google Scholar]
- 55.St. Geme, J. W., III. 2002. Molecular and cellular determinants of non-typeable Haemophilus influenzae adherence and invasion. Cell. Microbiol. 4:191-200. [DOI] [PubMed] [Google Scholar]
- 56.Tatusov, R. L., A. R. Mushegian, P. Bork, N. P. Brown, W. S. Hayes, M. Borodovsky, K. E. Rudd, and E. V. Koonin. 1996. Metabolism and evolution of Haemophilus influenzae deduced from a whole-genome comparison with Escherichia coli. Curr. Biol. 6:279-291. [DOI] [PubMed] [Google Scholar]
- 57.Tatusov, R. L., D. A. Natale, I. V. Garkavtsev, T. A. Tatusova, U. T. Shankavaram, B. S. Rao, B. Kiryutin, M. Y. Galperin, N. D. Fedorova, and E. V. Koonin. 2001. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 29:22-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tjaden, B. C., D. R. Haynor, S. Stolyar, C. Rosenow, and E. Kolker. 2002. Identifying operons and untranslated regions of transcripts using Escherichia coli RNA expression analysis. Bioinformatics 18:S337-344. [DOI] [PubMed] [Google Scholar]
- 59.Tuomanen, E. I., K. R. Powell, M. I. Marks, C. I. Laferriere, D. H. Altmiller, C. M. Sack, and A. L. Smith. 1981. Oral chloramphenicol in the treatment of Haemophilus influenzae meningitis. J. Pediatr. 99:968-974. [DOI] [PubMed] [Google Scholar]
- 60.VanBogelen, R. A., E. R. Olson, B. L. Wanner, and F. C. Neidhardt. 1996. Global analysis of proteins synthesized during phosphorus restriction in Escherichia coli. J. Bacteriol. 178:4344-4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.VanBogelen, R. A., E. E. Schiller, J. D. Thomas, and F. C. Neidhardt. 1999. Diagnosis of cellular states of microbial organisms using proteomics. Electrophoresis 20:2149-2159. [DOI] [PubMed] [Google Scholar]
- 62.Volz, K. 1999. A test case for structure-based functional assignment: the 1.2 A crystal structure of the yjgF gene product from Escherichia coli. Protein Sci. 8:2428-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Washburn, M. P., and J. R. Yates III. 2000. Analysis of microbial proteome. Curr. Opin. Microbiol. 3:2920-2927. [DOI] [PubMed] [Google Scholar]
- 64.Washburn, M. P., D. Wolters, and J. R. Yates III. 2001. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19:242-247. [DOI] [PubMed] [Google Scholar]
- 65.Washburn, M. P., R. Ulaszek, C. Deciu, D. M. Schieltz, and J. R. Yates III. 2002. Analysis of quantitative proteomic data generated via multidimensional protein identification technology. Anal. Chem. 74:1650-1657. [DOI] [PubMed] [Google Scholar]
- 66.Wilkins, M. R., C. Pasquali, R. D. Appel, K. Ou, O. Golaz, J. C. Sanchez, J. X. Yan, A. A. Gooley, G. Hughes, I. Humphery-Smith, K. L. Williams, and D. F. Hochstrasser. 1996. From proteins to proteomes: large scale protein identification by two-dimensional electrophoresis and amino acid analysis. Bio/Technology 14:61-65. [DOI] [PubMed] [Google Scholar]
- 67.Yi, E. C., M. Marelli, H. Lee, S. Purvine, R. Aebersold, J. D. Aitchison, and D. R. Goodlett. 2002. Approaching complete peroxisome characterization by gas-phase fractionation. Electrophoresis 23:3205-3216. [DOI] [PubMed] [Google Scholar]