Abstract
We describe here purification and biochemical characterization of the F1Fo-ATP synthase from the thermoalkaliphilic organism Bacillus sp. strain TA2.A1. The purified enzyme produced the typical subunit pattern of an F1Fo-ATP synthase on a sodium dodecyl sulfate-polyacrylamide gel, with F1 subunits α, β, γ, δ, and ɛ and Fo subunits a, b, and c. The subunits were identified by N-terminal protein sequencing and mass spectroscopy. A notable feature of the ATP synthase from strain TA2.A1 was its specific blockage in ATP hydrolysis activity. ATPase activity was unmasked by using the detergent lauryldimethylamine oxide (LDAO), which activated ATP hydrolysis >15-fold. This activation was the same for either the F1Fo holoenzyme or the isolated F1 moiety, and therefore latent ATP hydrolysis activity is an intrinsic property of F1. After reconstitution into proteoliposomes, the enzyme catalyzed ATP synthesis driven by an artificially induced transmembrane electrical potential (Δψ). A transmembrane proton gradient or sodium ion gradient in the absence of Δψ was not sufficient to drive ATP synthesis. ATP synthesis was eliminated by the electrogenic protonophore carbonyl cyanide m-chlorophenylhydrazone, while the electroneutral Na+/H+ antiporter monensin had no effect. Neither ATP synthesis nor ATP hydrolysis was stimulated by Na+ ions, suggesting that protons are the coupling ions of the ATP synthase from strain TA2.A1, as documented previously for mesophilic alkaliphilic Bacillus species. The ATP synthase was specifically modified at its c subunits by N,N′-dicyclohexylcarbodiimide, and this modification inhibited ATP synthesis.
In bacteria, mitochondria, and chloroplasts ATP synthesis is catalyzed by the membrane-bound F1Fo-ATP synthase fueled by an electrochemical gradient of protons (Δp) or, in some cases, Na+ ions (ΔpNa+) (3, 4, 29, 36, 47). F1Fo enzymes consist of a membrane-associated headpiece, F1, with the subunit composition α3β3γδɛ, and an integral membrane sector, Fo, with the subunit composition ab2c10-14. This molecular machine couples ion translocation through Fo to the synthesis of ATP at F1 by a rotational mechanism (30, 31, 43). The ion flux across the membrane induces the rotation of the rotor ring (c10-14), which is physically connected with central stalk subunits γ and ɛ (6, 17, 39-42, 46). Rotation of the asymmetrically bent γ subunit within the central cavity of the α3β3 cylinder elicits periodic structural changes in the catalytic β subunits which are instrumental in ATP synthesis (2).
The alkaliphiles are a unique group of microorganisms that thrive at high external pH values (pH 8.5 to 11.5) but maintain the cytoplasmic pH near neutral (e.g., pH 8.0). Because the total Δp is the sum of the membrane electrical potential (Δψ) (positive out) and the pH gradient (ΔpH) (acid out in neutrophiles), the large ΔpH generated by alkaliphiles in the opposite direction (acid in) has an adverse effect on the magnitude of the total Δp. It has been reported that bacteria growing at pH 10.5 have a Δp of −45 mV, and this value is considered submaximal for ATP synthesis (21). No bioenergetic problem would exist if the bacteria used Na+-coupled processes for solute transport, motility, and ATP synthesis because ΔpNa+ and Δψ are orientated in the same direction and thus add driving force to one another. Indeed, ion-solute transport systems and flagellar rotation often appear to depend on sodium as a coupling ion in alkaliphiles (21). However, the F1Fo-ATP synthases of the mesophilic alkaliphilic organisms Bacillus pseudofirmus and Bacillus alcalophilus have been shown to be exclusively proton coupled (7-9). A number of putative mechanisms have been proposed to explain how the ATP synthases in these bacteria remain proton coupled at low Δp values, but these models are still to be experimentally proven (21).
We recently isolated a facultative thermoalkaliphilic Bacillus species, designated strain TA2.A1, that grows aerobically at pH 7.5 to 10.0 and has a temperature optimum of 65 to 70°C (32, 33). Strain TA2.A1 is dependent upon sodium for growth, and we have demonstrated that solute transport (i.e., glutamate and sucrose transport) is Na+ dependent (33, 34). In this paper, we describe purification of the F1Fo-ATP synthase from Bacillus sp. strain TA2.A1 and the subunit composition of this enzyme. The purified enzyme was reconstituted into proteoliposomes, and ion transport studies showed that the ATP synthase utilizes protons as coupling ions for ATP synthesis.
MATERIALS AND METHODS
Bacterial strain and growth conditions.
Bacillus sp. strain TA2.A1 was grown at 65°C in an alkaline medium (pH 9.2) as described previously (32), except that in this study the growth medium was supplemented with 10 g of Tryptone Peptone (Difco) per liter to increase the final cellular growth yield.
Preparation of inverted membrane vesicles.
Cells of strain TA2.A1 (15 g, wet weight) were resuspended in 15 ml of 50 mM Tris-HCl buffer (pH 8.2) containing 0.1 mM diisopropylfluorophosphate (DFP). Lysozyme was added to a final concentration of 1.2 mg/ml, and the suspension was stirred for 45 min at room temperature. DNase I (2 mg) and 15 mM (final concentration) MgCl2 were then added, and this was followed by incubation for a further 15 min. All subsequent steps were performed at 4°C unless otherwise stated. The protoplasts were collected by centrifugation (180,000 × g for 1 h) and resuspended in 20 ml of extraction buffer (50 mM potassium phosphate buffer [pH 8.0], 0.1 mM DFP, 2 mM MgCl2,1 mM dithiothreitol). The protoplasts were broken by two passages through a precooled French pressure cell at 20,000 lb/in2. The extract was centrifuged at 8,000 × g for 20 min to remove unbroken cells. The cell-free supernatant was centrifuged at 180,000 × g for 45 min to harvest membrane vesicles. No ATPase activity was detected in the supernatant after centrifugation, and the cell membranes were red.
Preparation of the isolated F1-ATPase.
To dissociate the F1 portion of the ATP synthase from membrane vesicles, the procedure described by Laubinger and Dimroth (24) performed with a low-ionic-strength dissociation buffer (1 mM Tris-HCl [pH 8.0], 0.5 mM K2-EDTA, 1 mM dithioerythritol, 0.1 mM DFP, 10% glycerol) and high-speed centrifugation (180,000 × g for 45 min) was employed. F1 was purified by fractionated polyethylene glycol 6000 (PEG 6000) precipitation. After addition of 2.4% PEG 6000, precipitates were removed by centrifugation at 39,000 × g for 15 min, and the final concentration of PEG 6000 in the F1-containing supernatant was adjusted to 12%. After 15 min of incubation on ice, the F1 was collected by centrifugation (39,000 × g, 15 min) and dissolved in 10 mM Tris-HCl (pH 8.0) containing 1 mM dithiothreitol, 0.1 mM DFP, and 10% glycerol.
Solubilization and purification of the F1Fo-ATP synthase.
To extract the ATP synthase from the cytoplasmic membrane, membrane vesicles obtained from 15 g of cells were washed twice in extraction buffer and then homogenized with 15 ml of extraction buffer containing 10% glycerol and 5% Triton X-100. After 1 h of incubation at 4°C with constant stirring, the nonsolubilized material was removed by ultracentrifugation (180,000 × g for 45 min). The solubilized ATP synthase in the red supernatant was precipitated by ammonium sulfate fractionation. A saturated solution of ammonium sulfate at 5°C in 50 mM Tris-HCl buffer (pH 8.5) containing 5 mM MgSO4 was initially added to achieve a final saturation of 35%, and the suspension was stirred for 30 min at 4°C. The suspension was centrifuged at 17,000 × g for 20 min, and the pellet was discarded since it contained less than 5% of the total ATPase activity. The supernatant was then brought to 65 to 70% saturation and stirred for 30 min at 4°C. This suspension was centrifuged at 17,000 × g for 20 min. The pellet, which contained approximately 90% of the total ATPase activity, was resuspended in 5 ml of 50 mM Tris-HCl (pH 8.2)-0.05% Triton X-100.
Hydroxyapaptite was prepared as described by Bernardi (1) and was stored at 4°C under an equal volume of 1 mM potassium phosphate buffer (pH 8.0). The ATPase-containing solution was diluted fourfold with deionized water and loaded onto a hydroxyapatite column (bed volume, 70 ml; diameter, 3 cm), which had been equilibrated with 4 column volumes of buffer A (30 mM potassium phosphate buffer [pH 8.0], 10% glycerol, 0.05% Triton X-100, 1 mM dithiothreitol). The column was run at a hydrostatic pressure of 1.0 to 1.5 ml/min. After the enzyme was applied, the column was washed with 1 column volume of buffer A and 2 column volumes of buffer B (50 mM potassium phosphate buffer [pH 8.0], 10% glycerol, 0.05% Triton X-100, 1 mM dithiothreitol). The ATPase was eluted with approximately 30 ml of buffer C (150 mM potassium phosphate buffer [pH 8.0], 20% glycerol, 0.25% Triton X-100, 1 mM dithiothreitol), concentrated 15-fold by ultrafiltration with a YM100 membrane (Millipore), and stored under liquid N2.
All preparations of the ATP synthase were routinely analyzed on sodium dodecyl sulfate (SDS)-12% polyacrylamide gels (PAGE) by using the buffer system of Schägger et al. (38). Polypeptide bands were visualized by staining with Coomassie brilliant blue or silver staining (28). N-terminal sequencing of ATP synthase subunits was performed as described previously (35). To determine which bands corresponded to proteins soluble in organic solvents, the purified ATP synthase was extracted with chloroform-methanol (1:1) as previously described (45).
Labeling of subunit c with DCCD.
Subunit c of the F1Fo-ATP synthase from strain TA2.A1 was labeled with N,N′-dicyclohexylcarbodiimide (DCCD) and extracted with chloroform-methanol (1:1) as described previously (45). Purified ATP synthase (60 μg, 200 μl) from strain TA2.A1 in 50 mM potassium phosphate buffer (pH 7.5) was incubated with 20 μM DCCD for 20 min. The reaction was stopped by extraction with a 10 volumes of chloroform-methanol (1:1), and the c subunit was isolated in the organic layer after phase separation induced by the addition of 2 volumes of water. Sample preparation for matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectroscopy (MS) was performed as described by von Ballmoos et al. (45). The molecular masses of the DCCD-labeled and unlabeled subunit c were determined by using a Perspective Biosystems Voyager Elite system, a MALDI-TOF MS instrument with a reflector, as described previously (45). The measurements with the labeled subunit c were made in the linear positive mode to avoid decomposition of DCCD in the reflector mode. The instrument has an accuracy of ±0.1% in the linear mode.
Reconstitution of the purified ATP synthase into proteoliposomes.
A suspension of 30 to 60 mg of phosphatidylcholine (type II S; Sigma) in 2 ml of 10 mM Tricine-KOH (pH 8.0) containing 1 mM MgCl2 and 1 mM dithiothreitol was sonicated twice for 1 min in a water bath sonicator and cooled on ice for 1 h. Purified ATP synthase (0.2 to 0.3 mg of protein to obtain a lipid-to-protein ratio of 50:1 [wt/wt]) was added to the suspension; then 0.5% (final concentration) Triton X-100 was added, and the preparation was left on ice for 20 min. Liposomes were treated with Triton X-100 to favor insertion of the ATP synthase into the liposomes. The suspension was incubated for 45 min at room temperature with gentle stirring. To remove the Triton X-100, three aliquots of Bio-Beads (Bio-Rad) (50, 50, and 80 mg) were added at 0, 60, and 120 min, after which the proteoliposomes had formed. The suspension was carefully separated from the beads, and the proteoliposomes were collected by ultracentrifugation (50 min, 200,000 × g, 4°C). The supernatant was removed, and the pellet was resuspended in 10 mM Tricine-KOH (pH 8.0) containing 2 mM MgCl2 and stored at 4°C. Under these conditions, as little as 10% of the ATPase activity was found in the supernatant, indicating that 90% of the ATP synthase had been reconstituted into the liposomes. The proteoliposomes were used within 24 h. To load proteoliposomes with sodium, 100 mM NaCl was added to the reconstitution buffers under otherwise identical conditions. The concentration of sodium (total content) associated with the proteoliposomes was measured by atomic absorption at 588.9 nm with a Shimadzu AA646 atomic absorption-flame emission spectrophotometer.
ATPase activity measurements.
Purification of the F1Fo-ATP synthase was monitored by determining lauryldimethylamine oxide (LDAO)-stimulated ATPase activity at 45°C by using an ATP-regenerating assay with pyruvate kinase and lactate dehydrogenase. The standard assay mixture (final volume, 1 ml) contained 50 mM Tris-HCl (pH 8.4), 2 mM MgCl2, 3 mM phosphoenolpyruvate, 0.3 mM NADH, 0.4% LDAO, 15 U of lactate dehydrogenase, and 10 U of pyruvate kinase. The 50 mM Tris-HCl buffer was replaced with 50 mM morpholineethansulfonic acid (MES)-morpholinepropanesulfonic acid (MOPS)-Tris-HCl, and the effects of different pH values on ATPase activity were measured. The reaction was initiated by addition of 5 mM (final concentration) ATP and enzyme. The rate of NADH oxidation was monitored continuously at 340 nm. All enzyme activities were corrected for nonspecific NADH oxidation, which was observed in cell membranes. The pH limit of this coupled assay with ADP as a test substrate was approximately 9.2. The effect of Na+ ions on ATPase activity was determined by using the potassium salt of ATP prepared as previously described (24) and the cyclohexylammonium salt of NADH (<0.1% Na+) with the necessary precautions to reduce the endogenous Na+ level as much as possible, as described by Dimroth and Thomer (5). ATP hydrolysis at elevated temperatures (e.g., 65°C) was monitored by using a colorimetric assay that measures the amount of inorganic phosphate (Pi) liberated (20). The assay buffer used contained 100 mM Tris-HCl (pH 8.5), 2 mM MgCl2, and 4 mM ATP. The incubation time and the concentration of membrane protein were adjusted so that the ATPase assay was linear with time and less than 50% of the ATP was hydrolyzed. Nonenzymatic degradation of ATP under these conditions accounted for less than 10% of the total phosphate. One unit of ATPase activity was defined as the amount of enzyme that liberated 1 μmol of Pi or ADP per min.
ATP synthesis by proteoliposomes was determined with the luciferin-luciferase system by measuring the emitted light with a chemiluminometer as described previously (13). The 400-μl assay mixtures contained 10 or 100 mM Tricine-KOH (pH 8.0) as indicated below, 5 mM KH2PO4, 2.5 mM ADP, 2 mM MgCl2, and 200 mM KCl. The reaction was initiated by addition of valinomycin (2 μM) to apply a potassium diffusion potential of 100 mV, and samples (50 μl) were taken every 5 to 10 s. The reaction was stopped by addition of 10% trichloroacetic acid. To generate a Δp of approximately 310 mV (a ΔpH of 190 mV plus a ΔΨ of 120 mV), proteoliposomes were loaded with 100 mM maleinate (pH 5.0) and diluted 40-fold in 100 mM Tricine-KOH (pH 8.0) (13, 14). Fumarate (100 mM) was used to generate a ΔpH in the absence of ΔΨ. A ΔpNa+ of 100 mV was generated by reconstituting the enzyme in the presence of 100 mM NaCl under conditions identical to those described above with 100 mM maleinate.
ATP-dependent proton translocation was determined at 45°C by quenching of 9-amino-6-chloro-2-methoxyacridine (ACMA) in proteoliposomes. The 1.5-ml reaction mixture contained 5 mM potassium phosphate buffer (pH 7.5), 5 mM MgCl2, 1 μM ACMA, and 50 μl of reconstituted proteoliposomes. After the fluorescence signal had stabilized (100%), the reaction was initiated by addition of neutralized ATP (1.7 mM). Fluorescence was measured with an excitation wavelength of 410 nm and an emission wavelength of 480 nm (slit width, 10 nm).
Protein concentrations were determined by using a bicinchoninic acid protein assay kit from Pierce, with bovine serum albumin as the standard.
RESULTS
ATPase activity in inverted membrane vesicles.
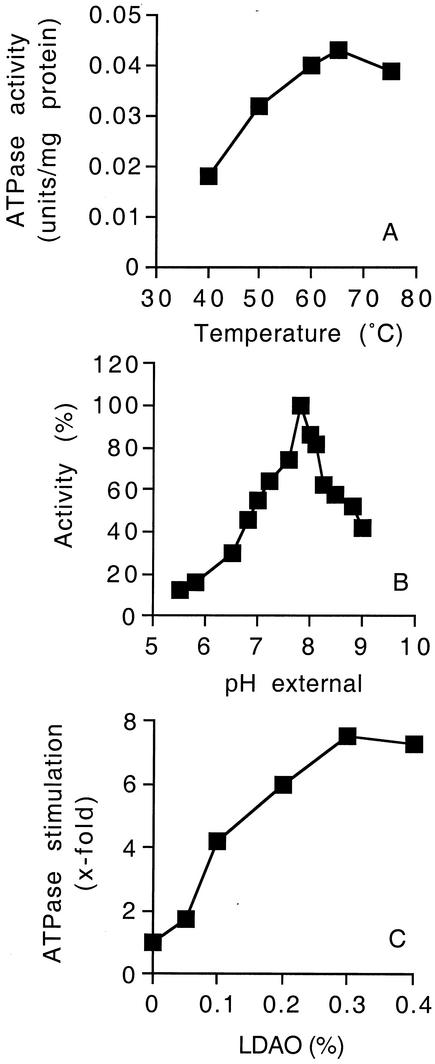
Initial characteristics of the F1Fo-ATP synthase of strain TA2.A1 were investigated by measuring the ATP hydrolysis activity of inverted membrane vesicles. The ATPase activity of inverted membrane vesicles had a temperature optimum of 65°C (Fig. 1A) and a narrow pH optimum around pH 8.0 (Fig. 1B). ATPase activity was strictly dependent on Mg2+ (data not shown). Calcium could be substituted for Mg2+; however, the ATPase activity was only 40% of the magnesium-dependent activity (data not shown). An unusual feature of the F1Fo-ATP synthases from alkaliphilic bacteria is their very low activities in the ATP hydrolysis direction (7, 8). This latent ATP hydrolysis is shared by the ATP synthase from strain TA2.A1, which is 0.045 U of ATPase/mg of protein, compared to the activity of Escherichia coli membranes, which is 2.9 U of ATPase/mg of protein (25). Various additives (e.g., methanol, octyl glucoside) have been reported to activate the ATP hydrolysis activities of alkaliphilic ATPases, and these additives were tested with the ATP synthase from strain TA2.A1. The following compounds were found to have no significant effect (less than 1.5-fold stimulation) on ATPase activity: glycerol, ethanol, and Na2SO3. Methanol (final concentration, 15%), octylglucoside (final concentration, 0.1%), and Triton X-100 (final concentration, 4%) stimulated ATP hydrolysis 1.5- to 2.5-fold (data not shown), whereas the amphipathic detergent LDAO at a final concentration of 0.4% stimulated ATP hydrolysis by the bacterial membranes 8-fold at 45°C (Fig. 1C). The stimulation was >15-fold with the purified enzyme (see below). The lower level of stimulation in membranes probably reflects the presence of additional ATPases which do not require activation by LDAO. LDAO stimulation was temperature dependent, and at 65°C the ATPase was stimulated only 1.8-fold by 0.4% LDAO (data not shown). Based on these results, LDAO (final concentration, 0.4%) was routinely used to monitor ATPase activity, which was measured spectrophotometrically by an ATP-regenerating assay coupled to NADH oxidation at 45°C (the maximum temperature for enzymes used in the coupled assay).
FIG. 1.
ATPase activity in inverted membrane vesicles of Bacillus sp. strain TA2.A1. (A) Effect of temperature on ATPase activity at pH 8.5. ATPase activity was determined by using a colorimetric assay that measured the amount of inorganic phosphate liberated at each temperature. (B) Effect of pH on ATPase activity. The buffer system used was 50 mM MES-MOPS-Tris-HCl containing 0.4% (final concentration) LDAO. (C) Effect of LDAO on ATPase activity at pH 8.5. For panels B and C, ATPase activity was measured at 45°C and pH 8.5 by using the ATP-regenerating assay described in Materials and Methods. The values are the means of duplicate determinations, and the experimental error associated with these values was less than 15%.
Purification and subunit composition of the F1Fo-ATP synthase from strain TA2.A1.
ATPase activity was solubilized from the bacterial membranes with 5% Triton X-100 and was purified by fractionated precipitation with ammonium sulfate followed by chromatography on hydroxyapatite, as described in Materials and Methods and summarized in Table 1. Overall, 2.1 mg of ATP synthase was obtained from 15 g (wet weight) of cells with a specific activity of 23 U/mg of protein, which is about 68-fold higher than the specific activity in bacterial membranes of strain TA2.A1.
TABLE 1.
Purification of the F1Fo-ATP synthase from strain TA2.A1a
| Step | Protein (mg) | Activity (U)b | Sp ac (U/mg)c | Purification (fold) |
|---|---|---|---|---|
| Membrane vesicles | 414 | 141 | 0.34 | 1.0 |
| Triton X-100 (5%) extraction | 172 | 126 | 0.73 | 2.1 |
| Ammonium sulfate | 40 | 102 | 2.55 | 7.5 |
| Hydroxyapatite | 2.1 | 49 | 23 | 68 |
The starting material consisted of 15 g (wet weight) of cells, and the values are averages for four preparations.
One unit equaled 1 μmol of ADP produced per min.
ATPase activity was measured by determining the LDAO (0.4%)-stimulated ATPase activity at 45 °C by the ATP-regenerating assay.
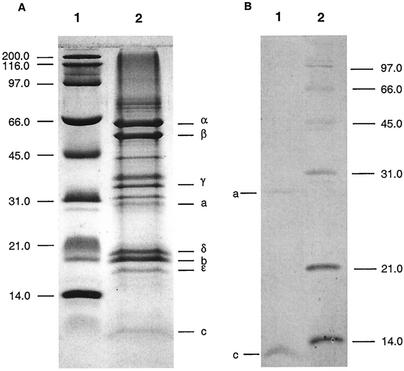
SDS-PAGE analysis of the purified enzyme revealed the typical subunit pattern of an F1Fo-ATP synthase (Fig. 2A), with the following putative subunits: α (55 kDa), β (50 kDa), γ (32 kDa), a (27 kDa), δ (20.3 kDa), b (19.5 kDa), ɛ (14.9 kDa), and c (7 kDa). Minor bands above the band for the a subunit and between the bands for the β and γ subunits were also resolved and represented trace contaminants. Upon treatment of the ATP synthase with chloroform-methanol hydrophobic subunits a and c were extracted into the organic solvent (Fig. 2B). To determine whether the band at approximately 45 kDa was an oligomer of subunit c, the ATP synthase was precipitated with trichloroacetic acid, since this procedure is known to dissociate even the strongest c oligomers (29). However, no change in the subunit pattern was observed after SDS-PAGE, indicating that a c oligomer that resists dissociation by SDS is not present in the ATP synthase from strain TA2.A1 (data not shown). By using N-terminal protein sequencing the identities of the 55-kDa (α), 50-kDa (β), 32-kDa (γ), 20.3-kDa (δ), and 19.5-kDa (b) bands were confirmed by the following sequences: α subunit, SIRPEEISA; β subunit, MNKGRIIQVM; γ subunit, MQGMREI; δ subunit, MAVAKRY; and b subunit, MILEL. No N-terminal sequences were obtained for the 27-kDa (a), 14.9-kDa (ɛ), and 7-kDa (c) subunits. For subunit c, MALDI-TOF MS in the reflector mode was used to confirm the sequence (see below). The c subunit was found to have an N-terminal substituent, thus explaining our inability to obtain N-terminal sequence data.
FIG. 2.
SDS-PAGE of the purified ATP synthase of Bacillus sp. strain TA2.A1. Samples were resolved on 12% polyacrylamide gels by the method of Schägger et al. (38) and were stained with Coomassie brilliant blue. (A) Lane 1, broad-range molecular mass marker (Bio-Rad) (the molecular masses [in kilodaltons] are indicated on the left); lane 2, purified Bacillus sp. strain TA2.A1 F1Fo-ATP synthase (30 μg) (the positions of the subunits are indicated on the right). (B) Solubilization of hydrophobic subunits a and c with chloroform-methanol (1:1).
Kinetic properties of the purified F1Fo-ATP synthase from strain TA2.A1.
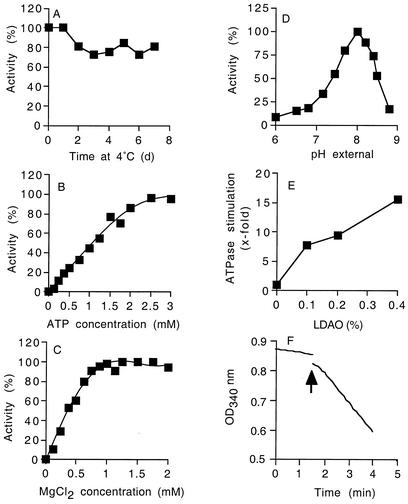
Characteristics of the purified F1Fo-ATP synthase were determined with the coupled spectrophotometric ATP hydrolysis assay in the presence of 0.4% LDAO at 45°C. In 150 mM potassium phosphate buffer (pH 8.0) and at 4°C, the enzyme was remarkably stable; approximately 80% of the activity was retained after 7 days, and 20% of the activity was lost during the first 2 days (Fig. 3A). Michaelis-Menten plots and kinetic analysis with the Lineweaver-Burk equation indicated that the purified ATP synthase had an apparent Km for ATP of 0.67 mM and a Vmax of 25 U/mg of protein when the Mg2+-to-ATP ratio was kept constant at 2:1 (Fig. 3B). The apparent Km for Mg2+ was 0.4 mM (Fig. 3C). The optimum pH for ATP hydrolysis was 8.0, and ATPase activity decreased markedly at pH values below 7.5 or above 8.5 (Fig. 3D). The optimum temperature for the ATPase was 65°C (data not shown). The ATPase was activated more than 2-fold by various detergents (e.g., l-α-diheptanoyl-phosphatidylcholine, dodecylmaltoside, cyclohexyl-hexylmaltoside, octylglucoside, and Triton X-100) and more than 15-fold in the presence of 0.4% LDAO (Fig. 3E). ATPase activity was not stimulated by Na+ or Li+ either in the presence or in the absence of LDAO. Since Na+-dependent F1Fo-ATP synthases are specifically activated by these alkali ions (23, 24, 29), the ATP synthase from strain TA2.A1 is apparently not a member of this group of enzymes. To address the question of whether the ATP hydrolysis activity of the ATP synthase is blocked within the F1 or Fo part of the complex, the F1 moiety was purified separately, and its ATP hydrolysis activity was measured (Fig. 3F). The ATPase activity of the F1 moiety was stimulated approximately eightfold upon addition of 0.4% (final concentration) LDAO (Fig. 3F). We concluded, therefore, that the inhibition of the ATP hydrolysis activity is an intrinsic property of the F1 moiety of the ATP synthase, as has been reported for other LDAO-activated F1Fo-ATPases (25).
FIG. 3.
Characterization of the purified F1Fo-ATP synthase from Bacillus sp. strain TA2.A1. (A) Stability of the ATP synthase after purification in 150 mM potassium phosphate buffer (pH 8.0) at 4°C. (B to D) Effects of ATP concentration (B), MgCl2 concentration (C), and pH (buffer system, 50 mM MES-MOPS-Tris-HCl) (D) on the activity of the purified ATP synthase. ATPase activity was measured at 45°C (pH 8.5) by using the ATP-regenerating assay, and the reaction buffer contained 0.4% LDAO; 100% ATPase activity was in the range from 20 to 25 U/mg of protein. (E) Effect of LDAO (final concentration in assay mixture) on ATPase activity. (F) Activation of the purified F1 ATPase of Bacillus sp. strain TA2.A1 by LDAO. The curve shift indicated by the arrow was due to addition of 0.4% (final concentration) LDAO. OD340 nm, optical density at 340 nm. For all experiments, the values reported are the means of two to four individual experiments, and the experimental error associated with the values was less than 15%.
Incubation of the purified F1Fo-ATP synthase with 50 to 250 μM DCCD in 50 mM potassium phosphate buffer (pH 7.5) led to less than 10% inhibition of the ATP hydrolysis activity after 20 min (data not shown). This result suggested that either the ATP synthase was not modified by DCCD or the enzyme was uncoupled in the assay mixture used for activity measurements.
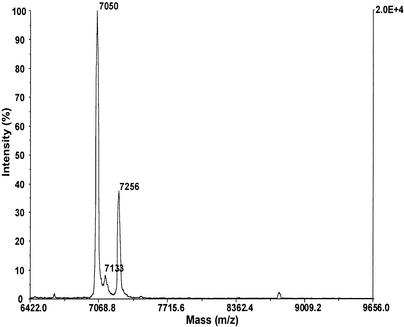
To substantiate that the purified F1Fo-ATP synthase was modified at the c subunits by DCCD, the enzyme was first incubated for 20 min with 20 μM DCCD in 50 mM potassium phosphate buffer (pH 7.5), and the c subunits were subsequently extracted into chloroform-methanol and analyzed by MALDI-TOF MS. The c subunit has a calculated mass of 7,023 Da based on the amino acid sequence of the c subunit (S. Keis, G. Kaim, P. Dimroth, and G. M. Cook, unpublished data), and a corresponding peak at m/z 7,050 Da was observed (Fig. 4). The difference between the observed and theoretical values (27 Da) indicates that there is N-terminal formylation (28 Da), like that found in the E. coli enzyme (11). When the preparation was labeled with DCCD, a second peak at m/z 7,256 Da was observed, representing the DCCD-modified c subunit (calculated mass, 7,257 Da) (Fig. 4). The peak at 7,133 Da was due to the decomposition of DCCD during MALDI, as has been observed previously (45). Hence, the F1Fo-ATP synthase is modified at the c subunits by incubation with DCCD, but no inhibition is observed in the subsequent ATP hydrolysis assay in the presence of 0.4% LDAO. This indicates that the enzyme is uncoupled under these conditions.
FIG. 4.
MALDI-TOF analysis of DCCD labeling of the F1Fo-ATP synthase from strain TA2.A1. DCCD labeling was performed with 20 μM DCCD in 50 mM potassium phosphate buffer (pH 7.5) for 20 min and was stopped by addition of 10 volumes of chloroform-methanol (1:1). Isolation of subunit c and sample preparation for MALDI-TOF MS were performed as described previously (45).
ATP synthesis by the F1Fo-ATP synthase reconstituted into proteoliposomes.
The purified enzyme was reconstituted into proteoliposomes to measure its vectorial ion-translocating activities. Due to the extremely low ATP hydrolysis activity in the absence of detergents, it was unlikely from the onset that we would detect proton pumping into the proteoliposomes, and indeed no ATP-dependent ACMA fluorescence quenching response was detected (data not shown). If the proteoliposomes were treated with Triton X-100, 90% of the ATPase activity was recovered, indicating that the enzyme had been incorporated into the lipid bilayer. The total Na+ concentration in the assay mixture was approximately 10 μM. Based on previous work (29), this should not have inhibited proton pumping if the enzyme belongs to the Na+-dependent F-type family.
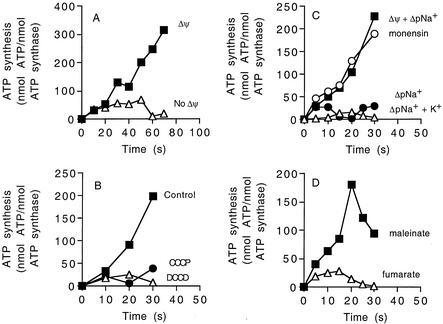
To assess the activity of the ATP synthase in a vectorially coupled functional state, ATP synthesis measurements were obtained (Fig. 5). When proteoliposomes were diluted 40-fold into 10 mM Tricine buffer (pH 8.0) containing the standard ADP, potassium phosphate, Mg2+, and 200 mM KCl, no ATP synthesis was detected (Fig. 5A). However, if 2 μM valinomycin was added to generate a potassium diffusion potential of 100 mV, ATP synthesis proceeded at a rate of 250 nmol of ATP/min (Fig. 5A). ATP synthesis under these conditions was completely eliminated by preincubation of the proteoliposomes with either 20 μM DCCD or 10 μM carbonyl cyanide m-chlorophenylhydrozone (CCCP) (Fig. 5B). Hence, the reconstituted ATP synthase is sensitive to inhibition by DCCD, and ATP synthesis stops if the protonophore CCCP collapses the Δψ.
FIG. 5.
ATP synthesis by the reconstituted ATP synthase in proteoliposomes. (A) Proteoliposomes energized by a valinomycin-induced potassium diffusion potential applied in the absence of ΔpNa+ at 45°C. Valinomycin (2 μM) was added at 5 s to create a diffusion potential of 100 mV (▪). In another experiment no valinomycin was added (▵). (B) Effect of CCCP (10 μM) (•) or DCCD (20 μM) (▵) on ATP synthesis by proteoliposomes energized by a valinomycin-induced potassium diffusion potential applied in the absence of ΔpNa+. Inhibitors were preincubated with proteoliposomes for 5 min prior to addition of valinomycin (5 s). (C) Effect of ΔpNa+ on ATP synthesis by proteoliposomes. To start the reaction, sodium-loaded proteoliposomes (100 mM NaCl) were diluted 40-fold into one of the following buffers and ATP synthesis was determined as described in the text: 10 mM Tricine-KOH containing 2.5 mM NaCl (inside NaCl concentration, 100 mM) to generate a ΔpNa+ of 100 mV (•); 2.5 mM NaCl and approximately 200 mM KCl to generate a ΔpNa+ of 100 mV (▵); 2.5 mM NaCl and approximately 200 mM KCl and 2 μM valinomycin to generate a ΔpNa+ of 100 mV and a K+/valinomycin diffusion potential of 100 mV (▪); 5 μM monensin, 2.5 mM NaCl, 200 mM KCl, and 2 μM valinomycin to generate a ΔpNa+ of 100 mV and a K+/valinomycin diffusion potential of 100 mV (○). (D) ATP synthesis energized by Δp generated by either maleinate (▪) (ΔpH of 190 mV and Δψ of approximately 120 mV) or fumarate (▵) (ΔpH of 190 mV) (13). For all experiments, the values are the means of two to six independent determinations, and the experimental error associated with these values was less than 15%.
The effect of ΔpNa+ on ATP synthesis was also studied (Fig. 5C). To do this, proteoliposomes were loaded with NaCl and diluted into assay buffer without NaCl to generate a ΔpNa+ of 100 mV. No ATP was synthesized under these conditions (Fig. 5C). However, ATP synthesis was observed under identical conditions if a valinomycin-induced potassium diffusion potential of 100 mV was also applied (Fig. 5C). The Na+-H+ antiporter monensin (5 μM) had no effect on the kinetics of ATP synthesis, as expected if ΔpNa+ was not a driving force for ATP synthesis by the ATP synthase from strain TA2.A1 (Fig. 5C). This coincides with previous findings demonstrating that the ΔΨ component is a kinetically indispensable driving force for ATP synthesis by various Na+- or H+-coupled F1Fo-ATP synthases (13, 14, 16). To further corroborate that this property also applies to the ATP synthase from strain TA2.A1, the proteoliposomes were energized for ATP synthesis by the acid bath procedure (12). To do this, the proteoliposomes were loaded with 100 mM dicarboxylic acid buffer (pH 5.0) and diluted subsequently into 100 mM Tricine-KOH buffer (pH 8.5). With both maleinate and fumarate in the dicarboxylate buffer, a ΔpH of about 190 mV was thus created, and an electrical diffusion potential of approximately 120 mV was also formed with maleinate but not with fumarate. Figure 5D shows that ATP synthesis was observed if the acid bath procedure was performed with maleinate but not if it was performed with fumarate, indicating that the ATP synthase from strain TA2.A1 requires a Δψ to drive ATP synthesis, like all other ATP synthases that have been investigated (13, 14, 16).
DISCUSSION
Bacteria are remarkably versatile in terms of adaptation to diverse environmental conditions. In order to understand the strategies adopted by the thermoalkaliphilic organism Bacillus sp. strain TA2.A1 to survive at an alkaline pH and high temperatures, studies aimed at elucidating the bioenergetics of this bacterium are warranted. We have recently shown that the Δp in strain TA2.A1 is suboptimal for ATP synthesis under normal growth conditions (32). Therefore, the question remained whether the ATP synthase from strain TA2.A1 utilizes H+ or Na+ ions as coupling ions, since the bioenergetic problem of a low Δp at a high environmental pH could be elegantly solved if Na+ serves as the coupling ion of the ATP synthase.
To study this, we purified and characterized the ATP synthase from strain TA2.A1. This enzyme had the usual subunit composition of a bacterial F1Fo-ATP synthase. Here, we provide compelling evidence that the F1Fo-ATP synthase from strain TA2.A1 is H+ coupled and not Na+ coupled. In contrast to Na+-coupled F1Fo-ATP synthases, neither the hydrolysis nor the synthesis of ATP by the ATP synthase from strain TA2.A1 was specifically affected by Na+ ions. ATP synthesis by the reconstituted ATP synthase was obligatorily coupled to Δψ, and ΔpNa+ had no effect. Moreover, ATP synthesis was not sensitive to the Na+-conducting ionophore monensin, but it was severely inhibited by the protonophore CCCP. While Na+-coupled ATP synthases from Propionigenium modestum or Ilyobacter tartaricus form a c11 oligomer with unusual stability due to the cross-bridging of individual subunits by Na+ ions (26), only the monomeric c subunit was found when we performed an SDS-PAGE analysis of the ATP synthase from strain TA2.A1. These data were corroborated by recent studies involving cloning and DNA sequencing of the atp operon of strain TA2.A1 (Keis et al., unpublished data). We found strong DNA sequence similarity to atp operons from alkaliphilic Bacillus species, thermophilic Bacillus species, and mesophilic Bacillus species, all of which code for proton-coupled F1Fo-ATP synthases. The lack of the sodium ion binding signature consisting of residues Q32, E65, and S66 (P. modestum numbering) (15, 18) in the c subunit of the strain TA2.A1 ATP synthase supports the notion that this is not an Na+-coupled enzyme. In this respect, our data complement the data for the well-characterized ATP synthases from the mesophilic alkaliphilic bacteria B. pseudofirmus OF4 and B. alcalophilus, which were found to be exclusively proton-coupled enzymes (7-9).
Another characteristic of the F1Fo-ATP synthases from mesophilic alkaliphilic bacilli is the selective blockage of ATP hydrolysis but not ATP synthesis (7, 8). This interesting feature of the enzyme is even more pronounced in the ATP synthase from strain TA2.A1. While we could readily observe Δp-driven ATP synthesis by reconstituted proteoliposomes, we were unable to detect any ATP-driven proton transport or proton-coupled ATP hydrolysis activity. In contrast, low but measurable ATP hydrolysis activities coupled to proton transport were reported for the ATP synthases from B. alcalophilus (9) and B. pseudofirmus OF4 (7). So far, there have been no studies aimed towards elucidating the molecular features responsible for the specific blockage of ATP hydrolysis by the ATP synthases from alkaliphilic bacteria, and no rationale from a physiological point of view has been provided. For a tenable explanation, the molecular features of the F1Fo-ATP synthases from alkaliphilic bacteria must be considered. These enzymes are known to consist of two motors, the soluble F1 motor, fueled by ATP hydrolysis, and the transmembrane Fo motor, fueled by the Δp. A rotating shaft consisting of the ɛ and γ subunits mechanically links these two motors. The direction in which the enzyme operates depends on whether the F1 motor or the Fo motor generates the larger torque. Under normal circumstances, the Fo motor generates the larger torque and drives the F1 motor in the ATP synthesis direction. However, when the Δp drops below the phosphorylation potential, the F1 motor hydrolyzes ATP, driving the Fo motor in reverse, whereupon it functions as a proton pump. Such a situation occurs frequently in anaerobic bacteria, where the main challenge is to keep the membrane potential at a significant level. For this purpose, the ATP synthase pumps protons outwards. Alkaliphilic bacteria are also confronted with situations in which the Δp is low (10, 21, 22, 32). This, however, is not caused by a low Δψ but by the inverted ΔpH at a high environmental pH. The main bioenergetic challenge for these alkaliphilic bacteria is to maintain the cytoplasmic pH near neutral when they are growing at an external pH of 10.5, and therefore ATPase-dependent proton pumping, if the Δp drops below a critical level, may be detrimental for this process. Thus, blocking of the ATP synthase of alkaliphilic bacteria in the ATP hydrolysis direction appears to be a necessary adaptation for growth and survival in highly alkaline environments in which the Δp generated by these bacteria is low.
From a mechanistic point of view the most intriguing question concerns the molecular details of how the F1Fo-ATP synthase from strain TA2.A1 manages to block the thermodynamically favorable ATP hydrolysis direction. In this respect it is important to understand how the enzyme disposes of potent cryptic ATPase activity that is unmasked in the presence of 0.4% LDAO (>15-fold stimulation). The classical ATPase inhibitor DCCD did not affect LDAO-activated ATPase activity, although this compound modified the c subunit DCCD binding sites, suggesting the presence of an uncoupled enzyme. Further analysis demonstrated that the preferential blockage of ATP hydrolysis activity and the uncoupling by LDAO were intrinsic to the F1 moiety.
The accumulated evidence indicates that in isolated F1 subunits from chloroplasts or bacteria, the ɛ subunit acts as an inhibitor of ATP hydrolysis activity and that this inhibition is elicited by the C-terminal helix-turn-helix motif of this subunit (19, 27, 37). The proposed regulatory role of the ɛ subunit in the entire F1Fo complex has been strongly supported by recent cross-linking studies that were based on two different conformations of the ɛ subunit, as determined by structural analyses (44). In the conformation with the C-terminal domain of the ɛ subunit facing toward F1, ATP hydrolysis is strongly inhibited, but ATP synthesis is not affected. In the other conformation of the ɛ subunit, the enzyme operates with the same efficiency in either direction (i.e., ATP synthesis or hydrolysis). Based on these data, it is tempting to propose that the ɛ subunit of the ATP synthase from strain TA2.A1 is permanently fixed in a conformation in which the rotational movement in the ATP hydrolysis direction is impaired.
Acknowledgments
This work and S.K. were supported by a Marsden grant from the Royal Society of New Zealand. G.M.C. acknowledges the financial support of the Eidgenössische Technische Hochschule during his sabbatical in Zürich.
The technical support of Fabienne Henzen and helpful discussions with Christopher Kastner and Thomas Meier are gratefully acknowledged.
REFERENCES
- 1.Bernardi, G. 1971. Chromatography of proteins on hydroxyapatite. Methods Enzymol. 22:325-339. [DOI] [PubMed] [Google Scholar]
- 2.Boyer, P. D. 1997. The ATP synthase—a splendid molecular machine. Annu. Rev. Biochem. 66:717-749. [DOI] [PubMed] [Google Scholar]
- 3.Capaldi, R. A., and R. Aggeler. 2002. Mechanism of the F1FO-type ATP synthase, a biological rotary motor. Trends Biochem. Sci. 27:154-160. [DOI] [PubMed] [Google Scholar]
- 4.Dimroth, P. 1997. Primary sodium ion translocating enzymes. Biochim. Biophys. Acta 1318:11-51. [DOI] [PubMed] [Google Scholar]
- 5.Dimroth, P., and A. Thomer. 1986. Kinetic analysis of the reaction mechanism of oxaloacetate decarboxylase from Klebsiella aerogenes. Eur. J. Biochem. 156:157-162. [DOI] [PubMed] [Google Scholar]
- 6.Hermolin, J., O. Y. Dmitriev, Y. Zhang, and R. H. Fillingame. 1999. Defining the domain of binding of F1 subunit epsilon with the polar loop of FO subunit c in the Escherichia coli ATP synthase. J. Biol. Chem. 274:17011-17016. [DOI] [PubMed] [Google Scholar]
- 7.Hicks, D. B., and T. A. Krulwich. 1990. Purification and reconstitution of the F1FO-ATP synthase from alkaliphilic Bacillus firmus OF4. Evidence that the enzyme translocates H+ but not Na+. J. Biol. Chem. 265:20547-20554. [PubMed] [Google Scholar]
- 8.Hoffmann, A., and P. Dimroth. 1990. The ATPase of Bacillus alcalophilus. Purification and properties of the enzyme. Eur. J. Biochem. 194:423-430. [DOI] [PubMed] [Google Scholar]
- 9.Hoffmann, A., and P. Dimroth. 1991. The ATPase of Bacillus alcalophilus. Reconstitution of energy-transducing functions. Eur. J. Biochem. 196:493-497. [DOI] [PubMed] [Google Scholar]
- 10.Hoffmann, A., and P. Dimroth. 1991. The electrochemical proton potential of Bacillus alcalophilus. Eur. J. Biochem. 201:467-473. [DOI] [PubMed] [Google Scholar]
- 11.Hoppe, J., and W. Sebald. 1984. The proton conducting FO-part of bacterial ATP synthases. Biochim. Biophys. Acta 768:1-27. [DOI] [PubMed] [Google Scholar]
- 12.Jagendorf, A. T., and E. Uribe. 1966. ATP formation caused by acid-base transition of spinach chloroplasts. Proc. Natl. Acad. Sci. USA 55:170-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaim, G., and P. Dimroth. 1999. ATP synthesis by F-type ATP synthase is obligatorily dependent on the transmembrane voltage. EMBO J. 18:4118-4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaim, G., and P. Dimroth. 1998. ATP synthesis by the F1FO ATP synthase of Escherichia coli is obligatorily dependent on the electric potential. FEBS Lett. 434:57-60. [DOI] [PubMed] [Google Scholar]
- 15.Kaim, G., and P. Dimroth. 1998. A triple mutation in the a subunit of the Escherichia coli/Propionigenium modestum F1FO ATPase hybrid causes a switch from Na+ stimulation to Na+ inhibition. Biochemistry 37:4626-4634. [DOI] [PubMed] [Google Scholar]
- 16.Kaim, G., and P. Dimroth. 1998. Voltage-generated torque drives the motor of the ATP synthase. EMBO J. 17:5887-5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaim, G., M. Prummer, B. Sick, G. Zumofen, A. Renn, U. P. Wild, and P. Dimroth. 2002. Coupled rotation within single FOF1 enzyme complexes during ATP synthesis or hydrolysis. FEBS Lett. 525:156-163. [DOI] [PubMed] [Google Scholar]
- 18.Kaim, G., F. Wehrle, U. Gerike, and P. Dimroth. 1997. Molecular basis for the coupling ion selectivity of F1FO ATP synthases: probing the liganding groups for Na+ and Li+ in the c subunit of the ATP synthase from Propionigenium modestum. Biochemistry 36:9185-9194. [DOI] [PubMed] [Google Scholar]
- 19.Kato, Y., T. Matsui, N. Tanaka, E. Muneyuki, T. Hisabori, and M. Yoshida. 1997. Thermophilic F1-ATPase is activated without dissociation of an endogenous inhibitor, epsilon subunit. J. Biol. Chem. 272:24906-24912. [DOI] [PubMed] [Google Scholar]
- 20.Kobayashi, H., and Y. Anraku. 1972. Membrane-bound adenosine triphosphatase of Escherichia coli. J. Biochem. 71:387-399. [PubMed] [Google Scholar]
- 21.Krulwich, T. A. 1995. Alkaliphiles: ′basic' molecular problems of pH tolerance and bioenergetics. Mol. Microbiol. 15:403-410. [DOI] [PubMed] [Google Scholar]
- 22.Krulwich, T. A., M. Ito, R. Gilmour, D. B. Hicks, and A. A. Guffanti. 1998. Energetics of alkaliphilic Bacillus species: physiology and molecules. Adv. Microb. Physiol. 40:401-438. [DOI] [PubMed] [Google Scholar]
- 23.Laubinger, W., and P. Dimroth. 1988. Characterization of the ATP synthase of Propionigenium modestum as a primary sodium pump. Biochemistry 27:7531-7537. [DOI] [PubMed] [Google Scholar]
- 24.Laubinger, W., and P. Dimroth. 1987. Characterization of the Na+-stimulated ATPase of Propionigenium modestum as an enzyme of the F1FO type. Eur. J. Biochem. 168:475-480. [DOI] [PubMed] [Google Scholar]
- 25.Lotscher, H.-R., C. deJong, and R. A. Capaldi. 1994. Interconversion of high and low adenosinetriphosphatase activity forms of Escherichia coli F1 by the detergent lauryldimethylamine oxide. Biochemistry 23:4140-4143. [DOI] [PubMed] [Google Scholar]
- 26.Meier, T., and P. Dimroth. 2002. Intersubunit bridging by Na+ ions as a rationale for the unusual stability of the c-rings of Na+-translocating F1Fo ATP synthases. EMBO Rep. 3:1094-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mendel-Hartvig, J., and R. A. Capaldi. 1991. Nucleotide-dependent and dicyclohexylcarbodiimide-sensitive conformational changes in the epsilon subunit of Escherichia coli ATP synthase. Biochemistry 30:10987-10991. [DOI] [PubMed] [Google Scholar]
- 28.Merril, C. R., M. L. Dunau, and D. Goldman. 1981. A rapid sensitive silver stain for polypeptides in polyacrylamide gels. Anal. Biochem. 110:201-207. [DOI] [PubMed] [Google Scholar]
- 29.Neumann, S., U. Matthey, G. Kaim, and P. Dimroth. 1998. Purification and properties of the F1FO ATPase of Ilyobacter tartaricus, a sodium ion pump. J. Bacteriol. 180:3312-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishio, K., A. Iwamoto-Kihara, A. Yamamoto, Y. Wada, and M. Futai. 2002. Subunit rotation of ATP synthase embedded in membranes: α- or β-subunit rotation relative to the c subunit ring. Proc. Natl. Acad. Sci. USA 99:13448-13452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noji, H., R. Yasuda, M. Yoshida, and K. Kinosita, Jr. 1997. Direct observation of the rotation of F1-ATPase. Nature 386:299-302. [DOI] [PubMed] [Google Scholar]
- 32.Olsson, K., S. Keis, H. W. Morgan, P. Dimroth, and G. M. Cook. 2003. Bioenergetic properties of the thermoalkaliphilic Bacillus sp. strain TA2.A1. J. Bacteriol. 185:461-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peddie, C. J., G. M. Cook, and H. W. Morgan. 1999. Sodium-dependent glutamate uptake by an alkaliphilic, thermophilic Bacillus strain, TA2.A1. J. Bacteriol. 181:3172-3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peddie, C. J., G. M. Cook, and H. W. Morgan. 2000. Sucrose transport by the alkaliphilic, thermophilic Bacillus sp. strain TA2.A1 is dependent on a sodium gradient. Extremophiles 4:291-296. [DOI] [PubMed] [Google Scholar]
- 35.Ploug, M., A. L. Jensen, and V. Barkholt. 1989. Determination of amino acid compositions and NH2-terminal sequences of peptides electroblotted onto PVDF membranes from tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis: application to peptide mapping of human complement component C3. Anal. Biochem. 181:33-39. [DOI] [PubMed] [Google Scholar]
- 36.Reidlinger, J., and V. Müller. 1994. Purification of ATP synthase from Acetobacterium woodii and identification as a Na+-translocating F1FO-type enzyme. Eur. J. Biochem. 223:275-283. [DOI] [PubMed] [Google Scholar]
- 37.Richter, M. L., B. Snyder, R. E. McCarty, and G. G. Hammes. 1985. Binding stoichiometry and structural mapping of the epsilon polypeptide of chloroplast coupling factor 1. Biochemistry 24:5755-5763. [DOI] [PubMed] [Google Scholar]
- 38.Schägger, H., H. Aquila, and G. von Jagow. 1988. Coomassie blue-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for direct visualization of polypeptides during electrophoresis. Anal. Biochem. 173:201-205. [DOI] [PubMed] [Google Scholar]
- 39.Schulenberg, B., R. Aggeler, J. Murray, and R. A. Capaldi. 1999. The γɛ-c subunit interface in the ATP synthase of Escherichia coli. Cross-linking of the ɛ subunit to the c subunit ring does not impair enzyme function, that of γ to c subunits leads to uncoupling. J. Biol. Chem. 274:34233-34237. [DOI] [PubMed] [Google Scholar]
- 40.Seelert, H., A. Poetsch, N. A. Dencher, A. Engel, H. Stahlberg, and D. J. Müller. 2000. Structural biology. Proton-powered turbine of a plant motor. Nature 405:418-419. [DOI] [PubMed] [Google Scholar]
- 41.Stahlberg, H., D. J. Müller, K. Suda, D. Fotiadis, A. Engel, T. Meier, U. Matthey, and P. Dimroth. 2001. Bacterial Na+-ATP synthase has an undecameric rotor. EMBO Rep. 2:229-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stock, D., A. G. Leslie, and J. E. Walker. 1999. Molecular architecture of the rotary motor in ATP synthase. Science 286:1700-1705. [DOI] [PubMed] [Google Scholar]
- 43.Tsunoda, S. P., R. Aggeler, M. Yoshida, and R. A. Capaldi. 2001. Rotation of the c subunit oligomer in fully functional F1FO ATP synthase. Proc. Natl. Acad. Sci. USA 98:898-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsunoda, S. P., A. J. W. Rodgers, R. Aggeler, M. C. J. Wilce, M. Yoshida, and R. A. Capaldi. 2001. Large conformational changes of the ɛ subunit in the bacterial F1FO ATP synthase provide a ratchet action to regulate this rotary motor enzyme. Proc. Natl. Acad. Sci. USA 98:6560-6564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.von Ballmoos, C., Y. Appoldt, J. Brunner, T. Granier, A. Vasella, and P. Dimroth. 2002. Membrane topography of the coupling ion binding site in Na+-translocating F1FO ATP synthase. J. Biol. Chem. 277:3504-3510. [DOI] [PubMed] [Google Scholar]
- 46.Vonck, J., T. K. von Nidda, T. Meier, U. Matthey, D. J. Mills, W. Kühlbrandt, and P. Dimroth. 2002. Molecular architecture of the undecameric rotor of a bacterial Na+-ATP synthase. J. Mol. Biol. 321:307-316. [DOI] [PubMed] [Google Scholar]
- 47.Yoshida, M., E. Muneyuki, and T. Hisabori. 2001. ATP synthase—a marvelous rotary engine of the cell. Nat. Rev. Mol. Cell. Biol. 2:669-677. [DOI] [PubMed] [Google Scholar]