Abstract
Lactococcus lactis NZ9010 in which the las operon-encoded ldh gene was replaced with an erythromycin resistance gene cassette displayed a stable phenotype when grown under aerobic conditions, and its main end products of fermentation under these conditions were acetate and acetoin. However, under anaerobic conditions, the growth of these cells was strongly retarded while the main end products of fermentation were acetate and ethanol. Upon prolonged subculturing of this strain under anaerobic conditions, both the growth rate and the ability to produce lactate were recovered after a variable number of generations. This recovery was shown to be due to the transcriptional activation of a silent ldhB gene coding for an Ldh protein (LdhB) with kinetic parameters different from those of the native las operon-encoded Ldh protein. Nevertheless, cells producing LdhB produced mainly lactate as the end product of fermentation. The mechanism underlying the ldhB gene activation was primarily studied in a single-colony isolate of the recovered culture, designated L. lactis NZ9015. Integration of IS981 in the upstream region of ldhB was responsible for transcription activation of the ldhB gene by generating an IS981-derived −35 promoter region at the correct spacing with a natively present −10 region. Subsequently, analysis of 10 independently isolated lactate-producing derivatives of L. lactis NZ9010 confirmed that the ldhB gene is transcribed in all of them. Moreover, characterization of the upstream region of the ldhB gene in these derivatives indicated that site-specific and directional IS981 insertion represents the predominant mechanism of the observed recovery of the ability to produce lactate.
Homolactic fermentation by lactic acid bacteria involves the classical Embden-Meyerhoff-Parnas pathway leading to pyruvate, which is converted to lactic acid by lactate dehydrogenase. This enzyme and the gene that encodes it have been studied in many lactic acid bacteria, including Lactococcus lactis (11, 34), Streptococcus thermophilus (19), and various lactobacilli (2, 15, 47, 51). L. lactis is the best-studied representative of this group, and the complete and partial genomes of several strains have been determined (4, 29). The gene encoding L. lactis Ldh was identified and characterized by Llanos and coworkers in 1992 (33, 34). The ldh gene is the last gene of the so-called lactic acid synthesis or las operon, which also encodes the glycolytic enzymes phosphofructokinase and pyruvate kinase. Transcription of the las operon was shown to yield a polycistronic transcript encompassing all three genes. But under some conditions, transcripts representing only two genes (pfk and pyk or pyk and ldh) or even a single gene (ldh) of the operon were also detected, which probably resulted from RNA processing upstream of the pyk and ldh genes (38). It has been shown that the las operon is subject to CcpA-mediated carbon catabolite transcriptional activation, and a CcpA target site (cre sequence) was found within the las promoter region (38). The las operon-encoded lactococcal Ldh protein converts pyruvate to lactate with high efficiency. Moreover, through the concomitant conversion of NADH to NAD+, this reaction provides the electron sink required to maintain redox balance, which has been shown to be a critical determinant in the control of pyruvate flux in L. lactis (9, 18, 35, 41).
Construction of defined ldh disruption mutants of L. lactis has allowed redistribution of the lactococcal pyruvate pool toward products other than lactate (20, 23, 26, 35, 43, 50). Under aerobic conditions, the ldh-deficient strains displayed an almost complete loss of lactate production and acetoin was found to be the main end product of fermentation, while the amounts of other metabolic end products like acetate, butanediol, ethanol, and formate appeared to depend on the fermentation conditions applied (23, 43). The presence of molecular oxygen allows the cells to maintain their redox balance through the activity of the endogenous NADH oxidase, thereby sustaining rapid sugar fermentation by the ldh-deficient cells under aerobic conditions (23, 35). However, under anaerobic conditions, the rate of sugar fermentation is reduced in these cells and the main metabolic end products observed were formate, ethanol, and butanediol (43). The conversion of pyruvate to ethanol and butanediol suggests that these pathways are used as an alternative electron sink in these cells, since the enzymatic conversions involved include reducing steps that use NADH as a cofactor (23). Production of mannitol and use of acetate imply that ldh-deficient L. lactis strains suffer from redox stress under anaerobic conditions and support an important role for the redox balance in the control of lactococcal pyruvate metabolism (25, 40).
Here we describe the observation that an ldh::ery mutant of L. lactis recovers the ability to produce lactate upon prolonged anaerobic subculturing. Concomitant with the recovery of lactate production, the growth rate of this mutant under anaerobic conditions is restored to wild-type levels. The recovered ability to produce lactate is shown to depend on an Ldh protein with enzymatic characteristics that are clearly distinct from those determined for the las operon-encoded enzyme. Genetic analysis of a single-colony isolate of a derivative that recovered the ability to produce lactate revealed that transcription of an otherwise silent, alternative Ldh-encoding gene, ldhB, is activated. Moreover, the activation of transcription of ldhB is shown to be the result of site-specific, oriented IS981 insertion in the upstream region of this gene generating an IS981-derived −35 promoter sequence at the correct spacing relative to a natively present −10 promoter region.
MATERIALS AND METHODS
Strains and growth conditions.
L. lactis NZ9010 (ldh::ery) (23) and its parental strains NZ9000 (pepN::nisRK derivative of L. lactis MG1363) (31), MG1363 (21), and IL1403 (8) have been described previously. L. lactis NZ9015 is a lactate-producing single-colony isolate obtained after anaerobic culturing of strain NZ9010 for more than 100 generations, after which complete recovery of the ability to produce lactate was achieved. All lactococcal strains were grown at 30°C in M17 broth (Oxoid, Basingstoke, England) supplemented with 0.5% (wt/vol) glucose. Aerobic growth conditions involved shaking (200 rpm) of small-volume cultures in large-volume culture flasks (ratio of >5). Anaerobic (or rather microaerophilic) growth conditions were accomplished by growing cultures statically. Escherichia coli MC1061 (7) was used as a cloning host and grown aerobically at 37°C in tryptone-yeast broth (46). When appropriate, media were supplemented with erythromycin (5 μg/ml), tetracycline (2 μg/ml), and ampicillin (50 μg/ml).
Metabolite profile analysis.
Lactococcal strains were grown overnight under aerobic or anaerobic conditions in liquid culture. After removal of bacterial cells by centrifugation (10 min, 20,000 × g), concentrations of lactate, formate, acetate, acetoin, 2,3-butanediol, ethanol, and pyruvate were determined in the culture supernatants by high-performance liquid chromatography analysis as described previously (49). d-Glucose concentrations were determined by using a photometric enzymatic assay in accordance with the manufacturer's (R-Biopharm, Darmstadt, Germany) protocol. Metabolite production was calculated relative to the amount of glucose consumed.
DNA manipulations and plasmids.
Plasmid DNA was isolated from E. coli as previously described (3) and then subjected to anion-exchange chromatography on JetStar columns (Genomed, Oberhausen, Germany). Recombinant DNA techniques were performed essentially as previously described (46). Restriction endonucleases, Klenow fragment of E. coli DNA polymerase, Taq DNA polymerase, and T4 DNA ligase were used in accordance with the manufacturers' (Amersham Pharmacia Biotech, Roosendaal, The Netherlands, and Gibco BRL Life Technologies, Breda, The Netherlands) protocols. PCR amplifications were performed with 10 pmol of each primer (Genset Oligos, Paris, France) and 10 to 100 ng of template DNA with amplification cycles designed in accordance with the Taq (Amersham Pharmacia Biotech) or Pwo (Roche Diagnostics, Mannheim, Germany) DNA polymerase manufacturer's protocol with a DNA thermocycler (Perkin-Elmer, Shelton, Conn.). Plasmid DNA was introduced into L. lactis by electroporation as previously described (13).
Cloning procedure and sequence analysis.
The L. lactis MG1363 ldh gene and homologues of ldhB and hicD were amplified by PCR with primers lasldhF and lasldhR (ldh), ldhBF and ldhBR (ldhB), or hicDF and hicDR (hicD) (Table 1) and L. lactis MG1363 chromosomal DNA as the template. Use of L. lactis IL1403 chromosomal DNA as the template with the same primer combinations yielded the L. lactis IL1403 ldh, ldhB, and hicD genes, and primers ldhXF and ldhXR (Table 1) were used to amplify the L. lactis IL1403 ldhX gene. The PCR amplification products obtained were cloned in pGEM-T (Promega Biotech, Roosendaal, The Netherlands). The DNA sequences of the cloned fragments were analyzed.
TABLE 1.
Primers used in this study
| Primer | Sequencea | GenBank accession no. or source (location or origin) |
|---|---|---|
| lasldhF | 5′-ATGGCTGATAAACAACGTAAG-3′ | L07920 (2906-2926) |
| lasldhR | 5′-GCAGAAGCAAATTCTTCTTTAGC-3′ | L07920 (3848-3870 complement) |
| ldhBF | 5′-ATGAAAATTACAAGCAGAAAAGTAG-3′ | AE006274 (7566-7589 complement) |
| ldhBR | 5′-TTACTTAATAGATTCTATTACCTC-3′ | AE006274 (6645-6668) |
| ldhXF | 5′-ATGAAAATTAATAACAAAAAAGTTG-3′ | AE006345 (5225-5249) |
| ldhXR | 5′-TTACAAAGTACATTTTTCTTTAATTG-3′ | AE006345 (6171-6196 complement) |
| hicDF | 5′-ATGCGTAAAGTAGGTCTAATTGGTTG-3′ | AE006284 (6723-6748) |
| hicDR | 5′-CTAAAAAAGATTGTCCCCAACTC-3′ | AE006284 (7285-7307 complement) |
| ldhBF41 | 5′-GGCGGAATTCTTTGATAGATGTCAATCAAGATAAAGC-3′ | L. lactisMG1363ldhB (this study) |
| ldhBR42 | 5′-GGCGGAATTCAATATATCCGTGAACACTCCGAGG-3′ | L. lactisMG1363ldhB (this study) |
| ldhBR1 | 5′-CTTGTTCCGACAAATCCTGTTCC-3′ | L. lactisMG1363ldhB (this study) |
| rlrDF3 | 5′-AACATTGTTCGTATGAATAGCAA(TC)(GC)-3′ | AE006274 (8080-8104 complement) |
| rlrDF46 | 5′-TAATAAAATAAAAAAAGAAACCG-3′ | L. lactisMG1363ldhB (this study) |
| rlrDF45 | 5′-Cy5-TCCTTATCAAGAACTTGG-3′ | L. lactisMG1363ldhB (this study) |
| ldhBR43 | 5′-Cy5-TCCGACAAATCCTGTTCC-3′ | L. lactisMG1363ldhB (this study) |
Restriction sites are underlined.
The rlrD-ldhB intergenic region was amplified by PCR with primers rlrDF3 and ldhBR1 (Table 1) and chromosomal DNAs of L. lactis stains NZ9000, NZ9010, and NZ9015 as templates. The PCR products obtained were cloned into pGEM-T (Promega Biotech) and subjected to sequence analysis.
For construction of the ldhB knockout, plasmid pUCAEryBTc (23) was digested with EcoRI and XbaI, thereby removing the AeryB region. After the cohesive ends of the 4.3-kb vector fragment were filled in with Klenow, this fragment was circularized by ligation, yielding pUCTet. An internal fragment of ldhB was amplified by PCR with L. lactis MG1363 chromosomal DNA as the template and ldhBF41 and ldhBR42 as primers (Table 1). After digestion with EcoRI, the PCR product was cloned in similarly digested pUCTet, generating plasmid pNZ2020. The orientation and identity of the insert were verified by restriction analysis.
Nucleotide sequencing reactions on both strands of cloned DNA fragments were accomplished with the AutoRead Sequencing kit and initiated by using fluorescein-labeled universal and reverse pUC primers (Genset Oligos) in accordance with the manufacturer's protocol (Amersham Pharmacia Biotech). Sequencing reactions on PCR-amplified rlrD-ldhB intergenic regions from various strains and cultures were performed with the Thermo Sequenase fluorescently labeled primer cycle sequencing kit (Amersham Pharmacia Biotech) initiated with fluorescein-labeled synthetic primers ldhBR43 and rlrDF45 (Table 1). DNA sequence analyses were performed with an automated ALF DNA sequencer in accordance with the manufacturer's (Amersham Pharmacia Biotech) protocol. Sequence data were assembled with Clone Manager 5.0 (Scientific & Educational Software, Durham, England) and analyzed with the CLUSTALW and BLAST programs available at the Centre for Molecular and Biomolecular Informatics (Nijmegen, The Netherlands).
Southern and Northern analyses.
Chromosomal DNA was isolated from overnight cultures of L. lactis IL1403, MG1363, and its derivatives used in this study as described previously (53). Southern analysis was performed by size fractionation of fully digested DNA on a 1% agarose gel with BstEII-digested λ DNA as a molecular size marker. Total RNA was isolated from exponentially growing L. lactis cultures by the Macaloid method as described previously (30). For Northern analysis, 10 μg of total RNA was denatured and size fractionated on a formaldehyde-containing 1% agarose gel as described previously (52). A 0.24- to 9.5-kb RNA molecular size marker was used (Invitrogen, Breda, The Netherlands). Gels were blotted onto GeneScreen plus membranes as recommended by the manufacturer (New England Nuclear Life Science Products, Boston, Mass.). Gene-specific DNA fragments of L. lactis MG1363 ldh, ldhB, and hicD and L. lactis IL1403 ldhX were digested from the pGEM-T vectors harboring these genes (see the description of cloning procedures and sequence analysis above). After radiolabeling with [α-32P]dATP (Amersham Biosciences, Little Chalfont, England) by nick translation, they were used as probes for hybridization (46). Blots were washed with 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) at 65°C prior to autoradiography.
Primer extension.
Primer extensions on total RNAs of L. lactis strains NZ9000, NZ9010, and NZ9015 were performed as described previously (30) with the modification that detection of the extension product was based on fluorescence instead of radioactivity. Therefore, 10 ng of fluorescein-labeled oligonucleotide ldhBR43 (Genset Oligos; Table 1) was annealed to 15 μg of total RNA and the formamide stop solution (AutoRead Sequencing kit; Amersham Pharmacia Biotech) was used as a loading buffer. The corresponding DNA sequence analysis was performed with the same fluorescein-labeled primer ldhBR43 (Table 1) and the pGEM-T vectors harboring the rlrD-ldhB intergenic regions (see the description of the cloning procedures and sequence analysis above). Two microliters of primer extension product was analyzed on a ReproGel Long-Read Sequence gel (Amersham Pharmacia Biotech) next to the rlrD-ldhB intergenic sequence. Postrun comparison of peak intensities (AlfWin Evaluation; Amersham Biosciences, Uppsala, Sweden) revealed a specific ldhB transcription start product in L. lactis NZ9015, compared to strain NZ9010, which was mapped on the rlrD-ldhB intergenic region sequence of the same strain.
Characterization of enzyme kinetics.
Cells were grown under pH-controlled conditions at a pH of 6.5 and harvested in early stationary phase. One hundred milliliters of culture was centrifuged (15 min, 10,000 × g, 4°C), and cell pellets were washed in 0.1 M triethanolamine buffer pH = 6.5 (22). After resuspension in 2 ml of the same buffer, 2 g of 0.1-mm zirconia-silica beads (Biospec Products, Bartlesville, Okla.) was added and cells were disrupted by bead beating two times for 20 s each time at 4 m/s2 in a FastPrep 120 (Savant Instruments, New York, N.Y.) and kept chilled on ice in between. After removal of cell debris by centrifugation (5 min, 20,000 × g, 4°C), supernatants were used. Protein concentrations were determined as described by Bradford, with bovine serum albumin as the standard (6). Ldh activity was determined essentially as described by Hillier and Jago (22). Ldh activity was measured as the NADH oxidation rate after subtraction of the NADH oxidase (36) activity, i.e., the rate before pyruvate addition. NADH oxidation was monitored at 340 nm. However, when NADH affinity was studied, the concentrations used were higher and outside the range where absorbance and concentration have a linear relationship. In that case, NADH was monitored at 380 nm (the millimolar extinction coefficient was 1.244). As a control, NADH oxidation rates were monitored up to 0.4 mM NADH at both 340 and 380 nm, establishing that these rates were essentially the same. The kinetic parameters K0.5 (substrate concentration at which conversion takes place at 50% of the maximum rate), Vmax (maximum conversion rate), and the Hill coefficient were estimated by nonlinear regression with Sigmaplot (Jandel Scientific, San Rafael, Calif.).
Nucleotide sequence accession number.
The nucleotide sequence of the L. lactis NZ9000 ldhB gene and its upstream region that is reported in this paper has been submitted to the GenBank database and assigned accession number AY230155.
RESULTS
Phenotypic instability in lactate dehydrogenase-deficient L. lactis NZ9010.
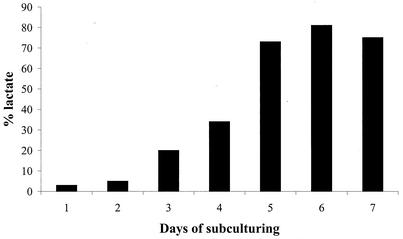
In L. lactis strain NZ9010, in which the las operon-encoded ldh gene has been replaced with an erythromycin resistance gene cassette, only very small amounts of lactate were detected upon aerobic subculturing (23). The main fermentation end products formed under this condition were found to be acetate and acetoin, thereby confirming previously described results obtained with independently constructed ldh-deficient strains (20, 43). Under these conditions, the growth rate of NZ9010 was comparable to that observed for parental strains NZ9000 and MG1363 (Table 2) and both the growth rate and the metabolic profile appeared stable upon subculturing (data not shown). Under anaerobic conditions, L. lactis NZ9010 did not produce significant amounts of lactate and the main metabolites observed were acetoin and ethanol (Table 2). Compared to wild-type cells (NZ9000), strain NZ9010 cells reached higher final turbidities and produced less acid. However, strain NZ9010 grew approximately fivefold slower under these conditions and serial subculturing led to increased amounts of lactate, finally stabilizing at approximately 85% of the total carbon conversion (Fig. 1). Concomitant with this recovery of lactate production, the growth rate of these cultures also increased to that of L. lactis NZ9000. Notably, the kinetics of lactate production and growth rate recovery varied in individual NZ9010 anaerobic subculturing experiments (data not shown), indicating that this phenotypic change is probably due to a mutation rather than the consequence of a regulatory phenomenon. To study this further, a lactate-producing colony was isolated from one of the NZ9010 subcultures that had completely recovered the ability to produce lactate and was designated L. lactis NZ9015. Lactate production in this strain appeared stable and independent of growth conditions. Both anaerobic and aerobic growth resulted in similar lactate production levels, indicating that lactate production in L. lactis NZ9015 is not subject to regulation by the availability of molecular oxygen. Furthermore, the growth rates observed for NZ9015 appeared to be comparable to those of the wild-type strain L. lactis NZ9000 under both aerobic and anaerobic conditions (Table 2).
TABLE 2.
Growth and fermentation characteristics of ldh-deficient L. lactis strainsa
| Strain | Aerobe | μ (h−1) | Final ODe | Final pH | Concn (mmol/liter) of product formed
|
% Recoveryd | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lactate | Formate | Acetate | Acetoin | Butanediol | Ethanol | Pyruvate | ||||||
| NZ9000 | + | 0.75 | 2.90 | 5.13 | 42.7 (72,1)b | NDc | 8.7 (14.7) | ND | 0.1 (0.4) | ND | ND | 87 |
| − | 0.78 | 2.53 | 4.87 | 52.0 (88.0) | ND | 1.2 (2.0) | ND | ND | ND | ND | 90 | |
| NZ9010 | + | 0.69 | 2.86 | 6.30 | 0.9 (1.5) | ND | 12.6 (21.3) | 18.0 (60.7) | ND | 0.9 (1.5) | 2.3 (3.8) | 89 |
| − | 0.17 | 3.14 | 5.59 | 1.9 (3.3) | ND | 9.8 (16.6) | 5.5 (18.7) | ND | 22.6 (38.3) | 0.3 (0.5) | 77 | |
| NZ9015 | + | 0.72 | 2.79 | 5.49 | 31.8 (53.7) | ND | 7.9 (13.3) | 7.5 (25.5) | 0.3 (0.9) | 0.0 (0.1) | ND | 93 |
| − | 0.72 | 2.50 | 4.91 | 47.3 (80.0) | ND | 2.8 (4.8) | ND | 0.2 (0.5) | 2.9 (4.9) | ND | 90 | |
| NZ9020 | + | 0.61 | 2.68 | 6.27 | 0.6 (1.0) | ND | 12.0 (20.3) | 17.9 (60.5) | 0.1 (0.4) | 1.4 (2.4) | 2.6 (4.5) | 89 |
| − | 0.14 | 2.89 | 5.54 | 1.5 (2.5) | ND | 10.0 (16.9) | 5.6 (18.9) | ND | 25.2 (42.7) | 0.5 (0.8) | 82 | |
Data represent the average of three experiments.
Values in parentheses are percentages of pyruvate converted into the product.
ND, not detected.
Total carbon recovery calculated relative to the total amount of glucose consumed.
OD, optical density.
FIG. 1.
Recovery of lactate production in ldh-deficient L. lactis NZ9010. L. lactis NZ9010 was subcultured anaerobically in GM17 for 7 days with 100-fold dilution (approximately 6.7 generations) each day. Lactate production relative to glucose consumption is shown (black bars). Recovery of lactate production occurs after a varying number of generations in independent NZ9010 cultures.
L. lactis NZ9015 produces an alternative lactate dehydrogenase activity.
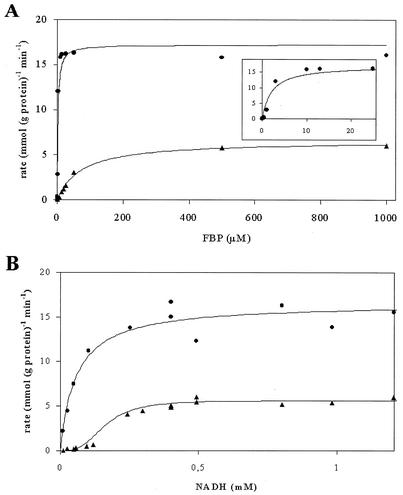
Primarily, it was established that the ability of L. lactis NZ9015 to produce lactate depends on the activity of an Ldh enzyme displaying enzymatic characteristics different from those of the Ldh protein produced by wild-type L. lactis NZ9000. The affinity of the Ldh protein produced by strain NZ9015 for the allosteric activator fructose-1,6-diphosphate (FBP) was much lower (K0.5 = 72 μM) than that of the Ldh protein of strain NZ9000 (K0.5 = 2 μM) (Fig. 2A). In contrast, its affinity for pyruvate was only slightly reduced (K0.5 = 4 and 1 mM, respectively). Notably, the enzyme kinetics for NADH of the Ldh protein produced by strain NZ9015 could not be described with simple Michaelis-Menten kinetics, which is in clear contrast to the Ldh protein of the wild-type strain (K0.5 = 0.2 and 0.06 mM, respectively). The relationship between NADH and the enzyme activity of the NZ9015 Ldh protein yielded a sigmoid curve (Fig. 2B), indicating cooperativity. The NADH kinetics of the Ldh enzyme produced by strain NZ9015 could be described by a Hill equation (10). The nonlinear regression yielded a Hill coefficient of 4, which may indicate that the enzyme has four subunits (10). The different substrate and activator kinetics show that the Ldh activity expressed in L. lactis NZ9015 is clearly distinct from the Ldh activity found in wild-type strain NZ9000. In addition, all kinetic analyses indicate that the limiting rate (Vmax) of the Ldh enzyme of L. lactis NZ9015 (5 U) is only 35 to 50% compared to the Ldh activity in NZ9000 (16 U). One unit is defined as 1 μmol/mg of protein/min.
FIG. 2.
Enzyme kinetics of LdhB and las operon-encoded Ldh. Affinity constants were determined by measuring NADH consumption as a function of the concentration of the activator FBP (A) and the cofactor NADH (B) with crude cell extracts of L. lactis NZ9000 and NZ9015, respectively. Activity curves of Ldh (filled dots) and LdhB are shown (filled triangles). The inset in panel A shows NADH consumption at low concentrations of FBP for las operon-encoded Ldh.
L. lactis NZ9015 produces an alternative Ldh-encoding gene designated ldhB.
In order to identify the gene that encodes the Ldh activity found in L. lactis NZ9015, the complete genome sequence of L. lactis IL1403 (4), which is closely related to L. lactis MG1363 and its derivatives (including NZ9000), was used as a template. Besides the ldh gene located within the las operon, the genome of L. lactis IL1403 contains three additional genes that were predicted to potentially encode NADH-dependent lactate-forming enzymes, ldhB, ldhX, and hicD. Both PCR and Southern approaches were used to evaluate the presence of homologues of ldhB, ldhX, and hicD in the chromosome of L. lactis MG1363 and its derivative NZ9000. Both strategies revealed that, indeed, for all three of the ldh-like genes found in IL-1403, a specific homologue could be detected in MG1363 (data not shown).
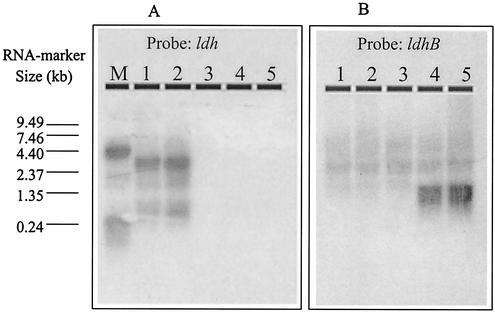
To investigate whether one of these ldh-like genes of L. lactis MG1363 was responsible for the recovery of lactate production observed in L. lactis NZ9015, the expression of these genes was analyzed by Northern blotting. Total RNA was isolated from L. lactis NZ9000 and NZ9015 grown under anaerobic and aerobic conditions, while RNA of strain NZ9010 was isolated from cells grown under aerobic conditions only. Northern analysis with the ldh gene-specific probe yielded similar levels of the las operon-encoded ldh mRNA in L. lactis NZ9000 grown under anaerobic and anaerobic conditions (Fig. 3A, lanes 1 and 2). The sizes of the ldh-specific las operon transcripts detected were in good agreement with the ldh-specific transcripts described previously (38). As anticipated, no ldh-specific transcript could be detected in either aerobically grown L. lactis NZ9010 (Fig. 3A, lane 3) or its Ldh recovery derivative NZ9015 (Fig. 3A, lanes 4 and 5). Hybridization of the same blots with the ldhX- and hicD-specific probes did not yield a detectable hybridization signal in any of the RNA samples used (data not shown). These results indicate that these genes are either not expressed in any of the strains or are expressed at a level below the Northern blot analysis detection limit. In contrast, the ldhB-specific probe hybridized with an mRNA of approximately 1.3 kb that could be detected in the RNA samples derived from strain L. lactis NZ9015 (Fig. 3, lanes 4 and 5) but not in the RNA samples of either strain NZ9000 or NZ9010 (Fig. 3, lanes 1 to 3). Moreover, the signal intensity appeared to be similar for aerobically and anaerobically grown cells (Fig. 3, lanes 4 and 5, respectively), indicating that ldhB transcription in strain NZ9015 is not subject to regulation by the availability of oxygen. Similarly, the Ldh activity level detected in strain NZ9015 was not influenced by the level of aeration of the culture (data not shown). Taken together, these data strongly suggest that the recovery of the lactate-producing ability observed in L. lactis NZ9015 is due to the activation of transcription of an alternative Ldh-encoding, ldhB gene that is not transcribed at a detectable level in parental strains NZ9000 and NZ9010.
FIG. 3.
Northern blot detection of ldh- and ldhB-specific transcripts in L. lactis NZ9000 and derivatives thereof. Total RNAs isolated from L. lactis NZ9000 grown under aerobic and anaerobic conditions, L. lactis NZ9010 grown under aerobic conditions, and L. lactis NZ9015 grown under aerobic and anaerobic conditions (lanes 1 to 5, respectively) were size fractionated on a formaldehyde denaturing 1% agarose gel and transferred to a membrane. ldh- and ldhB-specific mRNAs were detected with radioactively labeled ldh and ldhB gene probes (panels A and B, respectively). A 0.24- to 9.49-kb RNA marker (lane M) was used to determine the mRNA sizes.
Construction and characterization of an ldhB disruption mutant of L. lactis NZ9015.
To demonstrate that expression of the ldhB gene is responsible for the recovered lactate production in L. lactis NZ9015, an ldhB disruption mutant of this strain was constructed by single-crossover plasmid integration in this gene (see Materials and Methods for details). In one of the integrants obtained, the genetic conformation of the ldh::ery locus, as well as the ldhB locus, was confirmed by PCR and Southern blotting (data not shown), and this integrant was designated L. lactis NZ9020. Under anaerobic and aerobic conditions, strain NZ9020 did not produce significant amounts of lactate, and its fermentation profiles under both conditions were virtually identical to those observed for L. lactis NZ9010 (Table 2). Moreover, the final optical density and pH reached by NZ9020 cultures were almost the same as those observed for L. lactis NZ9010 (Table 2). Finally, the anaerobic growth rate of strain NZ9020 was reduced to the same extent as was observed for the initial Ldh-deficient mutant, NZ9010, compared to that observed for either of the Ldh-producing strains, NZ9000 or NZ9015 (Table 2). The slightly lower growth rate measured for L. lactis NZ9020 under both aerobic and anaerobic conditions relative to strain NZ9010 is probably due to the tetracycline selection that is required to maintain the genotype of this strain. These results provide good evidence for the role of the ldhB-encoded enzyme in restoration of the lactate-producing ability observed in L. lactis NZ9015.
Genetic analysis of the activation of the ldhB gene in L. lactis NZ9015.
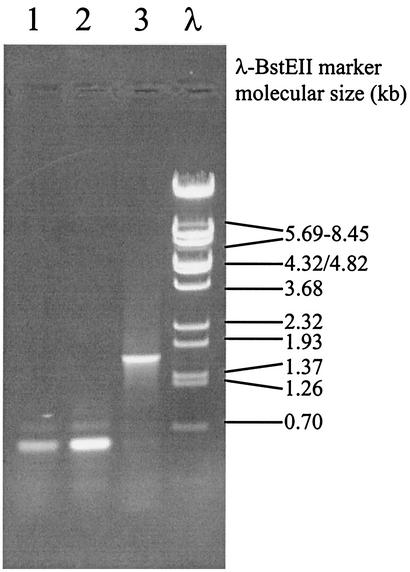
The ldhB gene appeared to be transcribed as a 1.3-kb mRNA, which is in good agreement with the expected size of a monocistronic ldhB transcript. Hence, it can be expected that the anticipated mutation that has caused its transcription activation would be located upstream of the ldhB gene. To elucidate the mechanism of transcription activation of the ldhB gene in L. lactis NZ9015, the upstream region of this gene was investigated. To this end, a degenerated primer, based on the protein sequence encoded by the gene located upstream of ldhB in the L. lactis IL1403 genome (rlrD), was designed and combined with a reverse ldhB primer (based on the MG1363 ldhB gene sequence) in PCRs with chromosomal DNAs from various lactococcal strains as templates. A 600-bp PCR product was obtained with either MG1363, NZ9000, or NZ9010 DNA as the template (Fig. 4, lanes 1 and 2). In contrast, chromosomal DNA of strain NZ9015 in the same PCR generated a product of approximately 1.8 kb (Fig. 4, lane 3), indicating genetic rearrangement of the ldhB locus in this strain relative to that of the parental strain. Both the 600-bp fragment obtained for NZ9000 and the 1.8-kb fragment obtained for NZ9015 were subjected to sequence analysis. The sequence of the upstream region of the L. lactis NZ9000 ldhB gene appeared highly similar to that found in strain IL1403 and confirmed the presence of an rlrD-like gene upstream of the ldhB gene in this strain. In addition to the sequences found in NZ9000, the upstream region of the NZ9015 ldhB gene contained sequences that displayed high homology to the L. lactis IS981 family sequence. Our results indicated that in strain NZ9015, an IS981-like element had been inserted 213 bp upstream of the ldhB start codon (Fig. 5A) and could have caused the activation of ldhB transcription. Primer extension performed on total RNA isolated from strain NZ9015 showed that the ldhB mRNA initiated 190 bp upstream of the ldhB start codon (Fig. 5B). Thereby, the −10 region (TAAAAT) of the ldhB promoter is apparently derived from the native ldhB upstream sequence, while the corresponding −35 region (TTGACT) of that promoter is derived from the IS981-like element (Fig. 5B). These data show that insertion of an IS element provides a consensus −35 region at the correct spacing (17 bp) relative to the already existing −10 region, thereby leading to activation of the normally silent ldhB gene.
FIG. 4.
L. lactis ldhB promoter region amplification. The rlrD-ldhB intergenic region was amplified by PCR with rlrD forward and ldhB reverse primers on chromosomal DNAs of L. lactis NZ9000, NZ9010, and NZ9015 (lanes 1 to 3, respectively). PCR products were size fractionated on a 1% agarose gel. BstEII-digested λ DNA was used as a reference (lane λ).
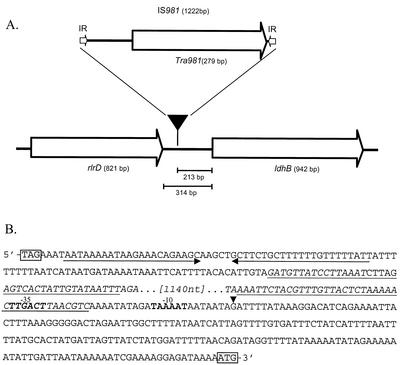
FIG. 5.
Schematic representation of the ldhB promoter region of L. lactis NZ9015. (A) Insertion position of IS981 (indicated by a filled triangle). The rlrD, ldhB, and tra981 genes and the positions of the IS981 imperfect inverted repeats (IR) are indicated by white arrows. (B) Nucleotide sequence of the ldhB promoter region of L. lactis NZ9015 from the rlrD stop codon to the ldhB start codon (light gray background). The upright sequence represents the native promoter region as it is also present in strains NZ9000 and NZ9010. Underlining arrows indicate the rlrD terminator. The IS981-originated sequence is in italics. The IS981 imperfect inverted repeats are underlined. The −35 and −10 regions are in bold. The vertical arrowhead indicates the ldhB transcription start site determined in L lactis NZ9015. nt, nucleotides.
IS981-mediated activation of ldhB is a predominant mechanism for the recovery of lactate production in ldh mutants of L. lactis.
To evaluate whether the observed activation of ldhB by insertion of an IS981-like element is a frequent event in ldh mutants of L. lactis, 10 individual cultures of NZ9010 were grown under anaerobic conditions for 100 generations. In all cultures, the specific growth and acidification rate appeared to be restored to wild-type levels at the end of these subculturing sequences, although the number of generations after which growth rate restoration was first observed appeared to be variable (data not shown). Northern analysis of the total RNA isolated from these cultures showed that ldhB appeared to be expressed in all cases. Although the absolute level of ldhB expression appeared to vary slightly among these cultures, in all cases, the ldhB transcripts appeared to be similar in size to that observed in strain NZ9015. These results suggest that activation of ldhB expression is the main mechanism of recovery of lactate production in stain NZ9010. Amplification of the rlrD-ldhB intergenic region with chromosomal DNA of the 10 cultures in which ldhB transcription had been activated generated, in four cases, an amplification product of approximately 1.8 kb. In contrast, in 5 of these cultures, no apparent change in the size of the rlrD-ldhB intergenic region was observed, while in the 10th culture, no amplification product could be obtained. Single-stranded sequence analysis of the amplified intergenic regions of cultures that did not display an apparent change in size revealed, in all cases, one or more mutations relative to the sequence of the parental strain, L. lactis NZ9010. Both the nature and the position of these mutations were variable, including single-base substitutions but also a 9-bp duplication (Fig. 6). However, no obvious explanation for the activation of ldhB expression in these mutants could be deduced since no reasonable match with the consensus promoter sequence regions of gram-positive bacteria (12, 27) was generated by these mutations. The observed activation of ldhB expression in these mutants possibly results from resolution of the putative high degree of secondary structure of the rlrD-ldhB intergenic region (data not shown) but was not further investigated in this study. Sequence analysis of the enlarged intergenic regions revealed that, in all of these cases, an IS981-like element had been inserted at precisely the same position as had been observed in strain NZ9015 (213 bp upstream of the ldhB start codon). Moreover, these IS981 insertions appeared to have taken place in a directional manner, since, in all cases, the transposase-encoding gene is oriented in the same direction as the ldhB gene. Thereby, in all cases, a −35-like region is generated at the exact 17-bp spacing relative to the preexisting −10 region, similar to what had been observed in strain NZ9015. Moreover, although the IS981-derived −35 region is part of the terminal inverted (imperfect) repeat of this insertion element, insertion of IS981 in the opposite orientation would not lead to a reasonable −35 region upstream of ldhB because of the sequence variation found in this region of the IS981 family. Furthermore, the small sequence deviations in the IS981 family-derived sequences observed among these cultures clearly indicated that these mutants are truly independent and result from individual IS981 insertions in this rlrD-ldhB intergenic region. Although these sequence deviations included variations in the −35 region, in all cases, a reasonable −35-like sequence was found, including TTGACT (as was found in NZ9015), TTGACA (perfect consensus, found in two of the cultures), and TTGATT. These data indicate that the mobility of the IS981-like elements provides L. lactis NZ9000 with a mechanism by which to activate the transcription of an otherwise silent Ldh-encoding gene, ldhB. Moreover, this event represents one of the predominant mechanisms of activation of this gene.
FIG. 6.
Schematic representation of mutations in the ldhB promoter regions of ldhB-expressing NZ9010 derivatives without IS981 integration. The nucleotide sequence of the ldhB promoter region of L. lactis NZ9010, from the rlrD stop codon to the ldhB start codon (light gray background), is shown. Mutations detected in the same region in five independent cultures that displayed ldhB expression are indicated (numbered 1 to 5) and include nucleotide substitutions (arrowheads) or insertions (light gray triangles). Cultures: 1, ATAAATTCA inserted after A91, A222G, T223A; 2, A187G; 3, G77A; 4, T131G, T205C; 5, A69G.
DISCUSSION
In this report, we describe the recovery of lactate production in L. lactis NZ9010 (23), which was only observed when this strain was grown under anaerobic conditions. This lactate production recovery coincided with a growth rate restoration to a level almost equal to that of parental strain NZ9000. This improved growth rate provides the selective advantage that allows rapid accumulation of lactate-producing NZ9010 derivatives. In wild-type lactococcal cells, Ldh provides the electron sink required for maintenance of the NADH/NAD+ ratio, which is an important control factor in lactococcal metabolism (9, 18, 35). Especially under anaerobic conditions, the Ldh enzyme provides the sole electron sink available, and ldh mutants of L. lactis have been shown to use alternative electron sink reactions to resolve the resulting redox balance problem (25, 40). In contrast, under aerobic conditions, molecular oxygen acts as an alternative electron sink through the activity of the lactococcal NADH oxidase (35), thereby explaining the observed stability of strain NZ9010 under these conditions.
To study the mechanism of the observed recovery of lactate production, a lactate-producing derivative of strain NZ9010 (designated L. lactis NZ9015) was analyzed in detail. The inferior characteristics observed for the Ldh enzyme present in this strain are reflected in the observation that the lactate-producing capacity in this strain never exceeded approximately 85% of the total carbon flux, which is significantly lower than that observed in wild-type cells (>95%) (20, 43). Previously, it has been shown that the las operon-encoded Ldh enzyme has a low degree of control over lactate formation rates in L. lactis (1), which is in agreement with the metabolic predictions generated by the kinetic model of the lactococcal pyruvate metabolism (23). However, replacement of the las operon-encoded Ldh enzyme with LdhB in that same model confirms the metabolic values reported here (data not shown; reference 23; jjj.biochem.sun.ac.za/wcfs.html). Intriguingly, an ldh mutant obtained by random mutagenesis appeared to produce an Ldh protein displaying enzyme characteristics similar to those of the Ldh protein produced by strain NZ9015 (5). Sequence analysis revealed mutations in the las operon-encoded ldh gene (17). However, on the basis of the results presented, the possibility cannot be excluded that the observed amino acid substitutions led to complete inactivation of this Ldh enzyme and subsequent LdhB production. Furthermore, an ldh deletion mutant of L. lactis MG1363 was shown to produce the end product lactate (22% of total carbon flux) under anaerobic conditions (41), which could be the result of analysis of an intermediate culture in the recovery process in which a part of the population expresses the ldhB gene.
With the complete genome sequence of L. lactis IL1403 as a template, the putative presence of alternative Ldh-encoding genes was examined in L. lactis MG1363. Initial analyses revealed that the L. lactis IL1403 genome displays a fourfold redundancy in predicted Ldh-encoding genes (ldh, ldhB, ldhX, and hicD). However, a more detailed analysis of the three alternative ldh-like genes of strain IL1403 raises doubts about the ldhX-encoded product as a true Ldh protein since it contains two, probably critical, deviations from the consensus Ldh active-site sequence. Moreover, the hicD gene is annotated as a pseudogene in L. lactis IL1403. Primarily, it was established by PCR and Southern blotting that all three alternative ldh-like genes of L. lactis IL1403 have a homologue in L. lactis MG1363. However, it should be noted that, on the basis of these results, the possibility could not be excluded that additional ldh-like genes are present in the L. lactis MG1363 genome. Nevertheless, subsequent Northern blot analysis showed that the ldhB gene homologue is transcribed in strain NZ9015 while it is not transcribed in parental strain NZ9000 or NZ9010. Importantly, the analysis of 10 independent NZ9010 mutants that had recovered the ability to produce lactate showed that the ldhB gene represents the preferred, if not the only, gene involved in lactate production recovery in NZ9010 derivatives.
Comparative analysis of the upstream region of the ldhB gene in the parental strains (NZ9000 and NZ9010) and strain NZ9015 revealed site-specific, oriented IS integration in the rlrD-ldhB intergenic region. Various studies with both gram-positive and gram-negative microbes indicate that IS elements play an important role in the adaptation to environmental (stressful) conditions (14, 32, 42, 45, 48). Furthermore, several IS-mediated mechanisms of gene expression modulation have been described (14, 28, 39). The presence of −35 regions located in the terminal inverted repeats has been described (16), and a mechanism of transcriptional activation of downstream genes by combination of those −35 regions with a natively present −10 region, as we have observed in strain NZ9015, has been reported (44). This mechanism of promoter improvement has been described before for the plasmid-located citP expression in L. lactis (37). However, IS insertion upstream of citP generated only 20% of its final expression level, while 80% of the citP transcript was still derived from the native citP promoter. Therefore, the predominant effect of this IS insertion event is deregulation of the citP promoter, rather than direct activation of citP expression (37). In contrast, in this study, we actually have shown that an inactive promoter turns to an active promoter after site-specific, oriented IS integration. Thus, clear evidence is given that gene activation of normally silent genes by IS integration could play an important role in adaptive evolution, especially when the selection pressure for such an event is high, as is the case in anaerobically growing L. lactis NZ9010 (ldh::ery). Moreover, genetic analysis of the rlrD-ldhB intergenic region of 10 independently obtained mutants with recovered lactate production revealed that IS981-mediated activation of ldhB represents a major mechanism of activation of this gene in L. lactis NZ9010.
In the present era of genomics, and thereby the discovery of a variable degree of gene redundancy in microbial genomes, it is important to understand what the function of these redundancies could be. The present study provides an example of a specific gene redundancy found in L. lactis that apparently allows this species to rapidly adapt to inactivation of the gene encoding one of its important metabolic enzymes by highly specific activation of a normally silent gene encoding the same function. With regard to the importance of Ldh-deficient mutants of L. lactis for metabolic engineering purposes, the identification of the most important (and possibly only) alternative Ldh-encoding gene in this organism allows the construction of stable and robust Ldh-deficient mutants, similar to strain NZ9020. Such a strain can be grown at an acceptable growth rate (similar to that observed for the wild type) under aerobic conditions. Subsequently, it can be used as an efficient cell factory in systems where no growth is required, like those described for the alanine- and diacetyl-producing mutants of L. lactis (24, 26).
Acknowledgments
We are grateful to Roelie Holleman for technical assistance in the metabolite analysis by high-performance liquid chromatography. We thank Arjan de Visser for critical discussions of IS-mediated adaptive evolution and for critically reading the manuscript. We thank Willem M. de Vos for critically reading the manuscript.
This work was in part supported by the EU-GEMOLAB project (contract BIO4-CT98-0118).
REFERENCES
- 1.Andersen, H. W., M. B. Pedersen, K. Hammer, and P. R. Jensen. 2001. Lactate dehydrogenase has no control on lactate production but has a strong negative control on formate production in Lactococcus lactis. Eur. J. Biochem. 268:6379-6389. [DOI] [PubMed] [Google Scholar]
- 2.Bhowmik, T., and J. L. Steele. 1994. Cloning, characterization and insertional inactivation of the Lactobacillus helveticus D(−) lactate dehydrogenase gene. Appl. Microbiol. Biotechnol. 41:432-439. [DOI] [PubMed] [Google Scholar]
- 3.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bolotin, A., P. Wincker, S. Mauger, O. Jaillon, K. Malarme, J. Weissenbach, S. D. Ehrlich, and A. Sorokin. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 11:731-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boumerdassi, H., C. Monnet, M. Desmazeaud, and G. Corrieu. 1997. Isolation and properties of Lactococcus lactis subsp. lactis biovar diacetylactis CNRZ 483 mutants producing diacetyl and acetoin from glucose. Appl. Environ. Microbiol. 63:2293-2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradford, M. M. 1976. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:52-58. [DOI] [PubMed] [Google Scholar]
- 7.Casadaban, M. J., and S. N. Cohen. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138:179-207. [DOI] [PubMed] [Google Scholar]
- 8.Chopin, A., M. C. Chopin, A. Moillo-Batt, and P. Langella. 1984. Two plasmid-determined restriction and modification systems in Streptococcus lactis. Plasmid 11:260-263. [DOI] [PubMed] [Google Scholar]
- 9.Cocaign-Bousquet, M., C. Garrigues, P. Loubiere, and N. D. Lindley. 1996. Physiology of pyruvate metabolism in Lactococcus lactis. Antonie Van Leeuwenhoek 70:253-267. [DOI] [PubMed] [Google Scholar]
- 10.Cornish-Bowden, A. 1995. Fundamentals of enzyme kinetics, 2nd ed. Portland Press, London, England.
- 11.Crow, V. L., and G. G. Pritchard. 1977. Fructose 1,6-diphosphate-activated l-lactate dehydrogenase from Streptococcus lactis: kinetic properties and factors affecting activation. J. Bacteriol. 131:82-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Vos, W. M., and G. F. M. Simons. 1994. Gene cloning and expression systems in lactococci. Blackie Academic & Professional, Glasgow, United Kingdom.
- 13.de Vos, W. M., P. Vos, H. de Haard, and I. Boerrigter. 1989. Cloning and expression of the Lactococcus lactis ssp. cremoris SK11 gene encoding an extracellular serine protease. Gene 85:169-176. [DOI] [PubMed] [Google Scholar]
- 14.Duwat, P., A. Cochu, S. D. Ehrlich, and A. Gruss. 1997. Characterization of Lactococcus lactis UV-sensitive mutants obtained by ISS1 transposition. J. Bacteriol. 179:4473-4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferain, T., D. Garmyn, N. Bernard, P. Hols, and J. Delcour. 1994. Lactobacillus plantarum ldhL gene: overexpression and deletion. J. Bacteriol. 176:596-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galas, D. J., and M. Chandler. 1989. Bacterial insertion sequences, p. 109-162. In D. E. Berg and M. M. Howe (ed.), Mobile DNA. American Society for Microbiology, Washington, D.C.
- 17.Garrigues, C., N. Goupil-Feuillerat, M. Cocaign-Bousquet, P. Renault, N. D. Lindley, and P. Loubiere. 2001. Glucose metabolism and regulation of glycolysis in Lactococcus lactis strains with decreased lactate dehydrogenase activity. Metab. Eng. 3:211-217. [DOI] [PubMed] [Google Scholar]
- 18.Garrigues, C., P. Loubiere, N. D. Lindley, and M. Cocaign-Bousquet. 1997. Control of the shift from homolactic acid to mixed-acid fermentation in Lactococcus lactis: predominant role of the NADH/NAD+ ratio. J. Bacteriol. 179:5282-5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garvie, E. I. 1978. Lactate dehydrogenases of Streptococcus thermophilus. J. Dairy Res. 45:515-518. [DOI] [PubMed] [Google Scholar]
- 20.Gasson, M., K. Benson, S. Swindell, and H. Griffin. 1996. Metabolic engineering of the Lactococcus lactis diacetyl pathway. Lait 76:33-40. [Google Scholar]
- 21.Gasson, M. J. 1983. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hillier, A. J., and G. R. Jago. 1982. l-Lactate dehydrogenase, FDP-activated, from Streptococcus cremoris. Methods Enzymol. 89:362-367. [DOI] [PubMed] [Google Scholar]
- 23.Hoefnagel, M. H., M. J. Starrenburg, D. E. Martens, J. Hugenholtz, M. Kleerebezem, S. Van II, R. Bongers, H. V. Westerhoff, and J. L. Snoep. 2002. Metabolic engineering of lactic acid bacteria, the combined approach: kinetic modelling, metabolic control and experimental analysis. Microbiology 148:1003-1013. [DOI] [PubMed] [Google Scholar]
- 24.Hols, P., M. Kleerebezem, A. N. Schanck, T. Ferain, J. Hugenholtz, J. Delcour, and W. M. de Vos. 1999. Conversion of Lactococcus lactis from homolactic to homoalanine fermentation through metabolic engineering. Nat. Biotechnol. 17:588-592. [DOI] [PubMed] [Google Scholar]
- 25.Hols, P., A. Ramos, J. Hugenholtz, J. Delcour, W. M. de Vos, H. Santos, and M. Kleerebezem. 1999. Acetate utilization in Lactococcus lactis deficient in lactate dehydrogenase: a rescue pathway for maintaining redox balance. J. Bacteriol. 181:5521-5526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hugenholtz, J., M. Kleerebezem, M. Starrenburg, J. Delcour, W. de Vos, and P. Hols. 2000. Lactococcus lactis as a cell factory for high-level diacetyl production. Appl. Environ. Microbiol. 66:4112-4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jensen, P. R., and K. Hammer. 1998. The sequence of spacers between the consensus sequences modulates the strength of prokaryotic promoters. Appl. Environ. Microbiol. 64:82-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jordan, E., H. Saedler, and P. Starlinger. 1968. O0 and strong-polar mutations in the gal operon are insertions. Mol. Gen. Genet. 102:353-363. [DOI] [PubMed] [Google Scholar]
- 29.Klaenhammer, T., E. Altermann, F. Arigoni, A. Bolotin, F. Breidt, J. Broadbent, R. Cano, S. Chaillou, J. Deutscher, M. Gasson, M. van de Guchte, J. Guzzo, A. Hartke, T. Hawkins, P. Hols, R. Hutkins, M. Kleerebezem, J. Kok, O. Kuipers, M. Lubbers, E. Maguin, L. McKay, D. Mills, A. Nauta, R. Overbeek, H. Pel, D. Pridmore, M. Saier, D. van Sinderen, A. Sorokin, J. Steele, D. O'Sullivan, W. de Vos, B. Weimer, M. Zagorec, and R. Siezen. 2002. Discovering lactic acid bacteria by genomics. Antonie Van Leeuwenhoek 82:29-58. [DOI] [PubMed] [Google Scholar]
- 30.Kuipers, O. P., M. M. Beerthuyzen, R. J. Siezen, and W. M. De Vos. 1993. Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis: requirement of expression of the nisA and nisI genes for development of immunity. Eur. J. Biochem. 216:281-291. [DOI] [PubMed] [Google Scholar]
- 31.Kuipers, O. P., P. G. de Ruyter, M. Kleerebezem, and W. M. de Vos. 1997. Controlled overproduction of proteins by lactic acid bacteria. Trends Biotechnol. 15:135-140. [DOI] [PubMed] [Google Scholar]
- 32.Lapierre, L., B. Mollet, and J. E. Germond. 2002. Regulation and adaptive evolution of lactose operon expression in Lactobacillus delbrueckii. J. Bacteriol. 184:928-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Llanos, R. M., C. J. Harris, A. J. Hillier, and B. E. Davidson. 1993. Identification of a novel operon in Lactococcus lactis encoding three enzymes for lactic acid synthesis: phosphofructokinase, pyruvate kinase, and lactate dehydrogenase. J. Bacteriol. 175:2541-2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Llanos, R. M., A. J. Hillier, and B. E. Davidson. 1992. Cloning, nucleotide sequence, expression, and chromosomal location of ldh, the gene encoding l-(+)-lactate dehydrogenase, from Lactococcus lactis. J. Bacteriol. 174:6956-6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lopez de Felipe, F., and J. Hugenholtz. 1999. Pyruvate flux distribution in NADH-oxidase-overproducing Lactococcus lactis strain as a function of culture conditions. FEMS Microbiol. Lett. 179:461-466. [DOI] [PubMed] [Google Scholar]
- 36.Lopez de Felipe, F., M. Kleerebezem, W. M. de Vos, and J. Hugenholtz. 1998. Cofactor engineering: a novel approach to metabolic engineering in Lactococcus lactis by controlled expression of NADH oxidase. J. Bacteriol. 180:3804-3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lopez de Felipe, F., C. Magni, D. de Mendoza, and P. Lopez. 1996. Transcriptional activation of the citrate permease P gene of Lactococcus lactis biovar diacetylactis by an insertion sequence-like element present in plasmid pCIT264. Mol. Gen. Genet. 250:428-436. [DOI] [PubMed] [Google Scholar]
- 38.Luesink, E. J., R. E. van Herpen, B. P. Grossiord, O. P. Kuipers, and W. M. de Vos. 1998. Transcriptional activation of the glycolytic las operon and catabolite repression of the gal operon in Lactococcus lactis are mediated by the catabolite control protein CcpA. Mol. Microbiol. 30:789-798. [DOI] [PubMed] [Google Scholar]
- 39.Moxon, E. R., P. B. Rainey, M. A. Nowak, and R. E. Lenski. 1994. Adaptive evolution of highly mutable loci in pathogenic bacteria. Curr. Biol. 4:24-33. [DOI] [PubMed] [Google Scholar]
- 40.Neves, A. R., A. Ramos, C. Shearman, M. J. Gasson, J. S. Almeida, and H. Santos. 2000. Metabolic characterization of Lactococcus lactis deficient in lactate dehydrogenase using in vivo 13C-NMR. Eur. J. Biochem. 267:3859-3868. [DOI] [PubMed] [Google Scholar]
- 41.Neves, A. R., R. Ventura, N. Mansour, C. Shearman, M. J. Gasson, C. Maycock, A. Ramos, and H. Santos. 2002. Is the glycolytic flux in Lactococcus lactis primarily controlled by the redox charge? Kinetics of NAD+ and NADH pools determined in vivo by 13C-NMR. J. Biol. Chem. 277:28088-28098. [DOI] [PubMed] [Google Scholar]
- 42.Papadopoulos, D., D. Schneider, J. Meier-Eiss, W. Arber, R. E. Lenski, and M. Blot. 1999. Genomic evolution during a 10,000-generation experiment with bacteria. Proc. Natl. Acad. Sci. USA 96:3807-3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Platteeuw, C., J. Hugenholtz, M. Starrenburg, I. van Alen-Boerrigter, and W. M. de Vos. 1995. Metabolic engineering of Lactococcus lactis: influence of the overproduction of α-acetolactate synthase in strains deficient in lactate dehydrogenase as a function of culture conditions. Appl. Environ. Microbiol. 61:3967-3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prentki, P., B. Teter, M. Chandler, and D. J. Galas. 1986. Functional promoters created by the insertion of transposable element IS1. J. Mol. Biol. 191:383-393. [DOI] [PubMed] [Google Scholar]
- 45.Riehle, M. M., A. F. Bennett, and A. D. Long. 2001. Genetic architecture of thermal adaptation in Escherichia coli. Proc. Natl. Acad. Sci. USA 98:525-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 47.Savijoki, K., and A. Palva. 1997. Molecular genetic characterization of the l-lactate dehydrogenase gene (ldhL) of Lactobacillus helveticus and biochemical characterization of the enzyme. Appl. Environ. Microbiol. 63:2850-2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schneider, D., E. Duperchy, E. Coursange, R. E. Lenski, and M. Blot. 2000. Long-term experimental evolution in Escherichia coli. IX. Characterization of insertion sequence-mediated mutations and rearrangements. Genetics 156:477-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Starrenburg, M. J. C., and J. Hugenholtz. 1991. Citrate fermentation by Lactococcus and Leuconostoc spp. Appl. Environ. Microbiol. 57:3535-3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Swindell, S. R., K. H. Benson, H. G. Griffin, P. Renault, S. D. Ehrlich, and M. J. Gasson. 1996. Genetic manipulation of the pathway for diacetyl metabolism in Lactococcus lactis. Appl. Environ. Microbiol. 62:2641-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taguchi, H., and T. Ohta. 1991. d-Lactate dehydrogenase is a member of the d-isomer-specific 2-hydroxyacid dehydrogenase family: cloning, sequencing, and expression in Escherichia coli of the d-lactate dehydrogenase gene of Lactobacillus plantarum. J. Biol. Chem. 266:12588-12594. [PubMed] [Google Scholar]
- 52.van Rooijen, R. J., and W. M. de Vos. 1990. Molecular cloning, transcriptional analysis, and nucleotide sequence of lacR, a gene encoding the repressor of the lactose phosphotransferase system of Lactococcus lactis. J. Biol. Chem. 265:18499-18503. [PubMed] [Google Scholar]
- 53.Vos, P., M. van Asseldonk, F. van Jeveren, R. Siezen, G. Simons, and W. M. de Vos. 1989. A maturation protein is essential for the production of active forms of Lactococcus lactis SK11 serine proteinase located in or secreted from the cell envelope. J. Bacteriol. 171:2795-2802. [DOI] [PMC free article] [PubMed] [Google Scholar]