Abstract
The staphylococcal accessory regulator locus (sarA) encodes a DNA-binding protein (SarA) that modulates expression of over 100 genes. Whether this occurs via a direct interaction between SarA and cis elements associated with its target genes is unclear, partly because the definitive characteristics of a SarA binding site have not been identified. In this work, electrophoretic mobility shift assays (EMSAs) were used to identify a SarA binding site(s) upstream of the SarA-regulated gene cna. The results suggest the existence of multiple high-affinity binding sites within the cna promoter region. Using a SELEX (systematic evolution of ligands by exponential enrichment) procedure and purified, recombinant SarA, we also selected DNA targets that contain a high-affinity SarA binding site from a random pool of DNA fragments. These fragments were subsequently cloned and sequenced. Randomly chosen clones were also examined by EMSA. These DNA fragments bound SarA with affinities comparable to those of recognized SarA-regulated genes, including cna, fnbA, and sspA. The composition of SarA-selected DNAs was AT rich, which is consistent with the nucleotide composition of the Staphylococcus aureus genome. Alignment of selected DNAs revealed a 7-bp consensus (ATTTTAT) that was present with no more than one mismatch in 46 of 56 sequenced clones. By using the same criteria, consensus binding sites were also identified upstream of the S. aureus genes spa, fnbA, sspA, agr, hla, and cna. With the exception of cna, which has not been previously examined, this 7-bp motif was within the putative SarA binding site previously associated with each gene.
Staphylococcus aureus is a versatile human pathogen capable of causing a wide variety of infections. Its capacity to cause disease arises from its production of a diverse array of virulence factors. When it is grown in vitro in liquid culture, the production of these virulence factors is coordinately regulated such that surface-associated virulence factors (e.g., adhesins) are produced primarily during the exponential growth phase while extracellular virulence factors (e.g., exotoxins) are preferentially produced as cultures enter postexponential growth (25). It has been suggested that this pattern of gene expression has an in vivo corollary that corresponds to before and after abscess formation (33). The primary regulatory locus modulating these events is the accessory gene regulator, or agr (26, 32, 35). The agr locus includes the chromosomal regions encoding two divergent transcripts designated RNAII and RNAIII. The RNAII-encoding region carries an operon (agrBDCA) containing the components of a quorum-sensing system. This system is induced in response to accumulating amounts of an octapeptide pheromone (20, 23, 29, 31). Induction results in increased transcription from both the RNAII (P2) and RNAIII (P3) promoters. RNAIII is the effector molecule responsible for repression of surface protein production and the concomitant increase in the production of exotoxins (27, 30).
Production of RNAIII is also influenced by the staphylococcal accessory regulator locus, or sarA (18). This locus includes three promoters (P1, P2, and P3) that are used to produce three overlapping transcripts (sarA, sarB, and sarC, respectively). All three transcripts include the sarA open reading frame, which encodes a DNA-binding protein that is required, at least under certain conditions, for maximal expression from the agr P2 and P3 promoters (6, 7). Mutation of sarA also alters expression from other promoters, including those for cna, fnbA, hla, spa, and sspA (4, 8, 9, 21, 24). In fact, transcriptional profiling experiments indicate that SarA influences expression of over 100 genes (14). Electrophoretic mobility shift assays (EMSAs) indicate that the regulatory impact of SarA on at least some of these genes involves a direct interaction between SarA and cis promoter elements (3, 11, 28, 34). Based on the alignment of these promoters, a 26-bp consensus SarA box has been proposed (5, 11). In cases in which this consensus is absent, it is presumed that the regulatory effects of SarA are indirect, possibly by virtue of the interaction between SarA and one of the other sar homologues (1). However, our data indicate that SarA represses cna transcription directly by virtue of its ability to bind cis elements associated with the cna promoter (3), and the cna promoter region does not contain a SarA box, at least as it is defined by Dunman et al. (14) [ATTTGTATTTAATATTT(T/A)T(T/A/G)TAATTG, with no more than seven mismatches; Ellen Murphy, personal communication]. This suggests that SarA may be capable of binding additional DNA motifs or that the present definition of a SarA box is inadequate.
To address these issues, we performed EMSAs using purified SarA and DNA fragments derived from the region immediately upstream of cna. The results were compared to those obtained with other DNA fragments reported to contain a SarA box. We also performed analysis by systematic evolution of ligands by exponential enrichment (SELEX) in an effort to obtain an unbiased definition of the DNA elements bound by SarA.
MATERIALS AND METHODS
EMSA.
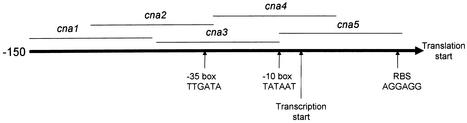
SarA was expressed and purified as described by Rechtin et al. (34). The concentration of SarA was determined spectrophotometrically at 280 nm by using an extinction coefficient of 7,740 M−1. Activity was determined by EMSA under stoichiometric conditions to be >95% (data not shown). EMSAs were performed as previously described (34). For the SELEX experiments (see below), the DNA targets were generated by synthesizing a 98-bp oligonucleotide in which the 24 bases on each end had a defined sequence while the 50 bases in the middle were entirely random (i.e., synthesized with an equimolar mixture of all four phosphoramidites). For direct comparison with the SELEX templates, we also generated 98-bp oligonucleotides corresponding to specific SarA targets (Table 1). In each case, the middle 50 bases were derived from the target gene with the putative SarA binding site centrally located. In the case of the cna gene, we generated five DNA targets, three of which were contiguous and collectively spanned the 150-bp upstream of the translational start codon. The other two spanned the junctions between these fragments (Fig. 1).
TABLE 1.
KD values of SarA target DNAsa
| DNA target | KD (pM) | Sequence |
|---|---|---|
| spa | 308 ± 27 | GCGTTTAAATTTAATTATAAATATAGATTTTAGTATTGCAATACATAACG |
| fnbB | 269 ± 117 | GCTGTTCAAGAGCTTTGTATGCAATATATATGTGAGTTTCAAATAATACG |
| fnbA | 228 ± 47 | GCGTTTCTGATGACTTGAATACAATTTATAGGTATATTTCAAATAATACG |
| sspA | 180 ± 82 | ATTTTTATTGTTATATTTAACTTGTAAATAAATTTTTTGGAGGTTTTTAG |
| agr | 145 ± 63 | GCATTTATTTTCCAATTTTTCTTAACTAGTCGTTTTTTATTCTTAACTGT |
| hla | 91 ± 25 | GCATATATAGTTAATTTTTATTTAATAGTTAATTAATTGATTTAATTCCG |
| cna1 | 182 ± 6 | ATGCACTTGTATTCGTTATACTGTATATATTTTGCATAATAAAATAATAA |
| cna2 | 104 ± 26 | TATATTTTGCATAATAAAATAATAATATGAATTTTTGATAAATTTCATTG |
| cna3 | 162 ± 68 | TATGAATTTTTGATAAATTTCATTGAATAAGAACTAAATTAGTTTATAAT |
| cna4 | 196 ± 13 | AATAAGAACTAAATTAGTTTATAATTTATTATTAGTATCCTGTGGATATG |
| cna5 | 541 ± 28 | TTATTATTAGTATCCTGTGGATATGACATAGAGTATAAGGAGGGGTTTTT |
| SELEX 1 | 118 ± 40 | TATGGGAGCATATACTATTCTATGATATTTCTATGCAATTCAAGAGTCTA |
| SELEX 2 | 134 ± 2 | CGTCTTGCATAATATAAAATTTTGTTTTGAGTTGTAATCGAATGCGGTCT |
| SELEX 3 | 162 ± 68 | GAAGTAGCATGAACATATACCCGTTGACTCAATTTTATTTTAGTCCAACA |
| SELEX 4 | 194 ± 30 | AGCATATAGTATAAAATTATCATATTGGTGTACGTGTACGGGGTACTTTT |
| trp | 524 ± 34 | CAATTAATCATCGAACTAGTTAACTAGTACGCAAGTTCACGTAAAAAGGG |
All oligonucleotides were synthesized with the same 24-bp extensions to allow amplification of the target region. The sequence of the 5′ extension was GACCTGTGAACTGCGTAGTCCCTG. The sequence of the 3′ extension was GGAAGCTTAGACCGTCAACGTCGG. Putative SarA boxes associated with spa, fnbA, fnbB, sspA, and hla (5, 11) are shown in boldface type. The agr target corresponds to the A1-A2 binding site identified by Rechtin et al. (34). Relative locations of the cna targets are shown in Fig. 1. Sites that match the ATTTTAT motif with no more than a single mismatch are underlined, and a double underline indicates that the motif is located on the opposite strand of the DNA target.
FIG. 1.
Schematic representation of the cna promoter region. The designation of promoter elements associated with cna is based on analysis of sequence data with Omiga software (GCG). The location of each cna fragment used for EMSA is shown relative to these promoter elements. The sequence of each fragment is given in Table 1. The fragments collectively span the 150-bp region upstream of the cna translational start. RBS, ribosome binding site.
For calculations of the equilibrium dissociation constant (KD), 10 pM 32P-labeled DNA was incubated with a variable amount of SarA (range, 0 to 800 pM) in a 20-μl reaction mixture containing 10 mM HEPES (pH 7.6), 1 mM EDTA, 2 mM dithiothreitol, 50 mM KCl, 0.05% Triton X-100, and 5% glycerol. Binding reactions were equilibrated for 20 min at room temperature before electrophoresis. Bound and unbound DNA species were separated on 6% native polyacrylamide (acrylamide-bisacrylamide, 50:1) in 0.5× Tris-borate-EDTA at 200 V with the temperature maintained at 16°C by a circulating water bath. The results were quantified by phosphorimaging so that the KD for each fragment could be calculated. KD was defined as the amount of SarA required to shift 50% of the input DNA as described by Riggs et al. (36) and Hurlburt and Yanofsky (19). Competition EMSA was done the same way except that a variable amount of competitor DNA was added to the reaction mixture. All EMSA experiments were repeated at least twice. KD values are reported as the average values of the results of all experiments done with each target.
SELEX.
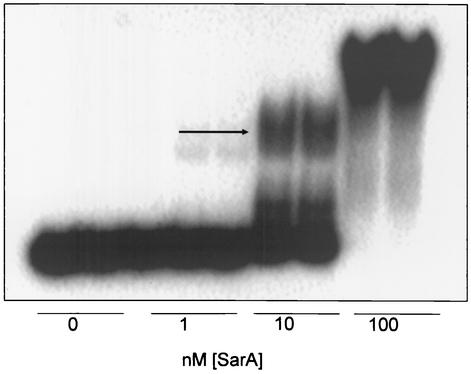
To generate the double-stranded template for SELEX, the 98-bp oligonucleotide pool containing random nucleotides at the middle 50 positions was used as a template in a primer extension reaction employing a labeled 24-bp primer (5′CCGACGTTTACGGTCTAAGCTTCC3′) complementary to the 3′ end of the 98-bp oligonucleotide. The primer was labeled with [32P]ATP and T4 polynucleotide kinase. Fifteen picomolar of labeled primer was mixed with 15 pM template, and the mixture was incubated for 5 min at 94°C, 1 min at 64°C, and 5 min at 72°C. The product was phenol extracted, ethanol precipitated, and resolved on a 12% native polyacrylamide gel electrophoresis (PAGE) gel (acrylamide-bisacrylamide, 19:1). The labeled product was excised from the gel and eluted in Tris-EDTA buffer (10 mM Tris-Cl [pH 7.5], 1 mM EDTA [pH 8.0]) overnight at 37°C. This process was followed by ethanol precipitation and quantification of DNA by spectrophotometry. EMSA was then performed with 10 nM DNA and 0, 1, 10, or 100 nM recombinant SarA. Because it shifted some, but not all, of the DNA pool (see Fig. 4), 10 nM SarA was used for all subsequent rounds of SELEX. After each round, the gel was exposed to X-ray film and the shifted protein or DNA bands were excised and eluted in Tris-EDTA buffer overnight at 37°C. A 15 pM concentration of the end-labeled primer was then used with a 15 pM concentration of an unlabeled primer complementary to the 5′ end of the 98-bp template (5′GACCTGTGAA CTGCGTAGTCCCTG3′) to amplify a pool of double-stranded DNA. Amplification was done for 1 min at 94°C, 1 min at 64°C, and 1 min at 72°C for a total of 15 cycles. The PCR product was purified as described above and used for EMSA. The process was then repeated for a total of seven rounds of SELEX.
FIG. 4.
EMSA results from first round of SELEX. A 10 nM concentration of the 32P-labeled SELEX template was incubated with increasing amounts of recombinant SarA and resolved by native PAGE. The region of the gel indicated by the arrow was excised to recover bound DNA fragments for the second round of SELEX. Ten nanomolar SarA was used in all subsequent SELEX experiments.
After the first and last rounds of SELEX, an aliquot of the amplification product was diluted to 1 pM and reamplified with unlabeled primers. The products from this reaction were cloned into pCR2.1 TOPO (Invitrogen, Carlsbad, Calif.) for DNA sequencing. Sequences obtained after the seventh round were compared by using the PILEUP program in the Genetics Computer Group (GCG) software package (13). Repetitive patterns were then identified by visual examination. Similar patterns in other clones were identified by using the GCG program FINDPATTERNS. The final alignment was then determined by using the GCG program LINEUP.
Mutagenesis of agr promoter region.
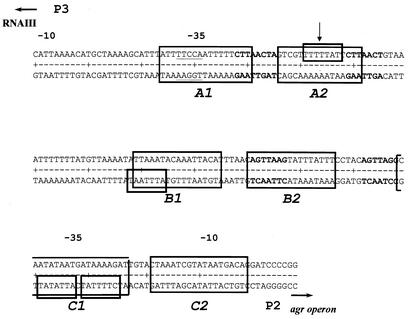
As part of ongoing experiments undertaken to further define the SarA binding site associated with the agr promoters, three sets of complementary 58-bp oligonucleotides that spanned the A1-A2 binding site defined by Rechtin et al. (34) were synthesized. In one case, the oligonucleotides corresponded precisely to the A1-A2 binding site and included the TTTTTAT motif (see Fig. 6). In the other two cases, oligonucleotides that altered a region that overlapped this motif at six of seven bases were synthesized. In one of these cases, the oligonucleotides were designed to maintain the overall G+C content by replacing TTTTTTA with AAAAAAT. In the other case, the oligonucleotides were designed to alter the G+C content by replacing TTTTTTA with CCCCCCG. Comparative EMSA experiments were done with each of these DNA targets as described above.
FIG. 6.
Putative SarA binding sites in the agr promoter region. The large boxes encompassing both strands indicate the protected regions identified by Rechtin et al. (34). The smaller boxes encompassing only one strand indicate sites that match at least six of seven bases in the ATTTTAT motif. The motifs on the upper strand are read left to right. The motifs on the lower strand are read right to left. The arrow indicates the binding motif in the A2 region that was chosen for mutagenesis. The bold nucleotides indicate the heptad repeats identified by Morfeldt et al. (28).
RESULTS
Binding of SarA to cis elements associated with cna.
Previous studies have established that sarA is the primary regulatory element controlling transcription of cna and that this regulatory effect is mediated in a direct manner via the binding of SarA to DNA sites upstream of cna (3, 17). To better define the SarA binding site associated with the cna promoter, we designed overlapping oligonucleotides that collectively span the 150 bp upstream of the cna start codon (Fig. 1 and Table 1). We also synthesized oligonucleotides that contain the putative SarA binding sites located cis to the spa, fnbA, fnbB, hla, sspA, and agr loci (Table 1). The Escherichia coli trp operon promoter-operator was used as a negative control. Binding affinity with each fragment was determined by EMSA by using recombinant SarA purified from E. coli (34).
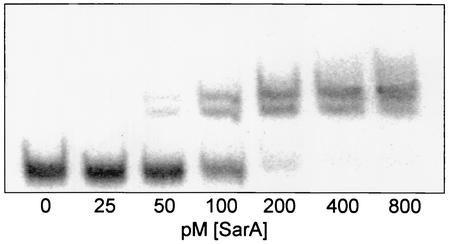
SarA bound all of the S. aureus target DNAs previously reported to contain a SarA binding site with an average KD ranging from 91 to 308 pM (Table 1). Four of the five DNAs derived from the cna promoter region bound SarA with comparable affinities. The exception was cna5, which bound SarA with an affinity similar to that of the trp promoter-operator (∼500 pM). This fragment corresponds to the first 50 bases immediately upstream of the cna translational start codon (Fig. 1). Because high-affinity binding was observed with cna fragments that do not overlap (Fig. 1), these results suggest that the region upstream of the cna promoter may contain multiple SarA binding sites. Importantly, none of the cna fragments that bound SarA with high affinity contained a SarA box as defined by Dunman et al. (14). These results are consistent with the hypothesis that SarA can bind DNA targets that do not contain a motif corresponding to a SarA box as it is currently defined. For all cases, including those of cna1 to cna4, we observed two shifted bands in our EMSA experiments (Fig. 2). This supports the hypothesis that SarA binding occurs at two half sites (34) and that binding alters the topology of the bound DNA (28, 37). The two shifted complexes presumably represent conformationally distinct complexes consisting of a single SarA dimer bound to its DNA target.
FIG. 2.
EMSA with hla promoter. A 10 pM concentration of the 32P-labeled hla template (Table 1) was incubated with increasing amounts of SarA and resolved by native PAGE. All promoters tested showed similar patterns, with two primary shifted products.
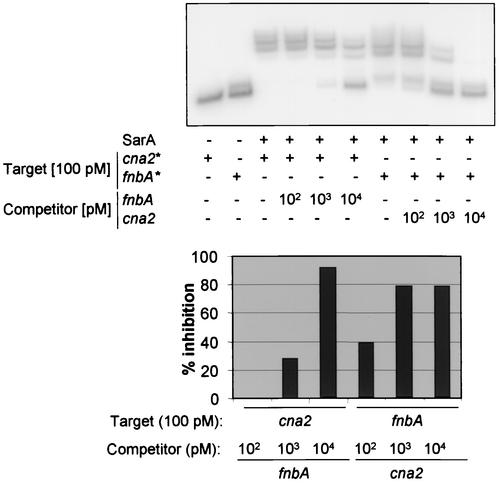
To confirm the presence of a high-affinity SarA binding site upstream of cna, we also did a competition EMSA experiment using the cna2 and fnbA fragments. When the experiment was done with 100 pM labeled cna2 (KD, ∼104 pM), competitive binding was observed only in the presence of an excess of unlabeled fnbA (Fig. 3). When the experiment was done with labeled fnbA (KD, ∼228 pM), competition was observed with an equimolar amount of unlabeled cna2 and was virtually complete with a 10-fold excess of the cna2 competitor. These results not only provide support for our KD calculations but also confirm that SarA can discriminate between alternative DNA targets.
FIG. 3.
Competitive EMSA. (Top) A 100 pM concentration of the labeled target DNA was mixed with 1,500 pM SarA in the presence of increasing amounts of unlabeled competitor. Bound and unbound DNAs were subsequently resolved by native PAGE. (Bottom) Percent inhibition based on phosphorimaging analysis of the gel shown in the top panel.
Use of SELEX to determine a consensus SarA binding site.
The DNA pool for our SELEX experiments consisted of 98-bp DNA molecules in which the 24 bases on each end were of known sequence and the central 50 bases were random. The presence of defined ends allowed us to generate 24-bp oligonucleotides that could be used as primers to amplify the bound DNA pool after each round of SELEX. Importantly, a sufficiently high concentration of SarA was capable of shifting the entire DNA pool (Fig. 4). This supports the hypothesis that SarA can bind DNA in an indiscriminate manner when present in a high concentration. A concentration of 10 nM SarA was chosen for our SELEX experiments because it shifted some, but not all, of the DNA pool in the first round of SELEX (Fig. 4). After the shifted band from the first round was excised, a sample of the bound DNA was cloned for sequencing. The remainder was amplified for use in subsequent rounds of SELEX with the 24-bp primers. After the first round of SELEX, the percent GC in 14 sequenced clones was 39% (data not shown). After seven rounds, the percent GC in 56 sequenced clones was 32% (Fig. 5A). This closely mimics the overall GC content of the S. aureus genome (2, 22). When the sequences of the 56 clones obtained after the seventh round of SELEX were aligned, we identified the 7-bp consensus ATTTTAT. Within this consensus, the degree of conservation at each position ranged from 73 to 95% (Fig. 5B). The average conservation across all seven bases in the consensus was 86%. In contrast, conservation in the flanking regions never exceeded 53%, with an average at any given position of only 37%. Within the consensus, there also appeared to be an exclusion of GC base pairs (Fig. 5B). This further emphasized the preference of SarA for AT-rich binding sites.
FIG. 5.
Consensus sequence identified by SELEX. (A) Alignment of 56 sequenced clones obtained by SELEX. The putative SarA binding motif is indicated in bold. (B) Numbers represent the percentages of the 56 sequenced clones with the indicated base at that position. Bold numbers correspond to the consensus region. The frequencies shown for regions immediately flanking the consensus are representative of the frequencies observed at more-distal regions as described in the text. This analysis was limited to those positions in which a minimum of 12 clones were represented at each position in the alignment.
To determine whether SarA recognized the SELEX templates with an affinity comparable to that of the S. aureus promoter regions previously reported to contain SarA binding motifs, four randomly selected SELEX clones were used as DNA targets for EMSA. The KD values for these clones ranged from 118 to 194 pM, which is very similar to the values obtained with DNA fragments derived from the promoter regions of other SarA-regulated genes (Table 1). Based on this finding, we searched for the ATTTTAT motif in each of these targets. When both strands were searched, allowing for no more than one mismatch, we identified this motif in every target DNA except those of fnbB, trp, and one of the five cna fragments (cna5) (Table 1). Importantly, trp and cna5 had the highest KD values of all the DNA targets we tested. However, it must also be noted that fnbB had a KD value similar to those of both fnbA and spa, both of which contained at least one site that differed from the ATTTTAT motif by no more than 1 bp (Table 1).
Mutational analysis of a SarA binding site in the agr promoter region.
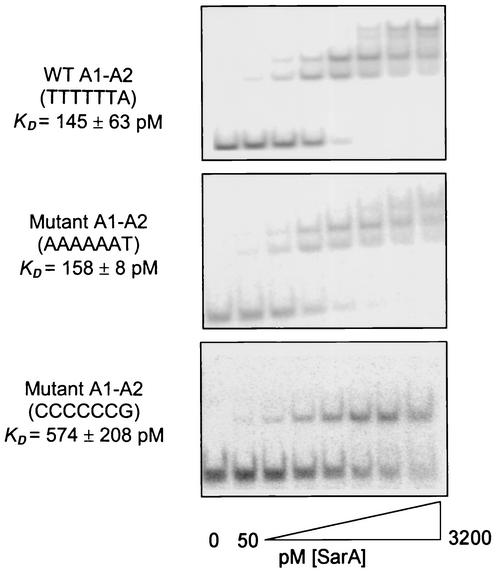
Previous DNase footprinting experiments indicated that the agr promoter region contains three SarA binding sites, each of which consists of two half sites (Fig. 6) (34). All three of these protected regions include an ATTTTAT motif that matches the consensus at no less than six of seven bases. Because it was protected with the lowest concentration of SarA, we based our EMSA experiments on the A1-A2 region (Table 1), which contains the motif TTTTTAT in the A2 half site (Fig. 6). To directly evaluate the significance of this motif, we synthesized oligonucleotides with mutations in which a region that overlaps this putative consensus at six of seven bases (TTTTTTA) was altered either in a fashion that conserved the overall GC content (AAAAAAT) or in a fashion that increased the overall GC content (CCCCCCG). The KD observed for the oligonucleotide with the conservative substitution (∼157 pM) was very similar to that observed for the wild-type A1-A2 fragment (∼145 pM) (Fig. 7). In contrast, the KD observed for the fragment with an increased GC content was increased to approximately 574 pM (Fig. 7).
FIG. 7.
EMSA analysis of wild-type and mutant SarA binding motifs. A 10 pM concentration of 32P-labeled DNA was incubated with an increasing concentration (twofold increments from 50 to 3,200 pM) of recombinant SarA and resolved by native PAGE. The upper panel illustrates the results obtained with the wild-type agr A1-A2 allele (Table 1). The middle panel illustrates the results obtained with a fragment in which the binding motif was altered in a manner that conserved the overall GC content. The bottom panel illustrates the results obtained with a fragment in which the binding motif was modified in a fashion that increased the overall GC content. WT, wild type.
DISCUSSION
Analysis of sarA mutants suggests that SarA modulates the expression of a number of S. aureus genes, including agr, clfA, clfB, geh, hla, hlb, fnbA, fnbB, spa, sspA, seb, sec, and tst (9). In most cases, it is unclear whether this occurs via a direct interaction between SarA and promoter elements of the target gene or via an indirect route involving some other regulatory factor. The laboratory of Cheung and coworkers has reported SarA binding data determined by using target DNAs derived from the agr, hla, spa, and fnbA promoter regions (11, 38). In all cases, binding was observed with a concentration of purified SarA of approximately 1.6 μM. The binding sites associated with each of these genes were also defined by DNase footprinting (10, 11, 38). In the case of the agr gene, other groups have proposed alternative binding sites. For instance, Morfeldt et al. (28) suggested on the basis of experiments done with deletion constructs that heptad repeats upstream of both the P2 and P3 promoters were necessary for the SarA-mediated regulation of agr transcription. The 26-bp binding site proposed by Chien and Cheung (10) is located between these sets of repeats. Our own DNase footprinting experiments identified three protected regions, each of which contains two half sites (34). The two regions that were protected at the lowest concentration of SarA (A1-A2 and B1-B2) overlapped with at least one of the heptads. The A1-A2 site is completely contained within the minimum region that Morfeldt et al. (28) found was required for the SarA-mediated regulation of RNAIII production (Fig. 6). The 26-bp protected region identified by Chien and Cheung (10) overlapped with the B1 half site but was not contained within the required region identified by Morfeldt et al. (28). Direct comparisons by EMSA (34) demonstrated that SarA bound the A1-A2 and B1-B2 sites with greater affinity than fragments containing the heptad repeats and that all of these fragments bound SarA with greater affinity than the 26-bp region identified by Chien and Cheung (10).
The 26-bp consensus defined by Chien et al. (11) was based on alignment of the promoter regions of six genes (agr, hla, spa, fnbA, fnbB, and sec) previously reported to be regulated at the transcriptional level by SarA. In this alignment, 19 bases were conserved in four of six genes, while 6 bases were conserved in three of six genes. Similarly, Chan and Foster (5) aligned the promoter regions of seven genes (tst, spa, agr [P3 promoter], hlb, seb, sspA, and hla) and identified a 29-bp consensus binding site. Of the 29 bases in this consensus, 5 were invariant, 4 had <50% identity, and the remaining 20 had >50% identity. The two alignments included three common genes (agr, hla, and spa). In two of these genes (agr and spa), the putative binding sites overlapped and included identical bases at 22 positions. In the other gene (hla), the putative binding sites identified by Chien et al. (11) and Chan and Foster (5) were in two entirely different regions. More specifically, the putative binding site used to derive the consensus defined by Chan and Foster (5) is located 270 bp upstream of the hla transcriptional start site, while the binding site used in the alignment of Chien et al. (11) is located only 32 bases upstream of the transcriptional start. Nevertheless, the two consensus sequences derived from each alignment included a 19-bp region that was identical at 14 positions. However, of these 19 bases, 18 were either A or T (5, 11), and the significance of these must be interpreted with caution, given the AT-rich nature of the S. aureus genome.
Dunman et al. (14) found that mutation of sarA altered the expression of 120 genes or operons. Using the 26-bp consensus sequence discussed above, and allowing for up to seven mismatches (Ellen Murphy, personal communication), only 19 of these genes were found to contain a putative SarA box upstream of the relevant gene or operon. This would suggest that transcription of at least some target genes may be mediated by SarA indirectly via an interaction between SarA and other regulatory elements. Alternatively, the current definition of a SarA box may be inadequate. Our experiments demonstrating that SarA binds the cna promoter region despite the absence of a SarA box (3) are suggestive of the latter explanation. It should also be noted that with the exception of agr, hla, fnbA, and spa (11, 34, 38), a detailed characterization of the SarA binding site(s) associated with individual target genes, including most of those used in the alignments discussed above, has not been reported. Indeed, while Chien et al. (11) included the fnbB promoter region in their alignment, Wolz et al. (38) concluded that mutation of sarA has no impact on fnbB promoter activity.
In an attempt to clarify the situation and further define the consensus SarA binding site, we performed quantitative DNA binding assays with purified SarA and DNA fragments previously reported to contain a SarA binding site. We also used the unbiased approach of SELEX analysis in an effort to further define the optimal binding site. The results obtained with each approach were consistent in that SarA bound the SELEX templates with an affinity similar to that observed with other SarA targets, including agr and cna. Alignment of 56 SELEX templates revealed the 7-bp consensus ATTTTAT. As noted above, it is difficult to ascribe much significance to a short, AT-rich consensus in a genome of such low GC content. However, of the 56 sequenced templates, 14 contained a region that matched this consensus perfectly and 32 contained a region that differed at only one position. Moreover, alignment of all 56 clones allowed us to identify 69 contiguous positions that were represented in at least 12 clones (Fig. 5A). The percent conservation at 62 of these bases averaged 37% and never exceeded 53%. In contrast, the percent conservation at the remaining seven positions averaged 86% and was never less than 73%. This difference, together with the observation that the seven bases that were most highly conserved among the 69 positions were contiguous in all 56 clones, strongly suggests that SarA prefers to bind DNA fragments that include this motif. Additionally, we found that this motif was present in all of the previously reported SarA targets with the exception of fnbB and one of the five cna fragments (cna5). Importantly, these two targets bound SarA with relatively low affinity. In fact, the only targets that bound SarA with a higher KD were spa and the trp promoter region included as a negative control. Interestingly, Arvidson and Tegmark (1) recently suggested that the SarA-mediated control of spa transcription occurs via an indirect mechanism. We also examined the SarA-regulated genes identified by Dunman et al. (14) and found that at least 72 of the 101 genes or operons that did not have a SarA box had a region that matched our ATTTTAT motif at no fewer than six of seven bases within 150 bp of the relevant translational start codon (data not shown). This suggests that a number of the genes previously thought to be indirectly regulated by SarA may in fact be regulated by a direct interaction between SarA and cis elements upstream of the target gene.
Finally, we also demonstrated that mutation of the ATTTTAT motif in at least one target (agr A1-A2) resulted in a reduced capacity to bind SarA. Mutation of this motif in a fashion that changed the overall GC content resulted not only in an increased KD but also in a change from two shifted complexes to one. This is reminiscent of the EMSA data from a previous work (34), in which DNA fragments with two high-affinity half sites generated two shifted complexes while those with only one site had only one shifted complex. The simplest explanation is that if SarA can bind the target firmly with both subunits of the dimer, it can impose conformational changes on the DNA. This notion is complicated by our SELEX results, in which each DNA fragment had only one consensus site. However, inspection of the regions flanking the consensus site reveals the possibility of lower homology or cryptic binding sites. This is very similar to the results of SELEX selection for the trp repressor of E. coli (12). In that work, one high-homology site was identified per DNA fragment selected and cryptic second sites were observed in the flanking regions.
Whether the putative binding site identified in these experiments will have predictive value in identifying genes that are directly regulated by SarA remains unclear. It will almost certainly depend on the location of the binding site relative to specific promoter elements. For instance, most of the putative binding sites associated with SarA-regulated genes overlap the −10 or −35 regions of the relevant promoter (5, 11). This is consistent with our demonstration that three of the four cna targets that bound SarA with high affinity overlap either the −10 or −35 regions of the cna promoter. This finding suggests that SarA may repress transcription of at least some target genes, including cna, by virtue of its ability to compete with RNA polymerase for binding sites. However, there is also evidence that SarA binding and its effect on transcription may depend on more general characteristics that are defined by local DNA topology. Indeed, Fujimoto et al. (15) suggested that SarA may preferentially bind bent DNA, and Morfeldt et al. (28) found that transcription of agr occurred in a SarA-independent manner when three bases located between the −10 and −35 regions of the agr P3 promoter were removed. Although our computer modeling suggests that some of the promoter regions we examined may be bent (data not shown), these regions were not bound with significantly higher affinity than other targets that were not predicted to bend. Still, an important role for DNA topology cannot be ruled out, especially since AT-rich sequences are often bent (16), and it is clear that SarA has a preference for AT-rich binding sites. Indeed, it seems apparent from both our studies and those of other investigators that SarA can bind essentially any DNA, particularly when mixed in relatively high concentrations with single DNA targets. This promiscuous binding is evident in the fact that SarA bound the trp operator with a KD of the same order of magnitude as that observed with high-affinity staphylococcal targets. This suggests that the more relevant issue is identification of those targets that are preferentially bound by SarA even when present in a complex mixture of potential targets. We believe that the SELEX approach we report addresses this issue and that the results we present collectively support the hypothesis that the preferential SarA binding site includes a motif identical to, or at least very similar to, the ATTTTAT consensus defined in this work.
Acknowledgments
This work was supported by grant AI43356 from the National Institute of Allergy and Infectious Diseases. K.M.S. was supported by a predoctoral fellowship from the American Heart Association and by funds supplied by the UAMS Committee for Allocation of Graduate Student Research Funds (CAGSRF).
We thank Piotr Czernik for his help with SELEX alignments.
REFERENCES
- 1.Arvidson, S., and K. Tegmark. 2001. Regulation of virulence determinants in Staphylococcus aureus. Int. J. Med. Microbiol. 291:159-170. [DOI] [PubMed] [Google Scholar]
- 2.Baba, T., F. Takeuchi, M. Kuroda, H. Yuzawa, K. Aoki, A. Oguchi, Y. Nagai, N. Iwama, K. Asano, T. Naimi, H. Kuroda, L. Cui, K. Yamamoto, and K. Hiramatsu. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819-1827. [DOI] [PubMed] [Google Scholar]
- 3.Blevins, J., A. F. Gillaspy, T. M. Rechtin, B. K. Hurlburt, and M. S. Smeltzer. 1999. The staphylococcal accessory regulator (sar) represses transcription of the Staphylococcus aureus collagen adhesin gene (cna) in an agr-independent manner. Mol. Microbiol. 33:317-326. [DOI] [PubMed] [Google Scholar]
- 4.Blevins, J. S., K. E. Beenken, M. O. Elasri, B. K. Hurlburt, and M. S. Smeltzer. 2002. Strain-dependent differences in the regulatory roles of sarA and agr in Staphylococcus aureus. Infect. Immun. 70:470-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan, P. F., and S. J. Foster. 1998. Role of SarA in virulence determinant production and environmental signal transduction in Staphylococcus aureus. J. Bacteriol. 180:6232-6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheung, A. L., and S. J. Projan. 1994. Cloning and sequencing of sarA of Staphylococcus aureus, a gene required for the expression of agr. J. Bacteriol. 176:4168-4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheung, A. L., M. G. Bayer, and J. H. Heinrichs. 1997. sar genetic determinants necessary for transcription of RNAII and RNAIII in the agr locus of Staphylococcus aureus. J. Bacteriol. 179:3963-3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheung, A. L., K. Eberhardt, and J. H. Heinrichs. 1997. Regulation of protein A synthesis by the sar and agr loci of Staphylococcus aureus. Infect. Immun. 65:2243-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheung, A. L., and G. Zhang. 2002. Global regulation of virulence determinants in Staphylococcus aureus by the SarA protein family. Front. Biosci. 7:1825-1842. [DOI] [PubMed] [Google Scholar]
- 10.Chien, Y., and A. L. Cheung. 1998. Molecular interactions between two global regulators, sar and agr, in Staphylococcus aureus. J. Biol. Chem. 273:2645-2652. [DOI] [PubMed] [Google Scholar]
- 11.Chien, Y., A. C. Manna, S. J. Projan, and A. L. Cheung. 1999. SarA, a global regulator of virulence determinants in Staphylococcus aureus, binds to a conserved motif essential for sar-dependent gene regulation. J. Biol. Chem. 274:37169-37176. [DOI] [PubMed] [Google Scholar]
- 12.Czernik, P. J, D. S. Shin, and B. K. Hurlburt. 1994. Functional selection and characterization of DNA binding sites for trp repressor of Escherichia coli. J. Biol. Chem. 269:27869-27875. [PubMed] [Google Scholar]
- 13.Devereaux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dunman, P. M., E. Murphy, S. Haney, D. Palacios, G. Tucker-Kellogg, S. Wu, E. L. Brown, R. J. Zagursky, D. Shlaes, and S. J. Projan. 2001. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J. Bacteriol. 183:7341-7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujimoto, D. F., E. W. Brunskill, and K. W. Bayles. 2000. Analysis of genetic elements controlling Staphylococcus aureus lrgAB expression: potential role of DNA topology in SarA regulation. J. Bacteriol. 182:4822-4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gabrielian, A., A. Simoncsits, and S. Pongor. 1996. Distribution of bending propensity in DNA sequences. FEBS Lett. 393:124-130. [DOI] [PubMed] [Google Scholar]
- 17.Gillaspy, A. F., C. Y. Lee, S. Sau, A. L. Cheung, and M. S. Smeltzer. 1998. Factors affecting the collagen binding capacity of Staphylococcus aureus. Infect. Immun. 66:3170-3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heinrichs, J. H., M. G. Bayer, and A. L. Cheung. 1996. Characterization of the sar locus and its interaction with agr in Staphylococcus aureus. J. Bacteriol. 178:418-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hurlburt, B. K., and C. Yanofsky. 1990. Enhanced operator binding by trp superrepressors of E. coli. J. Biol. Chem. 265:7853-7858. [PubMed] [Google Scholar]
- 20.Ji, G., R. C. Beavis, and R. Novick. 1995. Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc. Natl. Acad. Sci. USA 92:12055-12059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karlsson, A., P. Saravia-Otten, K. Tegmark, E. Morfeldt, and S. Arvidson. 2001. Decreased amounts of cell wall-associated protein A and fibronectin-binding proteins in Staphylococcus aureus sarA mutants due to up-regulation of extracellular proteases. Infect. Immun. 69:4742-4748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 23.Lina, G., S. Jarraud, J. Guangyong, T. Greenland, A. Pedraza, J. Etienne, R. P. Novick, and F. Vandenesch. 1998. Transmembrane topology and histidine protein kinase activity of AgrC, the agr signal receptor in Staphylococcus aureus. Mol. Microbiol. 28:655-662. [DOI] [PubMed] [Google Scholar]
- 24.Lindsay, J. A., and S. J. Foster. 1999. Interactive regulatory pathways control virulence determinant production and stability in response to environmental conditions in Staphylococcus aureus. Mol. Gen. Genet. 262:323-331. [DOI] [PubMed] [Google Scholar]
- 25.Lowy, F. D. 1998. Staphylococcus infections. N. Engl. J. Med. 339:520-532. [DOI] [PubMed] [Google Scholar]
- 26.Morfeldt, E., L. Janzon, S. Arvidson, and S. Lofdahl. 1988. Cloning of a chromosomal locus (exp) which regulates the expression of several exoprotein genes in Staphylococcus aureus. Mol. Gen. Genet. 211:435-440. [DOI] [PubMed] [Google Scholar]
- 27.Morfeldt, E., B. Taylor, A. von Gabain, and S. Arvidson. 1995. Activation of alpha-toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNA III. EMBO J. 14:4569-4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morfeldt, E., K. Tegmark, and S. Arvidson. 1996. Transcriptional control of the agr-dependent virulence gene regulator, RNAIII, in Staphylococcus aureus. Mol. Microbiol. 21:1227-1237. [DOI] [PubMed] [Google Scholar]
- 29.Novick, R. P., S. J. Projan, J. Kornblum, H. F. Ross, G. Ji, B. Kreiswirth, F. Vandenesch, and S. Moghazeh. 1995. The agr P2 operon: an autocatalytic sensory transduction system in Staphylococcus aureus. Mol. Gen. Genet. 248:446-458. [DOI] [PubMed] [Google Scholar]
- 30.Novick, R. P., H. F. Ross, S. J. Projan, J. Kornblum, B. Kreiswirth, and S. Moghazeh. 1993. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 12:3967-3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Otto, M. 2001. Staphylococcus aureus and Staphylococcus epidermidis peptide pheromones produced by the accessory gene regulator agr system. Peptides 22:1603-1608. [DOI] [PubMed] [Google Scholar]
- 32.Peng, H.-L., R. P. Novick, B. Kreiswirth, J. Kornblum, and P. Schlievert. 1988. Cloning, characterization, and sequencing of an accessory gene regulator (agr) in Staphylococcus aureus. J. Bacteriol. 170:4365-4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Projan, S. J., and R. P. Novick. 1997. The molecular basis of pathogenicity, p. 55-81. In K. B. Crossley and G. L. Archer (ed.), The staphylococci in human disease. Churchill Livingstone, New York, N.Y.
- 34.Rechtin, T. M., A. F. Gillaspy, M. A. Schumacher, R. G. Brennan, M. S. Smeltzer, and B. K. Hurlburt. 1999. Characterization of the SarA virulence gene regulator of Staphylococcus aureus. Mol. Microbiol. 33:307-316. [DOI] [PubMed] [Google Scholar]
- 35.Rescei, P., B. Kreiswirth, M. O'Reilly, P. Schlievert, A. Gruss, and R. P. Novick. 1986. Regulation of exoprotein gene expression in Staphylococcus aureus by agr. Mol. Gen. Genet. 202:58-61. [DOI] [PubMed] [Google Scholar]
- 36.Riggs, A. D., H. Suzuki, and S. Bourgeois. 1970. lac repressor-operator interaction: equilibrium studies. J. Mol. Biol. 48:67-83. [DOI] [PubMed] [Google Scholar]
- 37.Schumacher, M. A., B. K. Hurlburt, and R. Brennan. 2001. Crystal structures of SarA: a pleiotropic regulator of virulence in S. aureus. Nature 409:215-219. [DOI] [PubMed] [Google Scholar]
- 38.Wolz, C., P. Pohlmann-Dietze, A. Steinhuber, Y.-T. Chien, A. Manna, W. van Wamel, and A. Cheung. 2000. agr-independent regulation of fibronectin-binding protein(s) by the regulatory locus sar in Staphylococcus aureus. Mol. Microbiol. 36:230-243. [DOI] [PubMed] [Google Scholar]