Abstract
The activation of σG, a transcription factor, in Bacillus subtilis is coupled to the completion of engulfment during sporulation. SpoIIAB, an anti-sigma factor involved in regulation of σF, is also shown to form a complex with σG in vitro. SpoIIAA, the corresponding anti-anti-sigma factor, can disrupt the SpoIIAB:σG complex, releasing free σG. The data suggest the existence of an as-yet-unknown mechanism to keep σG inactive prior to engulfment.
Starvation induces the gram-positive bacterium Bacillus subtilis to initiate a simple, two-cell developmental process that results in the formation of dormant spores. Early in sporulation, the developing cell divides asymmetrically to produce a smaller compartment, the prespore, which becomes the spore, and a larger compartment, the mother cell, which participates in the maturation of the spore and finally lyses to release it. The different developmental fates of the two cells are governed by the sequential activation of four sporulation-specific transcription factors, beginning with σF in the prespore and then σE in the mother cell followed by σG and σK in the prespore and mother cell, respectively. To ensure the correct sequence of morphological events, the activation of each sigma factor is coupled to morphogenesis and/or to events occurring in the opposite cell (reviewed in references 11 and 22).
The late prespore-specific σ factor, σG, is regulated at at least three levels. First, its gene (sigG or spoIIIG) is transcribed from a promoter recognized by the first prespore-specific sigma factor, σF (and later by σG itself), thus restricting its localization to the prespore compartment (24). Second, unlike other σF-dependent genes, sigG is not transcribed in the presence of mutations in the spoIIG gene (20), which encodes the first mother cell-specific sigma factor, σE. Therefore, sigG transcription is also dependent on an as-yet-unidentified signal transduction pathway of which at least one component is expressed in the mother cell. The third regulatory mechanism exerted over σG acts at the level of protein activity. The sigG gene begins to be transcribed approximately 120 min after the initiation of sporulation. However, σG-dependent gene expression does not begin until 30 min later (20). Mutations in several different genes, including spoIIB, spoIID, spoIIM, spoIIIA, and spoIIIJ, prevent transcription of σG-dependent genes without affecting σG synthesis, implying that their products play a role in σG activation (1, 8, 9, 12, 15, 20, 21, 23). Three of the proteins, SpoIIB, SpoIID, and SpoIIM, are required for prespore engulfment, suggesting a link between activation of σG and the completion of engulfment (1, 21, 23). Little is known about how σG is held inactive prior to engulfment, but it has been suggested to involve the anti-sigma factor SpoIIAB (9). SpoIIAB is one of the proteins that regulate σF activation; it binds to σF and thereby prevents it from interacting with core RNA polymerase (2, 7, 16). In turn, SpoIIAB is antagonized by the anti-anti-sigma factor SpoIIAA. SpoIIAB and SpoIIAA can interact in two different ways. In the presence of ADP, the two proteins form a complex, resulting in the release of active σF (2, 5). However, SpoIIAB is also a protein kinase which (in the presence of ATP) phosphorylates SpoIIAA on a specific serine residue, rendering the product (SpoIIAA-P) unable to bind to SpoIIAB or to react with SpoIIAB:σF complexes (5, 6, 13, 16, 18). There is some evidence that SpoIIAB also regulates σG activity. Constitutive expression of SpoIIAB was found to repress σG activity in cells where σG was expressed from a σF-independent promoter (10). Biochemical cross-linking experiments using radiolabeled crude extracts from Escherichia coli strains overexpressing SpoIIAB and σG also indicated an interaction between the two proteins (9). Furthermore, σG activity is reduced drastically in a spoIIIA mutant that fails to degrade SpoIIAB in the prespore (9). The requirement for spoIIIA is bypassed partially by a σG mutant that is impaired in its interaction with SpoIIAB (9).
In this report, we show that purified σG and SpoIIAB proteins form a nucleotide-dependent complex, although the interaction is much weaker than that of σF and SpoIIAB. Furthermore, we demonstrate that purified SpoIIAA efficiently disrupts the SpoIIAB:σG complex, thereby releasing σG. Taken together, the data suggest that SpoIIAB interacts with σG in the same way as with σF and that there may be another mechanism to keep σG inactive at a time when σF is active.
Purified σG and SpoIIAB form a complex in the presence of nucleotide.
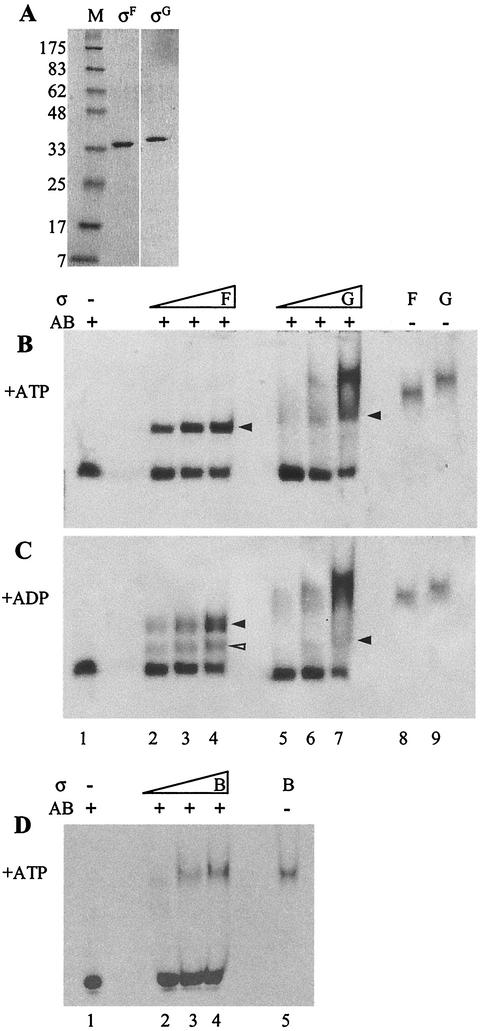
σG was purified using the IMPACT T7 system (New England BioLabs). The sigG gene of B. subtilis was amplified by PCR, and the product was cloned into the expression vector pTYB1, thereby fusing the sigG gene to an intein-chitin binding domain under the control of the IPTG-inducible T7 promoter. The fusion protein was overproduced in E. coli C41(DE3) (17) and purified on a chitin column as described in the New England BioLabs protocol. σG was purified further on a Superdex 75 gel filtration column equilibrated in 50 mM Tris-HCl (pH 8)-300 mM NaCl-1 mM dithiothreitol(DTT)-1 mM EDTA to remove copurified proteins. Results with the purified protein are shown in Fig. 1A. Using the IMPACT T7 system followed by a Superdex 75 gel filtration column, we also purified σF as a control (Fig. 1A) (I. Lucet, unpublished data). To examine whether σG forms a complex with SpoIIAB as σF does, we incubated increasing amounts of σG or σF (1, 2, or 4 μM) with purified SpoIIAB (2 μM) in a 30-μl mixture containing 50 mM Tris-HCl (pH 7.5), 50 mM KCl, 1 mM DTT, 3 mM MgCl2, and either ATP or ADP (1 mM) on ice for 30 min. The mixtures were subjected to 10% nondenaturing polyacrylamide gel electrophoresis (PAGE) with the appropriate nucleotide in the running buffer. The proteins were visualized by Coomassie blue staining. As shown in Fig. 1B, in the presence of ATP the SpoIIAB band decreased in intensity with increasing amounts of sigma factor and, in parallel, SpoIIAB(ATP):σF (lanes 2 to 4) and SpoIIAB(ATP):σG (lanes 5 to 7) complexes appeared that ran with a mobility intermediate between that of SpoIIAB and σF or σG. In contrast to the SpoIIAB(ATP):σF complexes, SpoIIAB(ATP):σG complexes ran in a smear rather than in a sharp band, suggesting that the complexes dissociated to some extent during the electrophoresis. In the presence of ADP, only a weak band corresponding to SpoIIAB(ADP):σG complexes was detected, suggesting that the complexes are unstable and dissociate rapidly during electrophoresis (Fig. 1C, lanes 5 to 7). In contrast, SpoIIAB and σF did form complexes in the presence of ADP (Fig. 1C, lanes 2 to 4), although the bands had a lower intensity than those formed in the presence of ATP. It was shown previously that SpoIIAB(ADP):σF complexes are less stable than those formed with ATP (14). A second band running just above the SpoIIAB band was also seen (open arrow head). This could be a monomer of SpoIIAB interacting with σF instead of a dimer. No complexes were formed in the absence of any nucleotide (data not shown), showing that nucleotide is required for this interaction just as it is for the interaction of σF with SpoIIAB (6). SpoIIAB was also incubated with purified σB (1, 2, or 4 μM), a closely related sigma factor of B. subtilis, which is known to be regulated by an anti-sigma factor that is similar to SpoIIAB (3). As shown in Fig. 1D (lanes 2 to 4), the two proteins did not form a detectable complex, demonstrating that the binding of SpoIIAB to σF and σG is specific. These results strongly suggested that in vitro, SpoIIAB binds to σG in the same nucleotide-dependent manner as that with which it binds to σF, although it appeared that the SpoIIAB:σG complexes are less stable on nondenaturing PAGE than SpoIIAB:σF complexes are. The data were also in agreement with experiments using chemical cross-linking of crude extracts of E. coli designed to express SpoIIAB and σG (9) that showed nucleotide-dependent binding between SpoIIAB and σG. In addition, it is apparent from the recently solved crystal structure of a SpoIIAB2:σF complex that the region of σF that interacts is highly conserved in σG (4).
FIG. 1.
SpoIIAB forms a complex with σG in the presence of nucleotide. (A) Purified σG and σF. Fractions containing the proteins were pooled after gel filtration and run on sodium dodecyl sulfate-12% polyacrylamide gels. M, molecular weight markers. (B and C) Increasing concentrations (1, 2, and 4 μM) of σF (lanes 2 to 4) or σG (lanes 5 to 7) were incubated with purified SpoIIAB (2 μM) in the presence of ATP (B) or ADP (C). (D) Increasing concentrations (1, 2, and 4 μM) of purified σB (lanes 2 to 4) were incubated with purified SpoIIAB (2 μM) in the presence of ATP. Running positions of the purified proteins alone are shown as follows: SpoIIAB (panels B to D, lanes 1), σF (panels B and C, lanes 8), σG (panels B and C, lanes 9), and σB (panel D, lane 5). Reactions were analyzed on 10% nondenaturing PAGE with the appropriate nucleotide added to the running buffer and were visualized by Coomassie blue staining. Complexes formed by σ and SpoIIAB are indicated by arrowheads.
One possible mechanism to ensure the inhibition of σG under conditions in which σF is fully active would be for SpoIIAB to have different affinities for σF and σG so that when σG is produced SpoIIAB binds to it in preference to σF. However, the results described above suggested that the interaction with σG was in fact the weaker of the two. To confirm this finding, surface plasmon resonance was used to study the interaction between SpoIIAB and σG. Surface plasmon resonance has been used previously to look at the SpoIIAB-σF interaction (14). SpoIIAB (the ligand) was dialyzed overnight at 4°C into phosphate-buffered saline to which 1 mM DTT had been added and was immobilized onto a matrix in the flow cell of a sensor chip (CM5; Biacore), as described in reference 14. Purified σF and σG (the analytes) were dialyzed into binding buffer (50 mM Tris-HCl [pH 7.5], 50 mM KCl, 1 mM DTT, 3 mM MgCl2) at 4°C overnight. Protein concentrations were determined by running samples on sodium dodecyl sulfate- PAGE gels stained with Sypro Orange (Amersham Bioscience) and comparing them with molecular weight markers of known concentrations. The dialyzed proteins were then diluted in binding buffer supplemented with 1 mM ATP to concentrations ranging from 0.1 to 5 μM. Using the Kinject function with a dissociation time of 150 ms, samples (20 μl) (containing 0.25 μM, 0.5 μM, 1 μM, and 2.5 μM protein) were injected into the flow cell with a flow rate of 20 μl min−1 in binding buffer containing ATP. After injection and dissociation, any undissociated analyte was removed by injection of 0.75 M NaCl in binding buffer without nucleotide until the sensorgram returned to the preanalyte baseline. Interactions with the ligand can be detected by an optical change at the gold surface onto which the matrix is attached. Dissociation from the ligand can also be measured through a reversal of the optical change once the flow of analyte has stopped. Analysis of the data was carried out using BIAevaluation software (Biacore); the sensorgram obtained from a control flow cell with no protein was subtracted from that obtained when the same concentration of σ factor was passed through the flow cell containing SpoIIAB. The resulting curves for at least two different concentrations of σ factor were then used to calculate the dissociation constant (Kd) for each σ factor. For the σF-SpoIIAB interaction, the Kd was found to be 8 nM, which is comparable to the value of 14 nM obtained previously (14). However, for the σG:SpoIIAB complex, a Kd value of 87 nM was obtained. Therefore, we can conclude that σG binds more weakly to SpoIIAB than does σF. The weaker interaction between the two proteins is probably responsible for the partial dissociation of the complexes during electrophoresis shown in Fig. 1. The fact that SpoIIAB forms a more stable complex with σF than with σG shows that the opposing regulation mechanisms of σG and σF activities are not modulated simply by different affinities of the complexes.
SpoIIAA dissociates the SpoIIAB:σG complexes.
Another possible explanation for a mechanism by which σG could be held inactive by SpoIIAB at a time when σF is active is that SpoIIAA is unable to interact with and dissociate the SpoIIAB:σG complex. To test this possibility, we used fluorescence spectroscopy to observe complex formation between σG and SpoIIAB in real time (Fig. 2; results for three independent experiments are shown). This technique takes advantage of the fact that neither SpoIIAB nor SpoIIAA contains the highly fluorescent amino acid tryptophan but σG contains a single tryptophan residue (W198). Tryptophan fluorescence can be selectively excited at wavelengths of around 290 nm. Therefore, using fluorescence spectroscopy we were able to observe changes in the chemical environment of the single tryptophan residue in σG. Exciting σG at 290 nm gave a maximum fluorescence emission at 350 nm; hence, this wavelength was used to observe changes in fluorescence intensity. Control experiments confirmed that SpoIIAA and SpoIIAB exhibited very weak fluorescence and that this fluorescence was unchanged by the addition of ATP. The fluorescence of σG by itself changed only slightly when ATP was added. For Fig. 2, appropriate corrections were made to account for these fluorescence values.
FIG. 2.
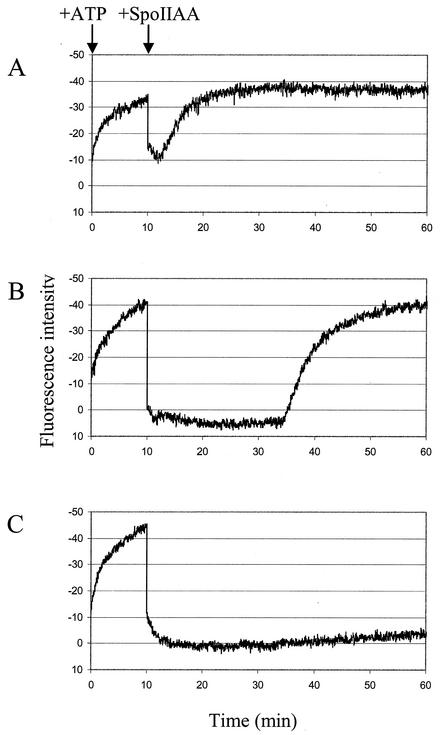
SpoIIAA induces the release of σG from SpoIIAB:σG complexes. Fluorescence spectroscopy was used to measure changes in the fluorescence intensity of the single tryptophan in σG. The results of three independent experiments are shown in panels A to C. Purified σG (0.8 μM) and SpoIIAB (1.25 μM) were mixed, and at time zero, ATP (100 μM) was added to the solution and the fluorescence intensity of the sample was determined. Different concentrations of SpoIIAA (5 μM [A], 10 μM [B], and 15 μM [C]) were added to the samples 10 min later.
Solutions of σG (0.8 μM) and SpoIIAB (1.25 μM) were preequilibrated at 25°C for 5 min in 50 mM Tris-HCl (pH 7.5)-50 mM KCl-1 mM DTT-3 mM MgCl2. The average basal fluorescence intensity was determined and set to 0. At time zero, ATP (100 μM) was added to the reaction mixture, which immediately resulted in a substantial decrease in fluorescence intensity (Fig. 2A to C). Since this decrease in fluorescence intensity was much larger than that seen when ATP was added to σG alone, it can be attributed to changes in the chemical environment of the tryptophan in σG on binding to SpoIIAB. Thus, these results again show that σG and SpoIIAB interact with each other and that the binding occurs rapidly. Presumably, the tryptophan (W198) in σG is either directly involved in binding to SpoIIAB or it moves as a result of the interaction. The crystal structure of the SpoIIAB:σF complex was solved recently; however, the location of the corresponding tryptophan (W190) in σF is still unknown, as most of the σF protein was disordered and the fold could not be determined (4).
Using this system we were then able to determine whether SpoIIAA was able to disrupt the complex. At 10 min, the complex was challenged with 5 (Fig. 2A), 10 (Fig. 2B), and 15 (Fig. 2C) μM SpoIIAA. An excess of SpoIIAA was required to mimic the effect of the phosphatase SpoIIE, which replenishes the pool of SpoIIAA in vivo by dephosphorylation of SpoIIAA-P. Control experiments omitting each of the components in turn showed that the changes in fluorescence are due to effects on the tryptophan fluorescence of σG and therefore to changes in the level of the SpoIIAB(ATP):σG complex (data not shown). The addition of SpoIIAA caused a large, immediate increase in fluorescence intensity, which may be attributable to the release of σG from SpoIIAB. With 10 and 15 μM SpoIIAA, the values of the fluorescence intensity fell to the basal level observed for σG alone, indicating that SpoIIAA disrupts all of the SpoIIAB(ATP):σG complexes. Addition of 5 μM SpoIIAA led to a smaller increase in fluorescence intensity, suggesting that this concentration of SpoIIAA was not enough to disrupt all of the complexes.
After the initial increase in fluorescence, the reaction mixture containing 5 μM SpoIIAA gradually rose again to reach the level attained before the addition of the SpoIIAA. A similar rise was seen, albeit after a long delay, with 10 μM SpoIIAA (about 25 min) and 15 μM SpoIIAA (data not shown). We assume that these changes can be attributed to the phosphorylation of SpoIIAA by SpoIIAB (2, 5, 6, 16) and that the reaction proceeds in the presence of ATP until all of the SpoIIAA is used up; at this point the SpoIIAB is free to rebind to the sigma factor. (The experiments were repeated several times, giving similar results.) These results showed that SpoIIAA can efficiently disrupt SpoIIAB:σG complexes, releasing free σG, just as it disrupts SpoIIAB:σF complexes (2, 5, 6, 16).
Previous work has shown that SpoIIAB is capable of regulating σG activity under some circumstances (9, 10). However, three lines of evidence now suggest that SpoIIAB alone is not normally responsible for the temporal control of σG activity until after the completion of engulfment. First, we have shown that binding of SpoIIAB to σG is weaker than that to σF. Thus, in the presence of a mixture of σF and σG, binding to σF would be strongly favored, so σG would be released preferentially over σF. Second, we have shown that SpoIIAB:σG complexes are dissociated efficiently by the presence of nonphosphorylated SpoIIAA. Therefore, the same mechanism that helps to release σF activity soon after septation in the prespore, which involves formation of SpoIIAA by the action of SpoIIE phosphatase, would also promote the release of σG at that time. Third, the amount of free SpoIIAB (at the time σG begins to be synthesized) is probably small, as it has been shown recently that SpoIIAB is selectively degraded in the prespore (19). So far, we cannot exclude the possibility that another factor is required for the interaction of SpoIIAB and σG which then enables SpoIIAB to participate in the regulation of σG activation in vivo and which is missing from our in vitro studies.
Taken together, all of these results suggest that SpoIIAB may not be the primary effector responsible for temporal control over σG activation and that at least one other factor remains to be discovered. The challenge now is to identify this putative factor so that the mechanism responsible for developmental regulation of σG activity can finally be resolved.
Acknowledgments
We thank I. Lucet for her help with the Biacore and Helen Prescott for technical assistance.
This work was supported by grants from the Biotechnology and Biological Sciences Research Council (BBSRC) (to M.D.Y.) and Medical Research Council (MRC) (to J.E.). L.E. was the recipient of a BBSRC postgraduate studentship.
REFERENCES
- 1.Abanes-De Mello, A., Y. L. Sun, S. Aung, and K. Pogliano. 2002. A cytoskeleton-like role for the bacterial cell wall during engulfment of the Bacillus subtilis forespore. Genes Dev. 16:3253-3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alper, S., L. Duncan, and R. Losick. 1994. An adenosine nucleotide switch controlling the activity of a cell type-specific transcription factor in B. subtilis. Cell 77:195-205. [DOI] [PubMed] [Google Scholar]
- 3.Benson, A. K., and W. G. Haldenwang. 1993. Bacillus subtilis σB is regulated by a binding protein (RsbW) that blocks its association with core RNA polymerase. Proc. Natl. Acad. Sci. USA 90:2330-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell, E. A., S. Masuda, J. L. Sun, O. Muzzin, C. A. Olson, S. Wang, and S. A. Darst. 2002. Crystal structure of the Bacillus stearothermophilus anti-sigma factor SpoIIAB with the sporulation sigma factor σF. Cell 108:795-807. [DOI] [PubMed] [Google Scholar]
- 5.Diederich, B., J. F. Wilkinson, T. Magnin, M. Najafi, J. Errington, and M. D. Yudkin. 1994. Role of interactions between SpoIIAA and SpoIIAB in regulating cell-specific transcription factor σF of Bacillus subtilis. Genes Dev. 8:2653-2663. [DOI] [PubMed] [Google Scholar]
- 6.Duncan, L., S. Alper, and R. Losick. 1996. SpoIIAA governs the release of the cell-type specific transcription factor σF from its anti-sigma factor SpoIIAB. J. Mol. Biol. 260:147-164. [DOI] [PubMed] [Google Scholar]
- 7.Duncan, L., and R. Losick. 1993. SpoIIAB is an anti-sigma factor that binds to and inhibits transcription by regulatory protein σF from Bacillus subtilis. Proc. Natl. Acad. Sci. USA 90:2325-2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Errington, J., L. Appleby, R. A. Daniel, H. Goodfellow, S. R. Partridge, and M. D. Yudkin. 1992. Structure and function of the spoIIIJ gene of Bacillus subtilis: a vegetatively expressed gene that is essential for σG activity at an intermediate stage of sporulation. J. Gen. Microbiol. 138:2609-2618. [DOI] [PubMed] [Google Scholar]
- 9.Kellner, E. M., A. Decatur, and C. P. Moran, Jr. 1996. Two-stage regulation of an anti-sigma factor determines developmental fate during bacterial endospore formation. Mol. Microbiol. 21:913-924. [DOI] [PubMed] [Google Scholar]
- 10.Kirchman, P. A., H. DeGrazia, E. M. Kellner, and C. P. Moran, Jr. 1993. Forespore-specific disappearance of the sigma-factor antagonist SpoIIAB: implications for its role in determination of cell fate in Bacillus subtilis. Mol. Microbiol. 8:663-671. [DOI] [PubMed] [Google Scholar]
- 11.Kroos, L., and Y. T. Yu. 2000. Regulation of sigma factor activity during Bacillus subtilis development. Curr. Opin. Microbiol. 3:553-560. [DOI] [PubMed] [Google Scholar]
- 12.Lopez-Diaz, I., S. Clarke, and J. Mandelstam. 1986. spoIID operon of Bacillus subtilis: cloning and sequence. J. Gen. Microbiol. 132:341-354. [DOI] [PubMed] [Google Scholar]
- 13.Magnin, T., M. Lord, J. Errington, and M. D. Yudkin. 1996. Establishing differential gene expression in sporulating Bacillus subtilis: phosphorylation of SpoIIAA (anti-anti-σF) alters its conformation and prevents formation of a SpoIIAA/SpoIIAB/ADP complex. Mol. Microbiol. 19:901-907. [DOI] [PubMed] [Google Scholar]
- 14.Magnin, T., M. Lord, and M. D. Yudkin. 1997. Contribution of partner switching and SpoIIAA cycling to regulation of σF activity in sporulating Bacillus subtilis. J. Bacteriol. 179:3922-3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Margolis, P. S., A. Driks, and R. Losick. 1993. Sporulation gene spoIIB from Bacillus subtilis. J. Bacteriol. 175:528-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Min, K. T., C. M. Hilditch, B. Diederich, J. Errington, and M. D. Yudkin. 1993. σF, the first compartment-specific transcription factor of B. subtilis, is regulated by an anti-sigma factor that is also a protein kinase. Cell 74:735-742. [DOI] [PubMed] [Google Scholar]
- 17.Miroux, B., and J. E. Walker. 1996. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 260:289-298. [DOI] [PubMed] [Google Scholar]
- 18.Najafi, S. M., A. C. Willis, and M. D. Yudkin. 1995. Site of phosphorylation of SpoIIAA, the anti-anti-sigma factor for sporulation-specific σF of Bacillus subtilis. J. Bacteriol. 177:2912-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pan, Q., D. A. Garsin, and R. Losick. 2001. Self-reinforcing activation of a cell-specific transcription factor by proteolysis of an anti-sigma factor in B. subtilis. Mol. Cell 8:873-883. [DOI] [PubMed] [Google Scholar]
- 20.Partridge, S. R., and J. Errington. 1993. The importance of morphological events and intercellular interactions in the regulation of prespore-specific gene expression during sporulation in Bacillus subtilis. Mol. Microbiol. 8:945-955. [DOI] [PubMed] [Google Scholar]
- 21.Perez, A. R., A. Abanes-De Mello, and K. Pogliano. 2000. SpoIIB localizes to active sites of septal biogenesis and spatially regulates septal thinning during engulfment in Bacillus subtilis. J. Bacteriol. 182:1096-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rudner, D. Z., and R. Losick. 2001. Morphological coupling in development: lessons from prokaryotes. Dev. Cell 1:733-742. [DOI] [PubMed] [Google Scholar]
- 23.Smith, K., M. E. Bayer, and P. Youngman. 1993. Physical and functional characterization of the Bacillus subtilis spoIIM gene. J. Bacteriol. 175:3607-3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun, D. X., R. M. Cabrera-Martinez, and P. Setlow. 1991. Control of transcription of the Bacillus subtilis spoIIIG gene, which codes for the forespore-specific transcription factor σG. J. Bacteriol. 173:2977-2984. [DOI] [PMC free article] [PubMed] [Google Scholar]