Abstract
An unusual type of gene expression from an integrase promoter was found in cultures of the bacterium Pseudomonas sp. strain B13. The promoter controls expression of the intB13 integrase gene, which is present near the right end of a 105-kb conjugative genomic island (the clc element) encoding catabolism of aromatic compounds. The enzymatic activity of integrase IntB13 is essential for site-specific integration of the clc element into the bacterial host's chromosome. By creating transcription fusions between the intB13 promoter and the gfp gene, we showed that integrase expression in strain B13 was inducible under stationary-phase conditions but, strangely, occurred in only a small proportion of individual bacterial cells rather than equally in the whole population. Integrase expression was significantly stimulated by growing cultures on 3-chlorobenzoate. High cell density, heat shock, osmotic shock, UV irradiation, and treatment with alcohol did not result in measurable integrase expression. The occurrence of the excised form of the clc element and an increase in the rates of clc element transfer in conjugation experiments correlated with the observed induction of the intB13′-gfp fusion in stationary phase and in the presence of 3-chlorobenzoate. This suggested that activation of the intB13 promoter is the first step in stimulation of clc transfer. To our knowledge, this is the first report of a chlorinated compound's stimulating horizontal transfer of the genes encoding its very metabolism.
Bacteria can adapt to changing environmental conditions by a variety of genetic mechanisms. Some of these mechanisms involve acquisition of foreign DNA through mobile DNA elements such as plasmids, transposons, and genomic islands. These processes of foreign DNA acquisition, also named horizontal gene transfer, have been the focus of extensive research because of their general importance for microbial evolution, for the formation of catabolic pathways and antibiotic resistance in particular (17, 45), and for pathogenicity determinants (16, 22). Little attention has been given until now to the possibility of regulation of horizontal gene transfer processes by signaling pathways, effector compounds, and environmental stimuli. Some exceptions include the transfer of the opine catabolism-encoding plasmids in Agrobacterium spp., which is regulated by both catabolism of opines and quorum sensing (12-14, 31), regulation of transfer of the Bacteroides conjugative transposons by tetracycline (4, 36), and the stimulation of conjugation competence formation by peptide pheromones in Enterococcus faecalis (reviewed in reference 8).
The possibility of regulation of horizontal gene transfer by environmental stimuli is intriguing because it implies that not only specialized signaling pathways (as for Agrobacterium plasmid transfer) but also the presence of chemicals in the environment introduced by human activities may influence rates and types of horizontal gene transfer. We can thus envision that chemical compounds interact directly with regulatory pathways of horizontal gene transfer elements, thereby influencing the rate of horizontal gene transfer and indirectly creating a selective environment for (adapted) microorganisms with improved resistance to and ability to biodegrade the chemical. The consequences of such processes might be that selection occurs at the level of those horizontal gene transfer elements which are responsive to environmental stimuli.
In the present study we describe a possible link between the transfer of the clc genomic island of Pseudomonas sp. strain B13 (9) and the presence of 3-chlorobenzoate (CBA). Metabolism of CBA is, at least, in part mediated by enzymes encoded on the clc element. The clc element has a size of 105 kb and is present in two copies on the chromosome of strain B13. The clc element has the typical structure of a genomic island (47), a somewhat loose term coined for unstable and potentially transferable genome regions flanked by a tRNA gene, an integrase gene, and a short target site duplication at the other end. In most cases, genomic islands confer pathogenicity determinants (5, 17, 22), but other functions, such as nitrogen fixation and aromatic compound degradation, are also found (43).
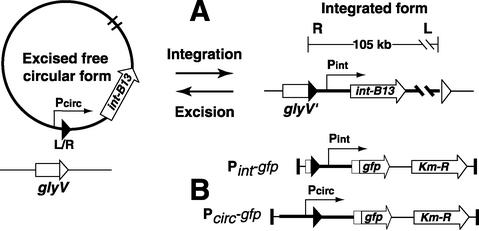
Pathogenicity islands can make up a substantial part of strain-specific differences, as was shown for Escherichia coli (32) and Pseudomonas aeruginosa (24). Through genome sequencing projects, it has now become apparent that genomic islands similar to the clc element exist in Xylella fastidiosa (41), Burkholderia fungorum, Ralstonia metallidurans, and P. aeruginosa (24). Unlike most known pathogenicity islands, the clc element is conjugative, and self-transmission to a number of different gram-negative γ- and β-Proteobacteria has been demonstrated (35, 42, 44, 49). Self-transfer starts with excision of the clc element. The result of excision is a circular intermediate which can be transferred to a new recipient cell and reintegrates site specifically into the chromosome at a gene for glycine tRNA (Fig. 1) (33). Integration is mediated by an integrase enzyme (IntB13), which for the clc element is about one-third larger than other P4-type integrases (34). Integration results in a short target site duplication at the other end (33, 34).
FIG. 1.
(A) Schematic presentation of the two forms of the clc genomic island and of the reactions catalyzed by the IntB13 integrase. During integration, the 18-bp 3′ end (depicted by open triangles) of the target glycine tRNA gene (glyV) is replaced by an identical 18-bp sequence (solid triangles) originating from the right (R) end of the clc element. Excision results in a closed junction between the left and right ends of the element (depicted as L/R). The promoter regions of the integrase gene (intB13) and of the integrated (Pint) and excised circular (Pcirc) forms of the clc element are depicted by thin arrows showing the direction of transcription (not drawn to scale). (B) Structures of the Pint-gfp and Pcirc-gfp fusions present in strain B13 derivatives. Solid vertical bars correspond to the I and O ends of the Tn5 delivery system.
In all recipient strains analyzed so far, the clc element has always integrated site specifically into one or more copies of the gene for glycine tRNA (33). The intB13 integrase gene (Fig. 1A) is located proximal to the right end of the element (34). Two different promoters control intB13 expression. One of these, named Pcirc, is present at the left end of the clc element facing outward and drives intB13 transcription in the excised circular form (39). Upon integration, the Pcirc promoter is physically separated from the intB13 gene (Fig. 1), and transcription of intB13 takes place from the Pint promoter, which could be roughly mapped upstream of intB13 (39). As in analogous systems, such as the conjugative transposons Tn916 (38) and CTnDOT (6, 7), the activity of the IntB13 integrase probably also catalyzes excision of the genomic island from the chromosome, although auxiliary recombination-directed factors may be needed (26). The complete cycle of integration, excision, and conjugal transfer makes the clc element a valuable model system with which to study the behavior of genomic islands.
In this work, we analyzed the physiological conditions stimulating transfer of the clc genomic island in Pseudomonas sp. strain B13. The analyses were based on three independent observation methods: (i) measurement of the transcriptional activity of the integrase gene from a single-copy integrase promoter-green fluorescent protein gene (gfp) fusion inserted in the B13 chromosome, (ii) analysis of the amount of circular intermediate by hybridization, and (iii) determination of transfer frequencies of the clc element in bacterial matings.
MATERIALS AND METHODS
Bacterial strains.
Escherichia coli strains DH5α, HB101 (37), and CC118 λpir (23) were used and grown as described previously (21). Pseudomonas sp. strain B13 (9) is the original host and donor of the clc element. Pseudomonas putida strain UWC1 (28) (kindly provided by Carole Newberry, Cardiff University, Wales, United Kingdom) was used as the recipient for the clc element. Luria-Bertani broth (LB) was routinely used for growing E. coli and Pseudomonas strains. As a defined mineral medium (MM), type 21C mineral medium (15) was used, supplemented with 10 mM CBA or fructose. When required, ampicillin, kanamycin, and/or rifampin was added at 50 μg/ml. Strains of Pseudomonas were grown at 30°C, and those of E. coli were grown at 37°C.
Promoter reporter constructions.
Integrase promoter reporters were constructed by amplifying a 282-bp fragment of the region upstream of the intB13 gene by PCR and fusing this fragment via several steps to a promoterless gfp gene, coding for the F64L, S65T enhanced green fluorescent mutant protein (29). Similarly, a 480-bp Pcirc promoter fragment corresponding to the region upstream of the intB13 gene in the circular form of the clc element was cloned from pRR146 (34) and fused to the gfp gene. The resulting Pint- and Pcirc-gfp fusions were further cloned as NotI fragments into the mini-Tn5 delivery vector pCK218 (23) and randomly inserted into the genome of Pseudomonas sp. strain B13 by mobilization and subsequent transposition in triparental filter matings (18). Strains with the correct insertion were verified by PCR, antibiotic resistance profiling, and Southern hybridization and named Pseudomonas sp. strain B13 (Pint-gfp) and strain B13 (Pcirc-gfp). For each construct, four independently derived exconjugant clones were analyzed in the induction experiments described below.
Fluorescence microscopy.
GFP fluorescence intensities of individual cells were examined with an Olympus BX50 epifluorescence microscope. Images of at least 200 cells per field were taken with a cooled black-and-white charge-coupled device camera (Photometrics SenSys:1401E, Roper Scientific Inc.), a 100/1.30 oil immersion lens (UPIanF1; Olympus), and an exposure time of 300 ms. Digital imaging of GFP fluorescence and quantification of fluorescence intensities of each individual cell were carried out by an automatic subroutine in the program Metaview (version 4.5; Universal Imaging Corporation, Visitron Systems) as described previously (21). Fluorescence intensities were expressed as cellular average gray values (AGVs).
Population analysis.
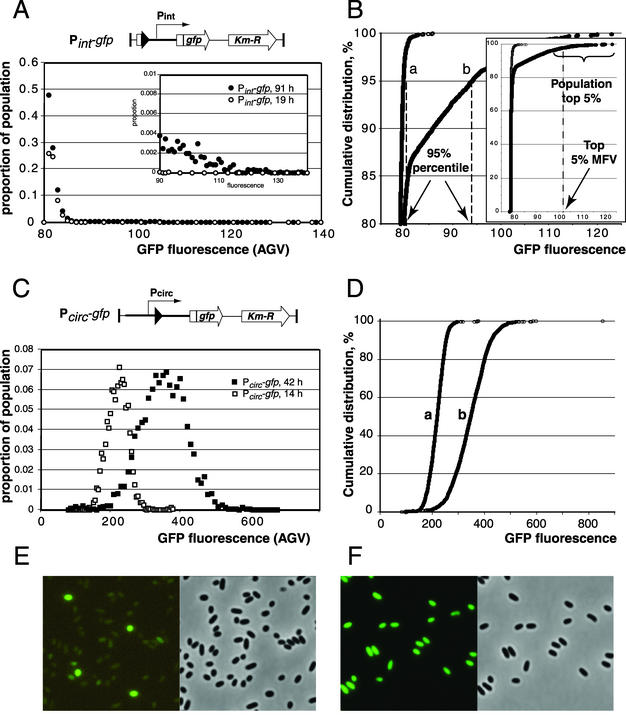
Differences in cellular AGVs among samples were analyzed by determining the distribution of AGVs for all cells in a population. For this purpose, the AGVs of all cells were sorted, ranked, and plotted against their position in the cumulative distribution curve (i.e., the ranking number divided by the total number of cells in the population, multiplied by 100). A statistical subroutine of the program R (20; http://cran.r-project.org) was written in order to analyze differences in the cumulative distribution curves based on distribution-free functions. The most prominent parameters for comparing statistical differences among samples were found to be the AGV (i.e., GFP fluorescence intensity) corresponding to the 95th percentile of the sample population (Fig. 2) and the arithmetic mean of the cellular AGV values for the 5% of the population with the highest single AGVs (indicated as the top 5% MFV). Intervals for 95% confidence were calculated for each of the derived 95th percentile fluorescence values based on 200 bootstrapping cycles. Examples of the untransformed distribution curves are shown in Fig. 2.
FIG. 2.
Distribution of GFP fluorescence intensities in cells of Pseudomonas sp. strain B13 (Pint-gfp) and B13 (Pcirc-gfp) taken at different stages of growth. (A and C) Untransformed distribution curves for the GFP fluorescence levels (expressed as AGV units per cell, calculated with Metaview) among individual cells of a population of Pseudomonas sp. strain B13 (Pint-gfp) and B13 (Pcirc-gfp), respectively. Each data point is the relative proportion of the population carrying fluorescence values in that particular range (i.e., bin size; for panel A, 1 fluorescence unit; for panel C, 5 and 10 units for the data series at 14 and 42 h, respectively). (B and D) Cumulative distribution plots from the data shown in A and C, respectively, in order to visualize the concepts of 95th percentile and top 5% mean fluorescence value (MFV). A fluorescence value of 79 corresponds to dark (nonfluorescent) cells and is an arbitrary value produced by the image recording system. Each data point represents the fluorescence value of a single cell. The inset in B shows the range of fluorescence values for the whole population, whereas the main panel focuses on the range above 80%. Note the strong aberration of the single-cell fluorescence distribution in populations of B13 (Pint-gfp) from the normal distribution as for strain B13 (Pcirc-gfp); see C and D. Cells of strain B13 (Pint-gfp) were grown in batch cultures with 10 mM CBA as the C source. Samples were taken at 19 h (exponential growth phase, curve a) and 91 h (stationary phase, curve b). Cells of Pseudomonas sp. strain B13 (Pcirc-gfp) were grown in batch cultures with 10 mM CBA as the C source, and samples were taken at 14 h (curve a, exponential growth phase) and 42 h (curve b, stationary phase). Note the different scales of fluorescence in panels A and B versus C and D. (E and F) Micrographs of typical population differences of strain B13 (Pint-gfp) and strain B13 (Pcirc-gfp), respectively, taken in stationary phase under GFP illumination (left) and the corresponding image in phase contrast (right). The fluorescence image in panel E was electronically enhanced to the same light intensity as that in panel F.
Detection and quantification of excised circular form of clc element.
Isolation of total genomic DNA was performed with the cetyltrimethylammonium bromide method (2). Genomic DNA samples were digested with EcoRI, size separated in a 0.8% agarose gel, Southern blotted to a Hybond-XL membrane (Amersham Bioscience), and hybridized with radioactively labeled DNA probes according to standard protocols (2). To detect the clc element in its integrated and excised circular form, a 1.7-kb SalI-NsiI fragment with the left end of the clc element from plasmid pRR104 (33) was used in hybridizations. Band intensities of the circular form and the two chromosomally integrated clc element copies were estimated from autoradiograms by scanning the X-ray films as described previously (3). Autoradiograms were scanned, processed in Adobe Photoshop 6.0, saved, and imported into Adobe Illustrator 8.0.1 as 8-bit TIFF files. In addition, the presence of the circular form was verified by PCR with two primers (RR316a and RR319) facing outwards from the ends of the clc element, as described previously (33).
Induction experiments.
Pseudomonas sp. strain B13 (Pint-gfp) was grown in batch in 200 ml of MM containing either 10 mM CBA or 10 mM fructose at 30°C with shaking at 200 rpm. Samples for optical density (OD, determined at a wavelength of 600 nm) measurements, GFP expression, and DNA extraction were taken at 1- to 6-h intervals and whenever appropriate. To ensure that the observed expression from the Pint-gfp and Pcirc-gfp fusions was not due to the chromosomal location of that particular insertion, we screened four independent insertions. All behaved the same with regard to single-cell GFP fluorescence intensity and distribution levels in the population.
For continuous cultivation, strain B13 (Pint-gfp) was grown in a 500-ml bioreactor with a working volume of 200 ml, fed with MM supplemented with either 0.1 mM or 10 mM CBA or 10 mM fructose. The bioreactor was operated at a constant dilution rate of 0.05 h−1. OD and GFP expression were analyzed daily on 2-ml samples taken aseptically from the reactor. To study the effects of starvation, 1-ml aliquots of the chemostat cultures were incubated in 15-ml closed polypropylene tubes shaken horizontally at 200 rpm at 30°C and sampled regularly.
A limited number of typical stress factors were tested on 2-ml aliquots of strain B13 (Pint-gfp) and wild-type B13 withdrawn from continuous cultures fed 10 mM fructose. To create a heat shock, aliquots were incubated for 2 or 5 min at 42°C. For UV treatment, the test culture was dispensed into a 90-mm-diameter plastic petri dish and exposed to UV radiation of 2 J/cm2 at 254 nm (for 90 s) in a UV Stratalinker 1800 (Stratagene, La Jolla, Calif.). High-osmolarity conditions were achieved by adding sodium chloride to the culture at final concentrations of 200 and 500 mM. Ethanol stress was performed by incubating cell aliquots at 1 and 10% ethanol (vol/vol). Monochlorobenzene was added as a 4% solution in heptamethylnonane. After UV treatment and heat shock and during alcohol and high-salt treatment, the samples were incubated on a shaker at 180 rpm and 30°C for 3 to 4 h and then divided into two portions, one for GFP expression measurements and another for total DNA isolation.
Transfer of clc genomic island on membrane filters.
To determine if the growth substrate would influence the frequency of clc element transfer, membrane filter matings were set up between exponentially grown cultures of Pseudomonas sp. strain B13 (Pint-gfp) as the donor and P. putida strain UWC1 as the recipient. Volumes of both donor and recipient cultures in the mating were chosen to give approximately 108 cells per filter each. Cells were collected by centrifugation at 3,000 × g for 5 min, resuspended gently in 50 μl of MM, and transferred to nitrocellulose filters (0.45-μm pore size, 25-mm diameter; Sartorius AG, Goettingen, Germany), which were placed on MM agar plates with 1 mM CBA, with 1 mM fructose, and without a C source. Directly after placement and after 4, 24, 48, and 72 h of incubation, the cells were washed from the filters with 0.5 ml of MM by vigorous vortexing, serially diluted, and plated on MM agar supplemented with 5 mM CBA and 50 μg of rifampin per ml to select for transconjugants. Colonies visible to the naked eye were scored after 4 days of incubation. The number of donor and recipient cells at the end of mating was determined from serial dilutions on LB plates supplemented with 50 μg of kanamycin per ml and 50 μg of rifampin per ml, respectively. The transfer frequency was calculated as the number of transconjugants per donor cell in the cell suspension washed from the filter.
RESULTS
Population-dependent expression of intB13 gene.
To determine the activity of the intB13 gene of the clc genomic island in Pseudomonas sp. strain B13, we first relied on observing transcriptional activity from an extra copy of the upstream region of the intB13 gene (as in the integrated clc element) fused to a promoterless gfp gene and inserted at random into the Pseudomonas sp. strain B13 chromosome by means of Tn5 delivery (Fig. 1). In this case, the production of GFP in individual cells served as an indicator of the integrase promoter transcription activity.
Surprisingly, only a small proportion of a population of B13 (Pint-gfp) cells containing the transcription fusion between the region upstream of the intB13 gene and gfp showed GFP production (Fig. 2A, B, and E). The proportion of induced cells and the amount of accumulated GFP in induced cells of B13 (Pint-gfp) were found to be dependent on the growth substrate and on the growth phase of the culture (see below) but remained small under all conditions tested. This extreme heterogeneity in GFP expression made it impossible to use common statistical descriptors such as mean fluorescence for the whole population with standard deviation as a measure of induction from the intB13 promoter. Therefore, we chose to use distribution-free statistical methods, such as the 95th percentile fluorescence value (Fig. 2B) and the mean fluorescence value (MFV) of the top 5% of expressing cells (Fig. 2B, inset) to compare expression from Pint under different conditions.
To ensure that the observed gfp expression from the Pint-gfp fusion was not a peculiarity of GFP in strain B13 in general, we determined GFP expression from a Pcirc-gfp fusion (Fig. 1) in single copy inserted on the chromosome by the same Tn5 delivery system (Fig. 1B). In this case, the distribution of GFP fluorescence intensities over the population could be described with a normal distribution function (Fig. 2C, D, and F). Furthermore, to ensure that the observed expression from the Pint-gfp fusion was not due to the chromosomal location of that particular insertion, we screened four independent insertions. All behaved the same with regard to single-cell GFP fluorescence intensity and distribution levels in the population (data not shown).
Induction of integrase promoter under stationary-phase conditions.
During the exponential phase of batch growth, GFP production in cultures of B13 (Pint-gfp) was very low on both fructose and CBA (Table 1). Transition to the stationary phase significantly increased expression from the Pint-gfp fusion, although at most 13% of the population had GFP intensities higher than 5 units above background (background meaning nonfluorescent dark cells, with an AGV of 79). In stationary phase, both the proportion of GFP-containing cells and their fluorescence levels were higher with CBA than with fructose as the carbon source (Table 1).
TABLE 1.
GFP fluorescence intensity in strain B13 (Pint-gfp) grown in batch culturea
| Incubation time (h) | Fructose
|
CBA
|
||
|---|---|---|---|---|
| GFP fluorescence intensity (AGV units) | Top 5% MFV | GFP fluorescence intensity (AGV units) | Top 5% MFV | |
| 14b | 79.6 (79.5-79.6) | 80.4 | 79.6 (79.5-79.7) | 80.3 |
| 40 | 81.8 (81.6-82.0) | 86.8 | 87.43 (86.2-88.4) | 94.9 |
| 64 | 84.0 (83.5-84.8) | 92.4 | 96.5 (94.9-97.9) | 104.4 |
| 88 | 83.3 (82.2-84.1) | 91.7 | 94.0 (93.1-94.9) | 102.5 |
Fructose and 3-chlorobenzoate were used at 10 mM. GFP fluorescence intensity is given as cellular average gray value units of the 95th percentile of the population. Values in parentheses represent the 95% confidence intervals, calculated with the R software. MFV values are the arithmetic means of the cellular AGVs for the 5% of the population with the highest fluorescence intensities.
At 14 h, both fructose- and 3-chlorobenzoate-grown cultures were growing exponentially and entered the stationary phase after about 24 h of incubation.
Cells taken from continuously cultivated chemostats at a constant growth rate of 0.05 h−1 (which is close to stationary-phase growth) displayed levels of GFP production similar to that of stationary-phase-grown cells in batch culture (Table 2). Likewise, the fluorescence of the cells grown with CBA was higher than that of the cells grown with fructose (95th percentile AGV corresponding to 100.8 and 84.2 units, respectively). Removing cells from fructose-grown chemostat cultures and starving them in subsequent batch of incubation did not further increase GFP fluorescence values or the number of induced cells (Table 2). In contrast, both the proportion of induced cells (from 7% in the chemostat at steady state to 13% after 64 h of starvation) and the 95th percentile AGV increased from 100.8 units in steady state to 105.6 after 64 h of starvation in B13 (Pint-gfp) bacteria taken from CBA-fed bioreactors (Table 2). Spiking cells taken from the CBA-fed chemostat in batch with an additional 0.1 mM CBA did not result in any further increase in the cells' fluorescence during another 70 h of incubation. Spiking with 10 mM CBA first caused a decrease in 95th percentile AGV during a short resumed phase of exponential growth and a subsequent increase in both AGV and the number of induced cells during the following stationary phase (data not shown). Batch and chemostat experiments were performed twice independently, with similar results.
TABLE 2.
GFP production in strain B13 (Pint-gfp) under carbon starvation conditionsa
| Incubation time (h) | Fructose
|
CBA
|
||
|---|---|---|---|---|
| GFP fluorescence intensity (AGV units) | Top 5% MFV | GFP fluorescence intensity (AGV units) | Top 5% MFV | |
| 0b | 84.2 (82.4-85.9) | 108.6 | 100.8 (97.8-103.8) | 115.6 |
| 16 | 85.9 (84.5-88.7) | 109.1 | 100.6 (98.3-103.2) | 117.4 |
| 40 | 84.7 (84.0-86.9) | 105.9 | 105.6 (102.5-107.0) | 121.5 |
| 64 | 84.0 (83.8-84.4) | 99.5 | 105.6 (104.1-107.7) | 119.9 |
| 112 | 83.9 (83.3-85.4) | 99.0 | 103.6 (101.3-105.0) | 116.7 |
Cultures growing continuously on 10 mM fructose or 10 mM CBA were taken from chemostats. For other details, see Table 1, footnote a.
Zero indicates the culture during steady-state chemostat growth; subsequent time points refer to cells taken from the chemostat and incubated in batch culture without any carbon.
Formation of circular form of clc element.
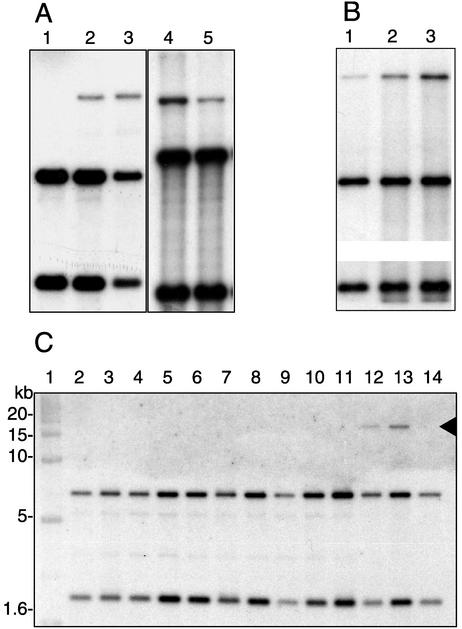
To determine if measurements of expression from the integrase promoter by GFP fluorescence were a proper indicator for stimulation of the excision and transfer cascade, we quantified the product of excision of the clc genomic island in strain B13 (Pint-gfp) cultures in the same growth experiments as described above. During exponential growth in batch with 10 mM CBA, the circular form of the clc element was not detectable by Southern hybridizations (Fig. 3A, lane 1) or by specific amplification in the PCR (not shown). Shortly after the culture entered stationary phase, the circular form appeared (Fig. 3A, lane 2), and within the next 24 h its proportion increased to about 16% of the total hybridizable clc DNA (i.e., circular form plus chromosomal copies) according to densitometric analysis. The appearance of the circular form was confirmed by PCR. Hence, Southern hybridizations proved sensitive enough to detect the appearance of the circular form. The relative level of excised form remained more or less constant for the next 48 h (not shown). In cells grown on fructose, the relative proportion of the circular form reached 7% (Fig. 3A, lane 5), which is half the amount after growth with CBA to the same growth phase.
FIG. 3.
Appearance of free circular form of clc element in starving cells of Pseudomonas sp. strain B13 (Pint-gfp) and wild-type strain B13 (C) on Southern blots of EcoRI-digested genomic DNA hybridized against a probe for the “left end” of the element (see Materials and Methods). The two chromosomal copies of the element appear as two lower bands, whereas the uppermost band (ca. 17 kb) represents the excised circular form. (A) Batch cultures. Percentages in brackets correspond to the measured proportion of the free circular form in total clc-specific DNA. Lane 1, mid-exponential stage of growth (14 h) with 10 mM CBA (0%); lanes 2 and 3, same culture 3 h (6%) and 7 h (11%) after entering stationary phase, respectively; lanes 4 and 5, 48-h-old batch cultures grown with 10 mM CBA (15%) and 10 mM fructose (7%), respectively. (B) Starving cultures taken from the chemostat with 10 mM CBA and a dilution rate of 0.05 h−1. Lane 1, chemostat culture in steady state (4%); lanes 2 and 3, same culture after 16 h (8%) and 64 h (15%) of starvation, respectively. (C) Appearance of the clc circular form during growth on 10 mM CBA and after different stress treatments (as described in Materials and Methods). Lanes: 1, markers; 2, LB-grown cells; 3, 42°C; 4, 37°C; 5, 4°C; 6, monochlorobenzene; 7, 5% ethanol; 8, 2% ethanol; 9, UV at 50 J/cm2; 10, UV at 10 J/cm2; 11, UV at 2 J/cm2; 12, 3 h in stationary phase; 13, 16 h in stationary phase; 14, exponential phase. Autoradiograms were scanned, processed in Adobe Photoshop 6.0, saved, and imported into Adobe Illustrator 8.0.1 as 8-bit TIFF files. For convenience of presentation, in panel B the middle sections of the two original images were left out in order to fit both chromosomal bands on the image. Panels A, B, and C were not run on the same gel.
In cultures taken from the chemostat and incubated under conditions of carbon starvation, the proportion of the excised form compared to both chromosomal copies increased from about 4% under steady-state conditions with 10 mM CBA and a growth rate of 0.05 h−1 to 15% after 64 h of starvation (Fig. 3B).
Effects of cell density and general stress conditions.
GFP fluorescence values of cells growing under steady-state conditions with 0.1 mM CBA and a dilution rate of 0.05 h−1 were slightly higher than those grown with 10 mM CBA (Table 3), despite the 80-fold cell density difference (OD600 of 0.01 and 0.83, respectively). When cells were taken from the chemostats and incubated for 60 h in batch with no carbon added (C starvation), the 95th percentile fluorescence values of both cultures became equal. When the culture originally fed with 10 mM CBA was concentrated 10-fold and incubated under conditions of carbon starvation as before, again no significant changes in GFP fluorescence values were observed (not shown). This suggested that expression of the integrase in both growing and starving cells was not a cell density-dependent effect.
TABLE 3.
GFP production in strain B13 (Pint-gfp) at different cell densitiesa
| Cell culture | GFP fluorescence intensity (AGV units) with 0.1 mM CBA
|
GFP fluorescence intensity (AGV units) with 10 mM CBA
|
||
|---|---|---|---|---|
| 95th percentile | Top 5% MFV | 95th percentile | Top 5% MFV | |
| Chemostat, 6 days | 83.8 (83.7-84.2) | 86.0 | 80.9 (80.8-81.0) | 84.7 |
| Chemostat, 14 days | 83.8 (83.5-84.1) | 88.8 | 81.0 (80.8-81.2) | 82.5 |
| Starvation, 60 h | 85.1 (83.8-87.4) | 94.5 | 85.3 (84.2-86.3) | 93.3 |
Cells were taken from the chemostat after 6 days or 14 days of continuous cultivation or taken on day 6 and then incubated in batch culture without any carbon for 60 h. Cells were grown with 0.1 mM (OD600 = 0.01) or 10 mM (OD600 = 0.83) CBA. GFP fluorescence intensity values are given as for Table 1. Note that absolute values of GFP fluorescence in this table cannot be compared to those in Table 2 due to slight variations in the brightness of the UV light source.
None of the following stress factors which are known to cause induction of prophages caused a significant increase in GFP fluorescence and accumulation of the excised clc form: UV irradiation, heat shock, osmotic stress in the presence of sodium chloride, and treatment with ethanol (Fig. 3C, Table 4).
TABLE 4.
Effect of various stresses on GFP production in strain B13 (Pint-gfp)a
| Cell culture conditionsb | GFP fluorescence (AGV units)
|
|
|---|---|---|
| 95th percentile | Top 5% MFV | |
| No treatmenta | 83.4 (83.2-83.6) | 86.3 |
| 1 mM CBAa | 83.4 (83.2-83.5) | 85.8 |
| 5 mM CBAa | 83.4 (83.1-83.6) | 85.8 |
| No treatmentb | 85.7 (85.5-86.0) | 89.1 |
| UVb | 85.1 (84.9-85.4) | 88.3 |
| 42°C, 5 minb | 83.7 (83.5-83.9) | 85.7 |
| 0.2 M NaClb | 84.5 (84.3-84.6) | 87.3 |
| 0.5 M NaClb | 86.7 (86.4-87.1) | 89.4 |
| 10% ethanolb | 84.3 (84.1-84.6) | 86.6 |
Cultures were taken from steady-state chemostat cultures growing on 10 mM fructose. Two-milliliter aliquots were treated as described in Materials and Methods. GFP fluorescence intensity is given as for Table 1.
Conditions with the same superscript are for the same batch of cells.
Transfer of clc element.
As a final determinant for stimulation of the clc element's transfer process, we analyzed transfer frequencies of the clc element in bacterial filter mating experiments. In matings between Pseudomonas sp. strain B13 (Pint-gfp) and P. putida UWC1, the occurrence of rifampin-resistant colonies growing on CBA (indicative of recipients which had acquired the clc element) was 2.0-, 5.6-, and 2.8-fold higher after 24, 48, and 72 h of mating time incubation, respectively, with 1 mM CBA added to the medium instead of fructose (Table 5). When the net increase in the number of transconjugant colonies was calculated for discrete incubation time intervals, the transfer rate of the clc element seemed to have increased between 48 and 72 h compared to the first 24 h.
TABLE 5.
Transfer of c/c element on membrane filters in the presence of CBA or fructosea
| Mating time (h) | Mean transfer frequency (SD)
|
Incubation period (h) | Transfer rate
|
||
|---|---|---|---|---|---|
| CBA | Fructose | CBA | Fructose | ||
| 24 | 1.27 × 10−3 (0.21 × 10−3) | 0.63 × 10−3 (0.34 × 10−3) | 0-24 | 5.28 × 10−5 | 2.61 × 10−5 |
| 48 | 50.8 × 10−3 (11 × 10−3) | 9.05 × 10−3 (3.0 × 10−3) | 24-48 | 2.06 × 10−3 | 0.35 × 10−3 |
| 72 | 249 × 10−3 (84 × 10−3) | 89.8 × 10−3 (42 × 10−3) | 48-72 | 8.27 × 10−3 | 3.36 × 10−3 |
Both CBA and fructose were present at 1 mM in the filter support medium. The mean transfer frequency (standard deviation) was calculated as the number of transconjugants per donor cell counted after 24, 48, and 72 h of mating time. The transfer rate was calculated as the net increase in the number of transconjugants per donor cell per hour during the corresponding incubation period.
Since the number of donor cells was in 1,000-fold excess compared to any transconjugants and the number of donor and recipient cells did not change significantly during mating, we assume that the observed increase in the number of transconjugants resulted from new gene transfer events and not from selective propagation of previously produced transconjugants on CBA present in the filter support. Matings performed with donor cells taken from exponentially growing cultures on MM agar without any C source resulted in at least 100-fold lower frequencies of transconjugant appearance. After 4 h of mating, 2.3 × 10−6 and 1.0 × 10−6 transconjugants arose per donor cell grown on CBA and fructose, respectively. Mating experiments were performed twice with similar outcomes.
DISCUSSION
The clc element of Pseudomonas sp. strain B13 is a representative of a growing class of genomic islands (17, 47). Although genomic islands have mostly been associated with pathogenicity determinants (22), the clc element is devoid of such factors and is now viewed as an ecological and catabolic genomic island (17). Current sequencing projects and other data from our own laboratory indicate that the clc-type element is more widely distributed in other bacteria. In the Xylella fastidiosa genome sequence (41), we identified by sequence comparisons a region of 71 kb that is highly related to the clc element, flanked on one end by a glycine tRNA gene and an integrase gene and on the other end by the 18-bp repeat (47), although the genes for chlorocatechol degradation are lacking in Xylella spp. (41). In Ralstonia sp. strain JS705, an environmental isolate from chlorobenzene-contaminated groundwater (46), an almost identical clc element was found, which, however, had acquired an extra insertion of about 12 kb (30). The unaligned Burkholderia fungorum genome sequence (GenBank accession no. NZ_AAAC01000413) also carries a region with more than 99% identity to the intB13 sequence and to other sequenced parts of the clc element (i.e., the clc genes [11] and the left-end region [39]). Interestingly, the downstream regions of both int in B. fungorum and intB13 carry a gene cluster similar to the ohb genes for ortho-halobenzoate degradation in Pseudomonas aeruginosa strain JB2 (19). Since the halobenzoate dioxygenase of strain JB2 is capable of converting 3-chlorobenzoate in addition to 2-chlorobenzoate (19), this would suggest that the complete degradation pathway for 3-chlorobenzoate might be encoded on the clc element.
Unlike most pathogenicity islands, the clc element not only excises but also mediates its own conjugal transfer and reintegration into a new recipient, making it a valuable model system with which to study the behavior of genomic islands. One of the central features of genomic islands certainly is the integrase, which mediates site-specific integration and is usually involved in the excision process as well. Although excisionase activity of IntB13 itself has not been demonstrated yet, evidence from related integrases (5-7, 26) gives sufficient basis for the working hypothesis that IntB13 is also involved in excision of the clc element. The integrase of the clc element has definite evolutionary and mechanistic relationships with the integrases of pathogenicity islands (47). From the work presented here, we can conclude that expression of intB13 from Pint in the integrated form of the clc element is tightly controlled and subject to regulation by environmental and growth conditions, with starvation and the stationary growth phase being the main trigger (Table 2). Quite surprisingly, one of the conditions causing further induction of the integrase was cultivation on CBA, which is the compound specifically metabolized by enzymes encoded on the clc element. This suggests a signaling and regulatory link between a specific growth substrate (i.e., CBA), integrase expression, and (consequently) gene transfer rates of the DNA region responsible for this metabolic trait (the clc element).
Levels of GFP fluorescence of single cells were representative of induction of the integrase gene itself and not an artifact from placing a separate Pint-gfp fusion elsewhere in the chromosome of strain B13. This was inferred from two independent types of assays: the appearance of the excised circular form of the clc genomic island by quantitative hybridization and the transfer rates of the clc element in bacterial conjugative matings. In exponentially growing cells, the excised form of the clc element was not detectable by Southern hybridization or by PCR (compared to the chromosomal copies), whereas during the stationary phase, the excised form emerged and accumulated. Both in stationary-phase cultures and under carbon starvation, the abundance of the free circular form was twice as high in cells grown on CBA as in those grown on fructose (Fig. 3). Conjugation transfer frequencies of the clc genomic island between B13 (Pint-gfp) as the donor and P. putida UWC1 also increased during matings for up to 72 h, suggesting that stationary-phase conditions and/or carbon starvation was a stimulus for the transfer process (Table 5). Furthermore, the transfer frequencies were up to five times higher in the presence of CBA as opposed to fructose, suggesting that the enhancement by CBA was also visible in the transfer process.
We have no clear clues yet as to how starvation and CBA exert their effects on the integrase promoter. Most likely, CBA itself does not act as signaling compound for induction, since spiking of CBA to starving cells of strain B13 (Pint-gfp) taken from the CBA-fed chemostat did not result in production of extra GFP. Autoinducer signaling of the acylhomoserine lactone type did not seem to be involved either, since at different cell densities no effects on GFP expression from the integrase promoter were observed (Table 3). None of the treatments that typically trigger the SOS/RecA response affected GFP expression and appearance of the circular form, suggesting that a RecA/LexA-type degradation is not directly involved in regulating integrase expression of the clc element (Table 4).
Despite detectable induction, transcription from the integrase promoter in the integrated form was generally tightly repressed, resulting in very low levels of GFP production in almost all cells under all growth conditions tested. Whenever induction was observed, it always occurred in only a small subpopulation of cells (at most 13%) (Fig. 2). This is very unusual and in contrast to what has been described for inducible gene expression, in which case induction will lead to a heterogenous but normal type of expression pattern (Fig. 2) in a bacterial population (25). It might be that the type of expression from the intB13 integrase is more common to related integrases of genomic islands and conjugative transposons but has not been seen before because LacZ was used as the reporter protein for integrase expression instead of GFP and single-cell analysis.
The expression of intB13 is reminiscent of stochastic gene expression observed in some prophages and in other experimental systems (1, 10). Stochastic gene expression is thought to be the result of slightly different amounts of transcriptional factors in each individual cell and of random microscopic events (10). A subpopulation-dependent type of expression could arise when two counteracting factors are working on the same promoter, with each at an individually different cellular level. If, due to stochastic cellular differences, the repressing factor were in excess in an individual cell, no expression would occur, whereas if the activating factor dominated, transcription would start (1). Evidence has been obtained from mutation analysis for the existence of both an activator and a repressor encoded near the left end of the clc element and acting on the integrase promoter (39). The population-dependent integrase expression may thus really be the result of individual differences in the amounts of such an activator and repressor.
Our present hypothesis for the role of CBA is that a metabolite of CBA at some point acts as an effector in modulating the interactions of this putative repressor and activator with the intB13 promoter and in the synthesis of the repressor and activator themselves by interfering with another “master” regulator acting on the promoters of those. A few other examples of (subtle) triggering on expression of conjugative elements have been described, such as the inducible effect of some antibiotics on integrase expression of prophages (48), of tetracycline on the Enterococcus conjugative transposon CTn916 (27, 40), and on transfer of the Bacteroides conjugative transposons CTnDOT and CTnERL (6, 7). However, until now there seems to be little in common with the biochemical mechanisms of such triggering. Therefore, we find it intriguing to discover if such environmental signaling mechanisms on conjugative transfer are more widespread and the result of direct effector-regulator interactions.
Acknowledgments
We thank Peter Reichert for help with the statistical program R and Sonja Studer for help with PCR detection of the clc circular form. We appreciate the critical input of Wolf Hardt, Johan Leveau, Steve Lindow, and Paolo Landini after reading the manuscript.
REFERENCES
- 1.Arkin, A., J. Ross, and H. H. McAdams. 1998. Stochastic kinetic analysis of developmental pathway bifurcation in phage lambda-infected Escherichia coli cells. Genetics 149:1633-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1996. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 3.Baumann, B., M. Snozzi, A. J. B. Zehnder, and J. R. Van Der Meer. 1996. Dynamics of denitrification activity of Paracoccus denitrificans in continuous culture during aerobic-anaerobic changes. J. Bacteriol. 178:4367-4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonheyo, G. T., B. D. Hund, N. B. Shoemaker, and A. A. Salyers. 2001. Transfer region of a Bacteroides conjugative transposon contains regulatory as well as structural genes. Plasmid 46:202-209. [DOI] [PubMed] [Google Scholar]
- 5.Burrus, V., G. Pavlovic, B. Decaris, and G. Guédon. 2002. Conjugative transposons: the tip of the iceberg. Mol. Microbiol. 46:601-610. [DOI] [PubMed] [Google Scholar]
- 6.Cheng, Q., B. J. Paszkiet, N. B. Shoemaker, J. F. Gardner, and A. A. Salyers. 2000. Integration and excision of a Bacteroides conjugative transposon, CTnDOT. J. Bacteriol. 182:4035-4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng, Q., Y. Sutanto, N. B. Shoemaker, J. F. Gardner, and A. A. Salyers. 2001. Identification of genes required for excision of CTnDOT, a Bacteroides conjugative transposon. Mol. Microbiol. 41:625-632. [DOI] [PubMed] [Google Scholar]
- 8.Clewell, D. B. 1999. Sex pheromone systems in enterococci, p. 10-20. In G. M. Dunny and S. C. Winans (ed.), Cell-cell signaling in bacteria. ASM Press, Washington, D.C.
- 9.Dorn, E., M. Hellwig, W. Reineke, and H. J. Knackmuss. 1974. Isolation and characterization of a 3-chlorobenzoate degrading pseudomonad. Arch. Microbiol. 99:61-70. [DOI] [PubMed] [Google Scholar]
- 10.Elowitz, M. B., A. J. Levine, E. D. Siggia, and P. S. Swain. 2002. Stochastic gene expression in a single cell. Science 297:1183-1186. [DOI] [PubMed] [Google Scholar]
- 11.Frantz, B., and A. M. Chakrabarty. 1987. Organization and nucleotide sequence determination of a gene cluster involved in 3-chlorocatechol degradation. Proc. Natl. Acad. Sci. USA 84:4460-4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuqua, C., M. Burbea, and S. C. Winans. 1995. Activity of the Agrobacterium Ti plasmid conjugal transfer regulator TraR is inhibited by the product of the traM gene. J. Bacteriol. 177:1367-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuqua, C., and S. C. Winans. 1996. Conserved cis-acting promoter elements are required for density-dependent transcription of Agrobacterium tumefaciens conjugal transfer genes. J. Bacteriol. 178:435-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuqua, W. C., and S. C. Winans. 1994. A LuxR-LuxI type regulatory system activates Agrobacterium Ti plasmid conjugal transfer in the presence of a plant tumor metabolite. J. Bacteriol. 176:2796-2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerhardt, P., R. G. E. Murray, R. N. Costilow, E. W. Nester, W. A. Wood, N. R. Krieg, and G. Briggs Phillips (ed.). 1981. Manual of methods for general bacteriology. American Society for Microbiology, Washington, D.C.
- 16.Hacker, J., and J. B. Kaper. 2000. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 54:641-679. [DOI] [PubMed] [Google Scholar]
- 17.Hacker, J., and E. Carniel. 2001. Ecological fitness, genomic islands and bacterial pathogenicity. A Darwinian view of the evolution of microbes. EMBO Rep. 2:376-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hickey, W. J., and G. Sabat. 2001. Integration of matrix-assisted laser desorption ionization-time of flight mass spectrometry and molecular cloning for the identification and functional characterization of mobile ortho-halobenzoate oxygenase genes in Pseudomonas aeruginosa strain JB2. Appl. Environ. Microbiol. 67:5648-5655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ihaka, R., and R. Gentleman. 1996. R: a language for data analysis and graphics. J. Comp. Graph. Stat. 5:299-314. [Google Scholar]
- 21.Jaspers, M. C. M., C. Meier, A. J. B. Zehnder, H. Harms, and J. R. van der Meer. 2001. Measuring mass transfer processes of octane with the help of an alkS alkB::gfp-tagged Escherichia coli. Environ. Microbiol. 3:512-524. [DOI] [PubMed] [Google Scholar]
- 22.Kaper, J. B., and J. Hacker (ed.). 1999. Pathogenicity islands and other mobile virulence elements. ASM Press, Washington, D.C.
- 23.Kristensen, C. S., L. Eberl, J. M. Sanchez-Romero, M. Givskov, S. Mølin, and V. de Lorenzo. 1995. Site-specific deletions of chromosomally located DNA segments with the multimer resolution system of broad-host-range plasmid RP4. J. Bacteriol. 177:52-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larbig, K., A. Christmann, A. Johann, J. Klockgether, T. Hartsch, R. Merkl, L. Wiehlmann, H.-J. Fritz, and B. Tümmler. 2002. Gene islands integrated into tRNAGly genes confer genome diversity on a Pseudomonas aeruginosa clone. J. Bacteriol. 184:6665-6680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leveau, J. H. J., and S. E. Lindow. 2001. Appetite of an epiphyte: quantitative monitoring of bacterial sugar consumption in the phyllosphere. Proc. Natl. Acad. Sci. USA 98:3446-3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lewis, J. A., and G. F. Hatfull. 2001. Control of directionality in integrase-mediated recombination: examination of recombination directionality factors (RDFs) including Xis and Cox proteins. Nucleic Acids Res. 29:2205-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manganelli, R., L. Romano, S. Ricci, M. Zazzi, and G. Pozzi. 1995. Dosage of Tn916 circular intermediates in Enterococcus faecalis. Plasmid 34:48-57. [DOI] [PubMed] [Google Scholar]
- 28.McClure, N. C., A. J. Weightman, and J. C. Fry. 1989. Survival of Pseudomonas putida UWC1 containing cloned catabolic genes in a model activated-sludge unit. Appl. Environ. Microbiol. 55:2627-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller, W. G., and S. E. Lindow. 1997. An improved GFP cloning cassette designed for prokaryotic transcriptional fusions. Gene 191:149-153. [DOI] [PubMed] [Google Scholar]
- 30.Müller, T. A., C. Werlen, J. C. Spain, and J. R. van der Meer. 2003. Evolution of a chlorobenzene degradative pathway among bacteria in a contaminated groundwater mediated by a genomic island in Ralstonia. Environ. Microbiol. 5:163-173d. [DOI] [PubMed]
- 31.Oger, P., and S. K. Farrand. 2002. Two opines control conjugal transfer of an Agrobacterium plasmid by regulating expression of separate copies of the quorum-sensing activator gene traR. J. Bacteriol. 184:1121-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perna, N. T., G. Plunkett, 3rd, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 33.Ravatn, R., S. Studer, D. Springael, A. J. B. Zehnder, and J. R. van der Meer. 1998. Chromosomal integration, tandem amplification, and deamplification in Pseudomonas putida F1 of a 105-kilobase genetic element containing the chlorocatechol degradative genes from Pseudomonas sp. strain B13. J. Bacteriol. 180:4360-4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ravatn, R., S. Studer, A. J. B. Zehnder, and J. R. van der Meer. 1998. IntB13, an unusual site-specific recombinase of the bacteriophage P4 integrase family, is responsible for chromosomal insertion of the 105-kilobase clc element of Pseudomonas sp. strain B13. J. Bacteriol. 180:5505-5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ravatn, R., A. J. B. Zehnder, and J. R. van der Meer. 1998. Low-frequency horizontal transfer of an element containing the chlorocatechol degradation genes from Pseudomonas sp. strain B13 to Pseudomonas putida F1 and to indigenous bacteria in laboratory-scale activated-sludge microcosms. Appl. Environ. Microbiol. 64:2126-2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salyers, A. A., N. B. Shoemaker, A. M. Stevens, and L. Y. Li. 1995. Conjugative transposons: an unusual and diverse set of integrated gene transfer elements. Microbiol. Rev. 59:579-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis (ed.). 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Scott, J. R., and G. G. Churchward. 1995. Conjugative transposition. Annu. Rev. Microbiol. 49:367-397. [DOI] [PubMed] [Google Scholar]
- 39.Sentchilo, V., A. J. B. Zehnder, and J. R. van der Meer. Characterization of two alternative promoters for integrase expression in the clc genomic island of Pseudomonas sp. strain B13. Mol. Microbiol., in press. [DOI] [PubMed]
- 40.Showsh, S. A., and R. E. Andrews, Jr. 1992. Tetracycline enhances Tn916-mediated conjugal transfer. Plasmid 28:213-224. [DOI] [PubMed] [Google Scholar]
- 41.Simpson, A. J., F. C. Reinach, P. Arruda, F. A. Abreu, M. Acencio, R. Alvarenga, L. M. Alves, J. E. Araya, G. S. Baia, C. S. Baptista, M. H. Barros, E. D. Bonaccorsi, S. Bordin, J. M. Bove, M. R. Briones, M. R. Bueno, A. A. Camargo, L. E. Camargo, D. M. Carraro, H. Carrer, N. B. Colauto, C. Colombo, F. F. Costa, M. C. Costa, C. M. Costa-Neto, L. L. Coutinho, M. Cristofani, E. Dias-Neto, C. Docena, H. El-Dorry, A. P. Facincani, A. J. Ferreira, V. C. Ferreira, J. A. Ferro, J. S. Fraga, S. C. Franca, M. C. Franco, M. Frohme, L. R. Furlan, M. Garnier, G. H. Goldman, M. H. Goldman, S. L. Gomes, A. Gruber, P. L. Ho, J. D. Hoheisel, M. L. Junqueira, E. L. Kemper, J. P. Kitajima, J. E. Krieger, E. E. Kuramae, F. Laigret, M. R. Lambais, L. C. Leite, E. G. Lemos, M. V. Lemos, S. A. Lopes, C. R. Lopes, J. A. Machado, M. A. Machado, A. M. Madeira, H. M. Madeira, C. L. Marino, M. V. Marques, E. A. Martins, E. M. Martins, A. Y. Matsukuma, C. F. Menck, E. C. Miracca, C. Y. Miyaki, C. B. Monteriro-Vitorello, D. H. Moon, M. A. Nagai, A. L. Nascimento, L. E. Netto, A. Nhani, Jr., F. G. Nobrega, L. R. Nunes, M. A. Oliveira, M. C. de Oliveira, R. C. de Oliveira, D. A. Palmieri, A. Paris, B. R. Peixoto, G. A. Pereira, H. A. Pereira, Jr., J. B. Pesquero, R. B. Quaggio, P. G. Roberto, V. Rodrigues, A. J. de M. Rosa, V. E. de Rosa, Jr., R. G. de Sa, R. V. Santelli, H. E. Sawasaki, A. C. da Silva, A. M. da Silva, F. R. da Silva, W. A. da Silva, Jr., J. F. da Silveira, et al. 2000. The genome sequence of the plant pathogen Xylella fastidiosa. Nature 406:151-157. [DOI] [PubMed] [Google Scholar]
- 42.Springael, D., K. Peys, A. Ryngaert, S. Van Roy, L. Hooyberghs, R. Ravatn, M. Heyndrickx, J. R. van der Meer, C. Vandecasteele, M. Mergeay, and L. Diels. 2002. Community shifts in a seeded 3-chlorobenzoate degrading membrane biofilm reactor: indications for involvement of in situ horizontal transfer of the clc-element from inoculum to contaminant bacteria. Environ. Microbiol. 4:70-80. [DOI] [PubMed] [Google Scholar]
- 43.Sullivan, J. T., and C. W. Ronson. 1998. Evolution of rhizobia by acquisition of a 500-kb symbiosis island that integrates into a Phe-tRNA gene. Proc. Natl. Acad. Sci. USA 95:5145-5149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thiem, S. M., M. L. Krumme, R. L. Smith, and J. M. Tiedje. 1994. Use of molecular techniques to evaluate the survival of a microorganism injected into an aquifer. Appl. Environ. Microbiol. 60:1059-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thomas, C. M. (ed.). 2000. The horizontal gene pool. Harwood Academic Publishers, Singapore.
- 46.van der Meer, J. R., C. Werlen, S. F. Nishino, and J. C. Spain. 1998. Evolution of a pathway for chlorobenzene metabolism leads to natural attenuation in contaminated groundwater. Appl. Environ. Microbiol. 64:4185-4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van der Meer, J. R., R. Ravatn, and V. Sentchilo. 2001. The clc element of Pseudomonas sp. strain B13 and other mobile degradative elements employing phage-like integrases. Arch. Microbiol. 175:79-85. [DOI] [PubMed] [Google Scholar]
- 48.Zhang, X., A. D. McDaniel, L. E. Wolf, G. T. Keusch, M. K. Waldor, and D. W. Acheson. 2000. Quinolone antibiotics induce Shiga toxin-encoding bacteriophages, toxin production, and death in mice. J. Infect. Dis. 181:664-670. [DOI] [PubMed] [Google Scholar]
- 49.Zhou, J. Z., and J. M. Tiedje. 1995. Gene transfer from a bacterium injected into an aquifer to an indigenous bacterium. Mol. Ecol. 4:613-618. [DOI] [PubMed] [Google Scholar]