Abstract
The phosphate (Pi) starvation stimulon of Corynebacterium glutamicum was characterized by global gene expression analysis by using DNA microarrays. Hierarchical cluster analysis of the genes showing altered expression 10 to 180 min after a shift from Pi-sufficient to Pi-limiting conditions led to identification of five groups comprising 92 genes. Four of these groups included genes which are not directly involved in P metabolism and changed expression presumably due to the reduced growth rate observed after the shift or to the exchange of medium. One group, however, comprised 25 genes, most of which are obviously related to phosphorus (P) uptake and metabolism and exhibited 4- to >30-fold-greater expression after the shift to Pi limitation. Among these genes, the RNA levels of the pstSCAB (ABC-type Pi uptake system), glpQ (glycerophosphoryldiester phosphodiesterase), ugpAEBC (ABC-type sn-glycerol 3-phosphate uptake system), phoH (unknown function), nucH (extracellular nuclease), and Cgl0328 (5′-nucleotidase or related esterase) genes were increased, and pstSCAB exhibited a faster response than the other genes. Transcriptional fusion analyses revealed that elevated expression of pstSCAB and ugpAEBC was primarily due to transcriptional regulation. Several genes also involved in P uptake and metabolism were not affected by Pi starvation; these included the genes encoding a PitA-like Pi uptake system and a putative Na+-dependent Pi transporter and the genes involved in the metabolism of pyrophosphate and polyphosphate. In summary, a global, time-resolved picture of the response of C. glutamicum to Pi starvation was obtained.
Phosphorus (P) is an indispensable component of all cells in living organisms. In bacteria, P typically is assimilated as inorganic orthophosphate (Pi), which is transported into the cell by specific uptake systems. Alternatively, organophosphates and phosphonates may serve as sole P sources; either these compounds are imported by specific uptake systems and degraded intracellularly or Pi that is liberated by extracellular degradation of the compounds is taken up into the cell. For several bacteria, particularly Escherichia coli and Bacillus subtilis, P metabolism and the regulatory mechanisms that permit adaptation to varying P availability have been well studied (22, 47).
E. coli possesses three Pi uptake systems (19). PitA is expressed constitutively and transports phosphate in a proton motive force-dependent manner (47). When the extracellular Pi concentration falls below about 4 μM, Pi is taken up primarily by the Pst system at the expense of ATP. The Pst system is an ABC transport system encoded by the pstSCAB genes of the pstSCAB-phoU operon, which is induced under Pi starvation conditions (47). When formed, the PitB transporter encoded by a cryptic homolog of pitA is able to transport Pi in a manner similar to the manner used by PitA (19). The genes constituting the Pi starvation stimulon were identified in screening analyses based on transcriptional lacZ fusions (32) and by proteome analysis (44). Induction of the Pi starvation genes is dependent on the PhoR-PhoB two-component regulatory system. Under Pi starvation conditions the sensor kinase PhoR phosphorylates PhoB, and PhoB∼P in turn activates transcription of at least 31 genes, which form the PhoB regulon (47). These genes include the phoBR operon, the pstSCAB-phoU operon encoding the high-affinity Pi ABC transport system and a regulatory protein, the ugpBAECQ operon encoding an sn-glycerol 3-phosphate ABC uptake system and glycerophosphoryl diester phosphodiesterase, the phoA-psiF operon encoding alkaline phosphatase and a protein of unknown function, phoE encoding a polyanion porin, phoH encoding an ATP-binding protein, psiE (function unknown), and phnCDEFGHIJKLMNOP (uptake of phosphonates and degradation via the C-P lyase pathway) (47). Thus, when Pi is scarce, E. coli takes up Pi by an ATP-driven high-affinity transport system, mobilizes Pi from extracellular sources by phosphatases, and induces systems for the uptake and degradation of organophosphates, such as glycerol 3-phosphate, or of phosphonates, such as ethylphosphonate (47). The response of B. subtilis to Pi starvation involves increased expression of two alkaline phosphatases (phoA and phoB), of the high-affinity Pi uptake system encoded by pstSCAB1B2, of the phoPR and resABCDE operons encoding the PhoP-PhoR and ResD-ResE two-component regulatory systems as well as proteins involved in cytochrome c synthesis, of glpQ coding for glycerolphosphoryl phosphodiesterase, of 13 hypothetical genes (ydhF, yfhM, yhaX, yhbH, yheK, yjbC, ykoL, ykzA, ysnF, yttB, yvgO, yxiE, and csbD), of the phosphodiesterase gene phoD having a role in teichoic acid turnover, and of the tuaABCDEFGH operon coding for teichuronic acid biosynthesis (4, 25, 35). Reduced expression was observed for the tagAB and tagDEF teichoic acid biosynthesis operons (22). Thus, upon Pi starvation B. subtilis takes up Pi by an ATP-driven high-affinity uptake system, mobilizes phosphate extracellularly by using phosphatases, and replaces teichoic acid in the cell wall with the non-phosphate-containing compound teichuronic acid. In B. subtilis Pi starvation activates a specific response involving PhoPR, Spo0A, and ResDE (7, 22, 42), as well as the general stress response (22). Upon Pi starvation the sensor kinase PhoR phosphorylates the response regulator PhoP, and PhoP∼P activates transcription of phoPR and resABCDE and activates or represses transcription of other Pho regulon genes. ResD-ResE is required for full induction of the Pho regulon genes, and Spo0A is required for termination of the Pi response and subsequent initiation of sporulation. The general stress response is mediated by σB, and genes of the σB regulon code for proteins that protect DNA, membranes, and proteins against oxidative stress and are required for survival under extreme environmental conditions, such as those imposed by heat, acid or osmotic stress (18, 36).
In Mycobacterium tuberculosis, a pathogenic member of the Corynebacterineae and the causative agent of tuberculosis, it was recognized that the most immunogenic antigen, Pab, is present at higher levels under Pi starvation conditions (3) and is a phosphate-binding protein similar to E. coli PstS (11). It was then realized that the genome of M. tuberculosis harbors three different phosphate-binding protein genes in three operons (pstBS1C1A2, pstS2, and pstS3C2A1) at one locus (12, 27) and that the phosphate-binding proteins are surface attached (27). By using translational fusions, two Pi starvation-responsive promoters were identified for the pstBS1C1A2 operon (43). In the related organism Mycobacterium smegmatis, alkaline phosphatase activity was shown to increase under Pi starvation conditions (43).
Corynebacterium glutamicum is a gram-positive bacterium, and its genome has a high G+C content. C. glutamicum is used for biotechnological production of more than 106 tons of amino acids, especially l-glutamate and l-lysine, per year (38). Amino acids are derived from intermediates of the central carbon metabolism. As P assimilation occurs mainly in the form of Pi in the energy and carbon metabolism reactions, P metabolism is closely intertwined with energy and central carbon metabolism. Thus, the interplay between P metabolism and C metabolism is of particular interest in amino acid-producing C. glutamicum strains. However, very little is known about P metabolism or its regulation in C. glutamicum. In this study, the Pi starvation stimulon of C. glutamicum was determined by monitoring global gene expression changes in response to Pi availability.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
C.glutamicum wild-type strain ATCC 13032 was used for all experiments. C. glutamicum was cultivated in CGXII minimal medium (23) containing 0.03 g of protocatechuic acid per liter and 40 g of glucose per liter as a carbon and energy source. As the sole P source, 13 mM Pi was used under Pi-sufficient conditions and 0.13 mM Pi was used under Pi-limited conditions. For the Pi up-shift experiment, C. glutamicum was precultured at 30°C with agitation at 120 rpm in CGIII medium (31). After washing, the cells were cultivated in CGXII medium under Pi-limiting conditions for 24 h, and then cells were inoculated into CGXII medium with various Pi concentrations at a starting optical density at 600 nm (OD600) of 0.6. For the Pi down-shift experiment, cells were first precultured in CGIII medium, then cultured for 24 h in CGXII medium under Pi-sufficient conditions, and finally inoculated into the same medium at a starting OD600 of 0.6. Exponentially growing cells from this Pi-sufficient culture were harvested and washed. One aliquot was used for RNA preparation (zero time), and another aliquot was shifted into the CGXII medium with a limiting Pi concentration (0.13 mM) (referred to as Pi down-shift). RNA was prepared 10, 30, 60, 90, 120, and 180 min after the Pi down-shift and was used for global gene expression analysis in a comparison with preshift conditions (zero time). E.coli JM109 was used as the host for construction of reporter plasmids and was cultivated in Luria-Bertani medium (39) at 37°C and 170 rpm.
Generation of C. glutamicum DNA microarrays.
DNA microarrays based on PCR products of C. glutamicum were generated for use in global gene expression analyses by using the procedures described previously (24, 34, 48, 50). The genes were amplified in 96-well plates with genomic DNA of C. glutamicum ATCC 13032 as the template and gene-specific primers purchased from degussa (Frankfurt, Germany). The identities and quality of the PCR products were checked by gel electrophoresis, and the PCR products were precipitated with isopropanol, resuspended in 3× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate, pH 7.0), and transferred to 384-well plates as described previously. The PCR products were printed onto poly-l-lysine-coated glass slides by using an arraying robot. The DNA microarrays were rehydrated in a 1× SSC atmosphere, UV cross-linked (650 μJ), and blocked in 230 ml of methyl pyrrolidinone containing 15 ml of 1 M boric acid (titrated to pH 8.0 with sodium hydroxide) and 4.4 g of succinic anhydride. The C. glutamicum whole-genome DNA microarray contained 3,673 PCR products covering 2,860 of the 2,994 genes (506 genes in duplicate) described for the genome according to the National Center for Biotechnology Information (NCBI) (accession no. NC003450) and 284 additional putative coding sequences (23 sequences in duplicate). In general, the PCR products were 500 ± 50 bp long and represented regions of the genes which facilitate specific hybridization. Additionally, 100 spots of C. glutamicum genomic DNA were used as normalization controls, and 16 spots of λ DNA, 16 spots of E. coli DNA, and one spot of the E. coli aceK gene were used as negative controls.
Preparation of total RNA.
Exponentially growing cells at an OD600 of about 5.0 were poured into ice-containing tubes precooled to −70°C and were harvested by centrifugation (5 min, 3,500 × g, 4°C) (48). The cell pellet either was directly subjected to RNA isolation or was immediately frozen in liquid nitrogen and stored at −70°C until it was used. For isolation of total RNA, the (frozen) cell pellet was resuspended in 350 μl of RLT buffer of the RNeasy system (Qiagen, Hilden, Germany). After this, 250 mg of 0.1-mm-diameter zirconia-silica beads (Roth, Karlsruhe, Germany) were added, and the cells were disrupted by 15 and 30 s of bead beating with a Silamat S5 (Vivadent, Ellwangen, Germany). After centrifugation (1 min, 13,000 × g), the supernatant was used for RNA preparation by using the RNeasy system with DNase I treatment according to manufacturer's instructions. Isolated RNA samples were checked for purity by denaturing formaldehyde agarose gel electrophoresis and spectrophotometrically and were kept at −70°C until they were used (34).
Gene expression analysis with C. glutamicum DNA microarrays.
Identical amounts (20 to 25 μg) of total RNA were used for random hexamer-primed synthesis of fluorescently labeled cDNA by reverse transcription with Superscript II (GibcoBRL/Life Technologies, Gaithersburg, Md.) and the fluorescent nucleotide analogues FluoroLink Cy3-dUTP (green) and Cy5-dUTP (red) (Amersham Pharmacia, Little Chalfont, United Kingdom) as described previously (24, 34, 48). The labeled cDNA probes were purified and concentrated by using Microcon YM-30 filter units (Millipore, Bedford, Mass.) (24). Fluorescently labeled cDNA probes containing 1.2 μg of poly(A) (Sigma, Munich, Germany) per ml as a competitor, 30 mM HEPES, and 0.3% sodium dodecyl sulfate in 3× SSC were hybridized to arrays in a humid chamber for 5 to 16 h at 65°C. After hybridization, the arrays were washed in 1× SSC-0.03% sodium dodecyl sulfate and finally in 0.05× SSC (24).
Immediately after stringent washing, the fluorescence intensities at 635 and 532 nm were determined with a GenePix 4000 laser scanner (Axon Inc., Union City, Calif.), and the images were processed as TIFF images. Raw fluorescence data were analyzed quantitatively by using GenePix 3.0 software (Axon Inc.). Data were normalized to the average ratio for C. glutamicum genomic DNA. The normalized ratio of the median was taken to reflect the relative RNA abundance for hybridization signals with a green or red fluorescence signal that was at least threefold greater than the median fluorescence background signal. For statistical analysis of the gene expression data (5, 20) P values for the independent replicate experiments were calculated based on the Student t test by using log-transformed fluorescence ratios for individual genes on the one hand and for genomic DNA on the other hand (28, 34). Of the genes that showed significantly changed RNA levels (P < 0.05), those with at least fourfold-increased or -decreased average RNA levels were considered further and subjected to a hierarchical cluster analysis by the average linkage clustering method (13).
Construction of transcriptional fusions.
The promoter regions upstream of the pstSCAB and ugpAEBC operons were amplified with primers PpstN (′-CG-GGATCC-TGCGGACTGCTGGGAAGATG-3′), PpstC (5′-CCC-AAGCTT-TAAGAATCGGTGATTTTCGTTCC-3′), PugpN (5′-CCC-AAGCTT-TTGGTGCGAAGGATTCCGATTC-3′), and PugpC (5′-CG-GGATCC-TCTGTCCGCCTTGATCTCTTGG-3′). Amplified fragments were subcloned into the pGEM-T vector (Promega, Madison, Wis.) and then into the corynebacterial promoter-probe pET2 vector (46). The pstS-cat and ugpA-cat fusion vectors pET2-pst and pET2-ugp were introduced into the C. glutamicum wild type by electroporation by using the following conditions: 25 μF, 600 Ω, and 2.5 kV/cm. After electroporation, 1 ml of Luria-Bertani medium was immediately added to each sample. Then the sample was exposed to 46°C for 6 min and incubated at 30°C for 90 min for regeneration. Each promoter activity was measured by determining the chloramphenicol acetyltransferase activity. The cultivation conditions used for C. glutamicum ATCC 13032(pET2-pst) and ATCC 13032(pET2-ugp) were the conditions described above for the Pi down-shift experiment. Chloramphenicol acetyltransferase activity was determined as described by Shaw (40), and protein concentrations were determined as described by Gornall et al.(16) with bovine serum albumin as the standard.
RESULTS
Phosphate as a sole P source for growth of C. glutamicum.
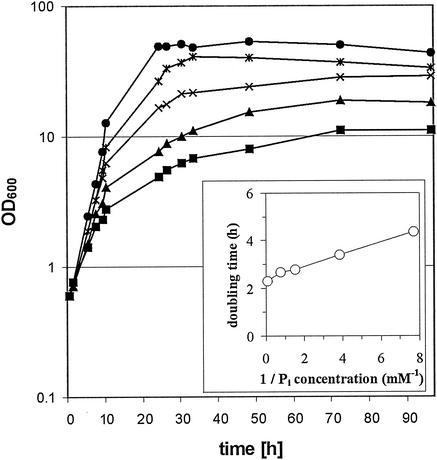
Initially, the growth characteristics of C. glutamicum with inorganic phosphate as the sole P source were determined. When C. glutamicum was grown in CGXII minimal medium with 40 g of glucose per liter, which contained 13 mM potassium phosphate as the sole P source, the cultures reached a final OD600 of 60. When the cells were washed and transferred to CGXII-glucose medium lacking a P source, they still grew and reached a final OD600 of 12 (data not shown), which indicated that there were internal P sources, such as polyphosphate (26). C. glutamicum cells precultured under Pi-limiting conditions showed almost no growth in the absence of a P source and proportional increases in the cell yield as well as the growth rate with increasing Pi concentrations when they were cultivated in CGXII-glucose medium (Fig. 1). The Pi concentration that supported growth of C. glutamicum with a half-maximal growth rate (Monod constant) was calculated to be 0.1 mM. Similar results were obtained when C. glutamicum was cultured with sodium phosphate instead of potassium phosphate as the sole P source (data not shown).
FIG. 1.
Growth of C. glutamicum ATCC 13032 in CGXII minimum medium with 40 g of glucose per liter and different concentrations of Pi. Cells were precultured in CGXII medium with 0.13 mM potassium phosphate for 24 h before inoculation into CGXII medium containing 0.13 mM (▪), 0.26 mM (▴), 0.65 mM (×), 1.3 mM (∗), or 13 mM (•) potassium Pi. (Inset) Doubling times plotted against reciprocal Pi concentrations.
Comparison of global gene expression during growth under Pi-limiting and Pi-sufficient conditions.
In order to identify genes that were differentially expressed in response to different Pi concentrations in the growth medium, the global gene expression patterns of C. glutamicum that was either maintained under Pi-limiting conditions or shifted from Pi-limiting to Pi-sufficient conditions were analyzed by using DNA microarrays. Cells were precultured in CGXII-glucose medium under Pi-limiting conditions (0.13 mM Pi) for 24 h and then used to inoculate fresh medium containing either a sufficient (13 mM) or limiting (0.13 mM) Pi concentration. After 7.5 h, exponentially growing cells (OD600, 4.3 and 2.1, respectively) were harvested, and RNA was prepared. Four independent DNA microarray experiments were carried out, two with potassium phosphate and two with sodium phosphate as the P source. Genes that showed reliable hybridization signals in at least three of four experiments and that were differentially expressed (RNA ratios higher than 4 or lower than 0.25) are listed in Table 1. Under Pi-limiting conditions, 14 genes showed decreased expression (RNA ratios less than 0.25) compared to the expression under Pi-sufficient conditions, and 14 genes showed increased expression (RNA ratios greater than 4) (Table 1). The former group included 12 ribosomal protein-encoding genes, the translation initiation factor gene (open reading frame [ORF] 2648, Cgl1378), and a gene whose function is unknown (ORF 2614). This finding reflected the slower growth under Pi-limiting conditions, as growth rates of 0.30 and 0.16 h−1 were observed under Pi-sufficient and Pi-limiting conditions, respectively. It was not clear whether expression of ORF 2614, which codes for a hypothetical protein containing 89 amino acids and does not seem to be part of an operon, was reduced as a consequence of slower growth. The group of genes which exhibited increased expression under Pi-limiting conditions included nine genes which are known to be induced by Pi starvation in other bacteria, one gene (ORF 2851, nucH) encoding a putative extracellular nuclease (a member of the endonuclease-exonuclease-phosphatase family, Pfam domain PF03372), and four genes without assigned functions (ORFs 1086, 1598, 1760, and 3082). The known phosphate starvation-inducible genes include phoH, glpQ, ugpAEBC (expression of ugpC increased only 2.5-fold), and pstSCAB (Table 1). An alkaline phosphatase gene (ORF 1400, Cgl0849) also showed increased expression, albeit to a lesser degree (3.4-fold).
TABLE 1.
Gene expression changes during growth of C. glutamicum under Pi-limiting and Pi-sufficient conditions
| ORF | NCBI no. | Annotation | Gene | Avg mRNA level under Pi-limiting conditions/avg mRNA level under Pi-sufficient conditionsa |
|---|---|---|---|---|
| 442 | Cgl0065 | Phosphate starvation-inducible protein | phoH | 4.6 |
| 976 | Cgl0485 | Ribosomal protein L10 | 0.1 | |
| 988 | Cgl0493 | Ribosomal protein S12 | 0.2 | |
| 989 | Cgl0494 | Ribosomal protein S7 | 0.1 | |
| 992 | Cgl0514 | Ribosomal protein S3 | 0.1 | |
| 993 | Cgl0515 | Ribosomal protein L16/L10E | 0.2 | |
| 994 | Cgl0516 | Ribosomal protein L29 | 0.2 | |
| 999 | Cgl0521 | Ribosomal protein L14 | 0.2 | |
| 1000 | Cgl0522 | Ribosomal protein L24 | 0.2 | |
| 1020 | Cgl0539 | Ribosomal protein L18 | 0.2 | |
| 1022 | Cgl0540 | Ribosomal protein S5 | 0.1 | |
| 1050 | Cgl0566 | Ribosomal protein L17 | 0.2 | |
| 1086 | Cgl0596 | Hypothetical protein | 9.6 | |
| 1421 | Cgl0869 | Ribosomal protein L28 | 0.2 | |
| 1598 | Cgl1021 | Hypothetical membrane protein | 4.1 | |
| 1760 | Cgl1170 | Hypothetical protein | 10.1 | |
| 2614 | (855659-855946) | Questionable ORF | 0.2 | |
| 2637 | Cgl1387 | Glycerophosphoryl diester phosphodiesterase | glpQ | 6.4 |
| 2639 | Cgl1385 | Glycerol-3-phosphate ABC-type transporter, periplasmic component | ugpB | 13.6 |
| 2640 | Cgl1384 | Glycerol-3-phosphate ABC-type transporter, permease component | ugpE | 10.7 |
| 2641 | Cgl1383 | Glycerol-3-phosphate ABC-type transporter, permease component | ugpA | 12.6 |
| 2648 | Cgl1378 | Translation initiation factor IF-3 | 0.2 | |
| 2851 | Cgl2592 | Predicted extracellular nuclease | nucH | 9.4 |
| 2871 | Cgl2575 | Phosphate ABC-type transporter, periplasmic component | pstS | 17.8 |
| 2873 | Cgl2574 | Phosphate ABC-type transporter, permease component | pstC | 12.1 |
| 2875 | Cgl2573 | Phosphate ABC-type transporter, permease component | pstA | 7.5 |
| 2876 | Cgl2572 | Phosphate ABC-type transporter, ATPase component | pstB | 9.3 |
| 3082 | Cgl2336 | Hypothetical protein | 4.2 |
The relative mRNA levels under Pi-limiting and Pi-sufficient conditions are averages from four experiments. Only ORFs with P values of <0.05 as determined by a Student's t test and with relative mRNA levels equal to or greater than 4 or equal to or less than 0.25 are shown.
Kinetics of gene expression changes after a shift from Pi-sufficient to Pi-limiting conditions.
In contrast to the set of experiments described above, in which gene expression was analyzed in cells grown for 7.5 h under Pi-limiting or Pi-saturating conditions, the experiments described below were performed to detect both the short-term and the long-term responses of C. glutamicum after a shift from Pi-excess conditions to Pi-limiting conditions (i.e., the kinetics of the response). Cells growing exponentially under Pi-sufficient conditions (13 mM) were harvested. RNA was prepared from one aliquot, and another aliquot was used to inoculate parallel cultures with medium containing a limiting Pi concentration (0.13 mM). From these cultures RNA was prepared 10, 30, 60, 90, 120, and 180 min after transfer. By using DNA microarrays the global gene expression at each of these times was compared to that of the culture before the transfer (zero time). Table 2 summarizes the observed expression differences and lists genes that were reliably detected and showed at least at one time point expression that was significantly altered by at least a factor of 4. Subsequently, hierarchical cluster analysis of these genes revealed five groups or subclusters of genes which showed similar expression profiles under the conditions used (Fig. 2).
TABLE 2.
Changes in gene expression at different times during the Pi starvation response of C. glutamicum
| ORF | NCBI no. | Annotation | Gene | Avg mRNA level after shift of Pi-limiting conditions/avg mRNA level before shift to Pi-limiting conditionsb
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| 10 min | 30 min | 60 min | 90 min | 120 min | 180 min | ||||
| 195 | Cgl2814 | Sulfate adenylate transferase subunit 1 | 2.2c | 0.3c | 0.2c | 0.4c | |||
| 312 | Cgl2917 | Putative integral membrane transport protein, similar to shikimate transport protein shiA | 10.0 | 0.4 | 0.6 | 0.5c | |||
| 348 | Cgl2946 | Hypothetical membrane protein | 1.2c | 0.7 | 0.3c | 0.2c | 0.2c | 0.1c | |
| 376 | Similar to heavy-metal-transporting ATPases | 2.2c | 5.7c | 3.4 | 2.5 | 1.3 | 1.0 | ||
| 378 | Cgl2965 | Two-component system, response regulator | 1.5 | 7.1c | 6.0c | 4.3c | 2.1c | 1.3 | |
| 380 | Cgl2966 | Hypothetical protein | 8.3c | 6.4c | 2.7c | 1.3 | 1.0 | ||
| 381 | Cgl2967 | Putative multicopper oxidase | cumA | 1.8c | 9.3c | 6.1c | 3.1c | 1.3 | 0.8 |
| 383 | Cgl2968 | Hypothetical protein | 1.8 | 5.7c | 3.4 | 1.0 | 0.6c | ||
| 399 | Cgl2979 | Hypothetical protein | 3.9c | 4.8c | 1.7 | 1.1 | 1.1 | 0.9 | |
| 403 | Cgl2982 | Single-stranded DNA-binding protein | ssb | 0.6c | 0.2c | 0.2c | |||
| 441 | Cgl0065 | Phosphate starvation-inducible protein | phoH | 1.1 | 1.8 | 3.3c | 4.4c | 4.7c | 4.5c |
| 442 | Cgl0065 | Phosphate starvation-inducible protein | phoH | 1.4 | 1.4 | 5.9c | 4.3c | 6.8c | 4.8c |
| 766 | Cgl0328 | 5′-Nucleotidase or related esterase | 1.2 | 7.3c | 6.5c | 19.3c | 10.3c | 7.2c | |
| 854 | Cgl0388 | Hypothetical membrane protein | 0.2c | 0.1c | 0.2c | 0.3c | 1.9 | 1.3 | |
| 856 | Cgl0390 | ABC-type transporter, permease component, similar to heme uptake systems | 0.4c | 0.2c | 0.3 | 0.5 | 1.6 | ||
| 857 | Cgl0391 | ABC-type transporter, ATPase component, similar to heme uptake systems | 0.3c | 0.2c | 0.3 | 0.5 | 1.5 | 0.9 | |
| 976 | Cgl0485 | Ribosomal protein L10 | 0.8 | 0.8 | 0.2c | 0.1c | 0.1c | 0.0c | |
| 978 | Cgl0486 | Ribosomal protein L7/L12 | 0.7 | 0.8 | 0.2c | 0.3c | 0.3 | 0.2c | |
| 988 | Cgl0493 | Ribosomal protein S12 | 1.5c | 0.8 | 0.3c | 0.3c | 0.2c | 0.2c | |
| 992 | Cgl0514 | Ribosomal protein S3 | 0.9 | 0.7 | 0.2c | 0.1c | 0.1c | 0.1c | |
| 993 | Cgl0515 | Ribosomal protein L16/L10E | 0.7 | 0.1c | 0.1c | ||||
| 994 | Cgl0516 | Ribosomal protein L29 | 0.9 | 0.7 | 0.2c | 0.1c | |||
| 999 | Cgl0521 | Ribosomal protein L14 | 0.3c | 0.2c | |||||
| 1000 | Cgl0522 | Ribosomal protein L24 | 1.2 | 0.8 | 0.2c | 0.2c | 0.1c | 0.1c | |
| 1020 | Cgl0539 | Ribosomal protein L18 | 1.0 | 0.7 | 0.2c | 0.2 | 0.2 | 0.1c | |
| 1044 | Cgl0560 | Translation initiation factor IF-1 | 1.1 | 0.4 | 0.3c | 0.4 | 0.2c | 0.4 | |
| 1047 | Cgl0562 | Ribosomal protein S11 | 1.1 | 0.7 | 0.2c | 0.2c | 0.2c | ||
| 1048 | Cgl0563 | Ribosomal protein S4 | 1.0 | 0.8 | 0.3c | 0.2c | 0.2c | ||
| 1050 | Cgl0566 | Ribosomal protein L17 | 1.0 | 0.7 | 0.3c | 0.2c | 0.2c | ||
| 1086 | Cgl0596 | Hypothetical protein | 10.8c | 18.8c | 17.6c | 15.0c | 8.9c | ||
| 1168 | Cgl0665 | Similar to vibriobactin utilization protein viuB | 0.1c | 0.3c | 1.3 | 0.8 | |||
| 1169 | Cgl0666 | ABC-type transporter, ATPase component, similar to iron uptake systems | 0.2c | 0.2c | 1.1 | 0.7 | |||
| 1170 | Cgl0667 | ABC-type transporter, permease component, similar to iron uptake systems | 0.1c | 0.2c | 1.0 | ||||
| 1173 | Cgl0669 | ABC-type transporter, periplasmic component, similar to iron uptake systems | 0.2c | 0.2c | 0.2c | 0.3c | 1.0 | 0.7 | |
| 1205 | Cgl0695 | Methylisocitrate lyase | prpB1 | 3.3c | 3.5c | 4.7c | 3.9c | ||
| 1206 | Cgl0696 | Methylcitrate synthase | prpC1 | 0.6 | 1.2 | 3.7c | 4.2c | 2.9c | |
| 1346 | Cgl0807 | Siderophore-interacting protein | 0.1c | 0.2c | 1.1 | ||||
| 1347 | Cgl0808 | ABC-type cobalamin/Fe3+-siderophore transport | 0.1c | 0.1c | 0.1c | 0.2c | 1.3 | 0.8 | |
| 1349 | Cgl0810 | ABC-type cobalamin/Fe3+-siderophore transport | 0.1c | 0.1c | 0.2c | 0.2c | 1.0 | 0.5 | |
| 1351 | Cgl0812 | ABC-type cobalamin/Fe3+-siderophore transport | 0.2c | 0.2c | 0.3c | ||||
| 1352 | Cgl0813 | ABC-type cobalamin/Fe3+-siderophore transport | 0.1c | ||||||
| 1420 | Similar to ribosomal protein L33 of E. coli (putative sequencing error) | 1.1 | 0.6 | 0.2c | 0.4 | 0.2c | |||
| 1421 | Cgl0869 | Ribosomal protein L28 | 1.0 | 0.8 | 0.2c | 0.2c | 0.2c | 0.2c | |
| 1558 | Cgl0982 | AraC-type DNA-binding domain-containing protein | 0.2c | 0.2c | 0.2c | 0.4c | 2.6 | 1.1 | |
| 1598 | Cgl1021 | Hypothetical membrane protein | 0.8 | 1.6c | 4.5c | 7.8c | 7.8c | 6.9c | |
| 1645 | Cgl1067 | Cysteine sulfinate desulfinase | 1.1c | 1.2 | 0.5c | 0.3c | 0.2c | 0.3c | |
| 1646 | Cgl1068 | Nicotinate-nucleotide pyrophosphorylase | 1.0 | 1.0 | 0.3c | 0.1c | 0.1c | 0.1c | |
| 1647 | Cgl1069 | Quinolinate synthase | 0.8 | 1.1 | 0.3c | 0.1c | 0.1c | 0.1c | |
| 1655 | Cgl1077 | 2-Polyprenyl-6-methoxyphenol hydroxylase | 4.6c | 1.6 | 0.9c | 1.0 | 1.0 | 0.9 | |
| 1722 | Cgl1139 | Methionine synthase II | 0.7 | 0.2c | 0.2c | 0.6c | |||
| 1855 | Cgl1248 | Siderophore-interacting protein | 0.4c | 0.3c | 4.3 | ||||
| 1875 | Cgl2996 | myo-Inositol-1-phosphate synthase | 0.8c | 1.3c | 0.8 | 0.4c | 0.2c | 0.5c | |
| 2146 | Cgl2035 | ABC-type transport systems, periplasmic protein, similar to iron(III) dicitrate uptake systems | 0.3c | 0.2c | 0.3c | 1.6 | 1.0 | ||
| 2168 | Questionable ORF | 1.6c | 1.1 | 0.3c | 0.2c | 0.2c | |||
| 2261 | Cgl2133 | Predicted Co/Zn/CD cation transporter | 0.8 | 1.2 | 2.0c | 2.7c | 3.5c | 4.8c | |
| 2262 | Cgl2134 | Dehydrogenase | 0.6c | 1.2 | 2.3c | 2.5c | 3.3c | 4.4c | |
| 2553 | Cgl1460 | ABC-type transporter, ATPase component | 1.0 | 2.8 | 3.0c | 5.0c | 2.7c | 2.3/PICK> | |
| 2554 | Cgl1459 | ABC-type transporter, permease component | 1.2 | 6.3 | 3.6c | 3.4c | 2.9c | 2.1 | |
| 2614 | Questionable ORF | 1.2c | 0.7 | 0.3c | 0.2c | 0.2c | 0.1c | ||
| 2624 | Cgl1399 | Arginine repressor | argR | 5.0c | 0.7 | 0.7c | 0.8 | 0.8 | 0.8 |
| 2625 | Cgl1398 | Ornithine carbamoyltransferase | argF | 5.5c | 0.6 | 0.6c | 0.6c | 0.7 | 0.7 |
| 2637 | Cgl1387 | Glycerophosphoryl diester phosphodiesterase | glpQ | 1.5 | 2.8c | 4.7c | 5.0c | 9.2c | 7.5c |
| 2638 | Cgl1386 | Glycerol-3-phosphate ABC-type transporter, ATPase component | ugpC | 1.1 | 4.2c | 3.9c | 4.3c | 4.5c | 4.5c |
| 2639 | Cgl1385 | Glycerol-3-phosphate ABC-type transporter, periplasmic component | ugpB | 2.2 | 12.9c | 22.8c | 18.7c | 17.2c | 14.5c |
| 2640 | Cgl1384 | Glycerol-3-phosphate ABC-type transporter, permease component | ugpE | 1.6c | 9.8c | 22.3c | 15.0c | 11.7c | 18.1c |
| 2641 | Cgl1383 | Glycerol-3-phosphate ABC-type transporter, permease component | ugpA | 2.1 | 15.1c | 16.8c | 15.0c | 11.0c | 12.5c |
| 2647 | Cgl1379 | Ribosomal protein L35 | 1.6c | 1.0 | 0.4c | 0.3c | 0.2c | 0.2c | |
| 2648 | Cgl1378 | Translation initiation factor IF3 | 2.0c | 1.1 | 0.3c | 0.2c | 0.1c | 0.2c | |
| 2779 | Cgl1281 | Co/Zn/Cd efflux system component | 3.8c | 4.7c | 1.9c | 1.0 | 0.6c | 0.7c | |
| 2831 | Cgl2607 | Two-component system, response regulator | 8.1c | 2.3 | 1.4 | 1.1 | 1.1 | 1.5c | |
| 2832 | Cgl2606 | Two-component system, sensory transduction | 4.7c | 1.7 | 1.0 | 0.8 | 0.8 | 0.9 | |
| 2849 | Cgl2594 | Predicted deacetylase | 4.7 | 2.0c | 2.1 | 2.1c | |||
| 2851 | Cgl2592 | Predicted extracellular nuclease | nucH | 4.8c | 19.5c | 31.7c | 25.8c | 16.9c | |
| 2871 | Cgl2575 | Phosphate ABC-type transporter, periplasmic component | pstS | 5.0c | 18.0c | 23.2c | 21.5c | 19.1c | 13.8c |
| 2873 | Cgl2574 | Phosphate ABC-type transporter, permease component | pstC | 4.4c | 19.3c | 23.6c | 22.8c | 20.9c | 19.1c |
| 2875 | Cgl2573 | Phosphate ABC-type transporter, permease component | pstA | 1.9 | 5.8c | 12.5c | 8.6 | 7.1c | 6.9c |
| 2876 | Cgl2572 | Phosphate ABC-type transporter, ATPase component | pstB | 2.7c | 9.0c | 12.4c | 14.5 | 10.3c | 8.3c |
| 2895 | Questionable ORF | 1.3 | 0.7 | 0.3c | 0.2c | 0.2c | 0.1c | ||
| 3081 | Cgl2336 | Hypothetical protein | 1.0 | 2.8c | 3.8c | 4.2c | 4.6c | 4.2c | |
| 3082 | Cgl2336 | Hypothetical protein | 4.3c | 5.3 | 5.0c | 5.3c | |||
| 3086 | Questionable ORF | 0.5c | 0.2 | 0.4c | 0.3c | 0.5c | |||
| 3326 | Cgl1067 | Cysteine sulfinate desulfinase | 0.6 | 0.2c | |||||
| 3347 | Cgl1714 | Hypothetical protein | 0.2c | 0.2 | 0.3c | 4.2 | 2.5 | ||
| 3362 | Questionable ORF | 1.8c | 2.7 | 4.7c | |||||
| 3442 | Cgl3064 | Hypothetical membrane protein, similar to phosphoesterases | 2.3 | 5.3 | 7.3 | 11.9c | 6.3 | ||
| 3458 | Cgl3075 | ABC-type transport systems, periplasmic component, similar to iron uptake systems | 0.1c | 0.1c | 0.1c | 0.1c | 0.5 | 0.3 | |
| 3472 | Cgl2526 | Ferritin-like protein | 10.3c | 10.3c | 5.8c | 5.5c | 1.8 | 1.6 | |
| 64185 | Cgl2389 | Acyl-CoA:acetate CoA transferase beta subunit | 7.3c | 1.5 | 0.4 | ||||
| 64187 | Cgl2393 | Predicted hydrolase/acyltransferase | 7.7c | 1.7 | 0.8 | ||||
| 66073 | Cgl2397 | Protocatechuate 3,4-dioxygenase beta subunit | 5.2c | 1.8 | 1.3 | 1.1 | 1.1 | 1.0 | |
| 66513 | Cgl2392 | Acetyl-CoA acetyltransferase | 4.4c | 1.8 | 0.6 | 0.5 | 0.7 | ||
| 69134 | Cgl2642 | Phosphotransferase system IIC component | 1.3 | 0.5 | 0.4c | 0.2c | 0.3c | 0.2 | |
The relative mRNA levels 10, 30, 60, 90, 120, and 180 min after the shift to Pi-limiting conditions compared to the levels under preshift conditions are averages from three experiments. Only ORFs whose mRNA levels were equal to or greater than 4 or equal to or less than 0.25 at at least one time are shown.
Boldface type indicates relativemRNA levels that were equal to or greater than 10.
P < 0.05 as determined by a t test.
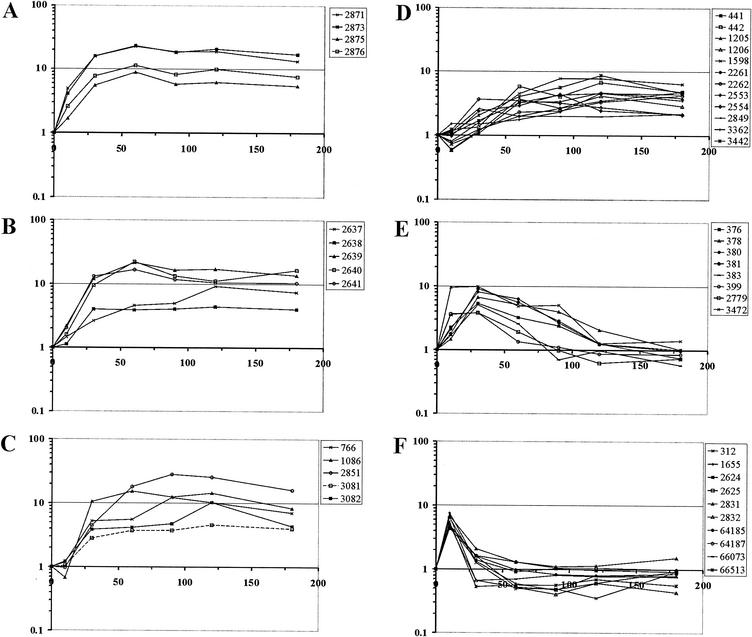
FIG. 2.
Hierarchical cluster analysis of gene expression changes during the response of C. glutamicum ATCC 13032 to Pi starvation. Gene expression data from 12 microarray experiments (columns) and 92 genes (lines) are represented. The microarray experiments included comparing gene expression of C. glutamicum ATCC 13032 before and 10, 30, 60, 90, 120, and 180 min after a shift from Pi-sufficient conditions to Pi-limiting conditions. Subclusters 1 to 5 are indicated by red, blue, pink, green, and brown, respectively. The scale bar indicates the color coding of the relative RNA levels.
Subcluster 1 included 25 genes which showed increased expression at a relatively early stage of adaptation and exhibited high expression levels until 180 min after the onset of Pi limitation. Except for ORF 1760, a gene encoding a hypothetical protein similar to yceI in E. coli, this subcluster contained all of the genes that showed increased expression in the experiments described above (Table 1): pstSCAB, glpQ, ugpAEBC, nucH, and the hypothetical genes ORFs 1086, 3081, and 3082 (Table 2). In addition, subcluster 1 comprised genes presumably involved in P metabolism, including the phosphate starvation-inducible gene phoH (represented by two PCR products), the Cgl0328 gene putatively encoding a 5′-nucleotidase or related esterase, the genes encoding an ABC transporter of unknown function (ORFs 2553 and 2554), and a gene encoding a phosphoesterase (ORF 3442). Furthermore, a number of genes not obviously related to P metabolism showed increased RNA levels after the shift to Pi limitation; these genes included the genes encoding methylcitrate lyase (prpB1), methylcitrate synthase (prpC1), ferrochelatase (ORF 2849), two hypothetical proteins (ORFs 3081 and 3362), a permease of unknown function (ORF 2261), and glucose-1-dehydrogenase (ORF 2262). It is not yet clear whether these proteins play a major role, direct or indirect, in adaptation of C. glutamicum to Pi starvation. With regard to the kinetics of expression changes, the genes of subcluster 1 can be ordered. Expression of the high-affinity Pi uptake system genes pstSCAB increased first after the shift to phosphate starvation conditions (Fig. 3A); this was followed by increases in expression of the sn-glycerol 3-phosphate uptake genes ugpAEBC and glpQ (Fig. 3B) and of the genes encoding extracellular nuclease (ORF 2851, nucH), 5′-nucleotidase or the related esterase/5′-nucleotidase (Cgl0328, ORF 766), and the hypothetical genes ORFs 1086, 3081, and 3082 (Fig. 3C). All other genes in subcluster 1 exhibited smaller and slower increases in expression (Fig. 3D).
FIG. 3.
Kinetics of changes in gene expression during the response of C. glutamicum ATCC 13032 to Pi starvation. Changes in gene expression 10, 30, 60, 90, 120, and 180 min after a shift from Pi-sufficient to Pi-limiting conditions compared to preshift conditions taken from Table 2 are shown for selected ORFs (see text for details). (A to D) Kinetics of changes in gene expression for subcluster 1 genes (Fig. 2): genes encoding the putative high-affinity phosphate transporter pstSCAB (ORFs 2871 to 2876) (A), the glycerophosphoryl diester phosphodiesterase and the sn-glycerol 3-phosphate transport system (glpQ and ugpAEBC, ORFs 2637 to 2641) (B), and the putative extracellular nuclease (ORF 2851, nucH), 5′-nucleotidase or a related esterase (Cgl0328, ORF 766), and the hypothetical proteins (ORFs 1086, 3081, and 3082) (C) and all other subcluster 1 genes (D). (E and F) Kinetics of changes in gene expression for the subcluster 2 genes (Fig. 2) except ORF 2831 (E) and for subcluster 3 genes and ORF 2831 (F).
Subcluster 2 (Fig. 2) included nine genes whose expression was transiently increased after the Pi down-shift, but the RNA levels slowly reached the same levels as before the shift (Fig. 3E and Table 2). Five of these genes are part of a chromosomal locus encoding a copper-exporting ATPase, an associated protein, a two-component regulatory system, a hypothetical protein, a multicopper oxidase, and a thiol-disulfide interchange protein (ORFs 376 to 383). Additionally, genes encoding a response regulator (ORF 2831), a nonheme ferritin (ORF 3472), a putative cation efflux protein, and two hypothetical proteins (ORFs 399 and 2779) belonged to subcluster 2.
Subcluster 3 contained nine genes whose expression rapidly increased after the Pi down-shift, but as soon as 30 min after the Pi down-shift preinduction RNA levels were reached (Fig. 3F). Five of these genes (ORFs 1655, 64185, 64187, 66073, and 66513) encode enzymes or subunits of enzymes predicted to be involved in the degradation of protocatechuate, which is present in CGXII medium to facilitate iron uptake, and similar compounds to yield acetyl coenzyme A (acetyl-CoA) and succinyl-CoA. Additionally, the genes encoding a putative transporter (ORF 312), the arginine repressor and ornithine carbamoyltransferase (ORFs 2624 and 2625), and a sensor kinase (ORF 2832) showed short-term expression increases after the onset of Pi limitation. Interestingly, expression of the gene encoding the corresponding response regulator (ORF 2831) transiently increased in a similar manner (Table 2 and Fig. 3F).
Subcluster 4 (Fig. 2) comprised 29 genes that showed decreased RNA levels 60 min after the onset of Pi limitation and continued to exhibit low levels at later times. Most of these genes code for proteins involved in translation or DNA replication; 15 genes code for ribosomal proteins (ORFs 976, 978, 988, 992 to 994, 999, 1000, 1020, 1047, 1048, 1050, 1420, 1421, and 2647), two genes code for translation initiation factors (ORFs 1044 and 2648), and one gene codes for the single-stranded DNA-binding protein (ORF 403). Reduced expression of these genes, as well as four hypothetical genes (ORFs 348, 2168, 2614, and 2895) and seven other genes (ORFs 1645 to 1647, 1722, 1875, 3326, and 69134), was correlated with reduced growth after the shift to Pi-limiting conditions.
Subcluster 5 included 18 genes which exhibited transiently reduced expression after the Pi down-shift. The RNA levels were lower 10, 30, 60, and 90 min after the onset of Pi limitation, but after 120 and 180 min the RNA levels were the same as those before the shift. Fifteen of these genes are presumably involved in iron metabolism and putatively encode a heme transport system and associated proteins (ORFs 854 to 857), two ferric siderophore transport systems and an associated protein (ORFs 1168 to 1173, 1346 to 1348, and 1349 to 1352), two ferric dicitrate-binding proteins (ORFs 3458 and 2146), and a siderophore utilization protein (ORF 1855). Besides these putative iron metabolism genes, two genes encoding hypothetical proteins (ORFs 3347 and 3086) and a gene encoding a transcriptional regulator gene (ORF 1558) belong to subcluster 5.
Construction and analysis of pstS-cat and ugpA-cat transcriptional fusions.
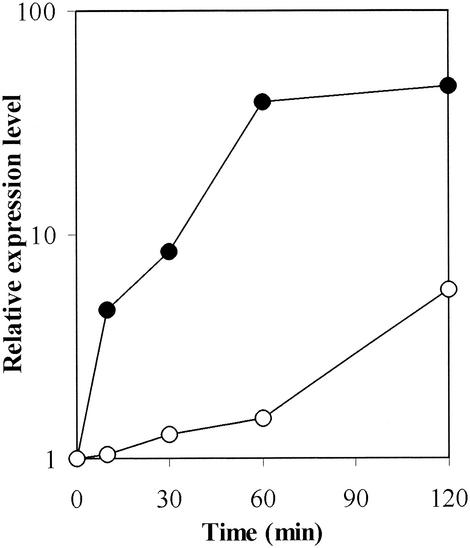
To determine whether the increased RNA levels of the pstSCAB and ugpAEBC operons are due to transcriptional control and in order to confirm the expression changes observed in the DNA microarray experiments, we constructed and analyzed transcriptional fusions of the promoter regions upstream of pstS and ugpA. The pstS and ugpA promoter regions were fused to the promoterless chloramphenicol acetyltransferase gene on the promoter-probe vector pET2 (46), and the resulting plasmids were introduced into C. glutamicum wild-type strain ATCC 13032. Using the cultivation protocol used for the kinetic analysis of gene expression changes, we determined expression of the plasmid-borne transcriptional fusions pstS-cat and ugpA-cat. Expression of pstS-cat increased immediately after the Pi down-shift and reached maximal levels about 60 min after the shift. The temporal expression profile of pstS-cat is in very good agreement with the profile obtained in the DNA microarray experiments (Fig. 4). Similarly, expression of the transcriptional fusion ugpA-cat increased after the Pi down-shift. Thus, we suggest that increased expression of the pstSCAB and ugpAEBC operons in response to reduced Pi availability is due primarily to transcriptional regulation. It is noteworthy that both expression of the ugpA-cat fusion and the RNA levels of the ugpAEBC operon genes showed a delayed increase when they were compared to the profile obtained for the pstSCAB operon.
FIG. 4.
Expression of the pstS-cat and ugpA-cat transcriptional fusions in C. glutamicum ATCC 13032(pET2-pst) and ATCC 13032(pET2-ugp) during the response to Pi starvation. The activities obtained from two or more independent cultivations varied less than 20% and were normalized to the activity before the shift to Pi starvation conditions. Symbols: •, pstS-cat fusion; ○, ugpA-cat fusion.
DISCUSSION
We identified the Pi starvation stimulon of C. glutamicum, and we determined the kinetics of the response to Pi starvation. A number of genes involved in phosphorus metabolism showed increased expression upon Pi starvation. The predicted functions of the proteins encoded by this group of genes include high-affinity uptake of inorganic phosphate, uptake of organophosphates, such as glycerol 3-phosphate, and hydrolysis of organophosphates and nucleic acids. Thus, the Pi starvation stimulons of C. glutamicum and E. coli are similar. However, 21 of 38 genes known to belong to the Pi starvation stimulons of E. coli and Salmonella enterica serovar Typhimurium are involved in phosphonate metabolism (47), whereas C. glutamicum lacks homologs of genes for phosphonate degradation (NCBI accession no. NC003450), as well as the capability to utilize phosphonates as P sources (unpublished results). On the other hand, a C. glutamicum gene encoding a putative 5′-nucleotidase or related esterase showed increased expression upon Pi starvation, but its E. coli homolog, ushA, is not a known member of the Pi starvation stimulon (47). The role of the C. glutamicum 5′-nucleotidase or related esterase (Cgl0328) in adaptation to low-Pi conditions remains to be studied.
On the gene expression level, the first response of C. glutamicum to Pi starvation is increased expression of pstSCAB encoding the putative high-affinity Pi uptake system (Fig. 3). As the pstSCAB expression kinetics determined by DNA microarray analysis correlated well with the expression kinetics of a plasmid-borne pstS′-′cat transcriptional fusion, control of pstSCAB expression in response to reduced Pi availability is due primarily to transcriptional regulation. The increases in expression of pstSCAB after Pi starvation were the most pronounced both in C. glutamicum and in E. coli, in which transcripts of the pstSCAB-phoU operon were shown to be rapidly processed posttranscriptionally (1). The E. coli Pst system, which transports Pi at the expense of ATP (10, 47), is composed of the periplasmic Pi-binding protein PstS, two integral membrane proteins (PstC and PstA), and the ATP-binding protein PstB. The protein encoded by phoU, which is part of the operon, is not required for Pi transport, and its function is unclear (41). The high affinity of the Pst system and coupling transport to ATP hydrolysis allow concentrative Pi uptake when extracellular Pi is scarce.
The increases in expression of ugpQ, predicted to code for a glycerophosphoryl diester phosphodiesterase, and increases in expression of the oppositely oriented ugpAEBC operon, encoding an ABC transporter for glycerol 3-phosphate, were less pronounced but occurred shortly after the increases in expression of pstSCAB (Fig. 3). In E. coli, the corresponding genes form a single operon, ugpBAECQ (6, 9), which is controlled in response to the Pi availability. In E. coli and S. enterica serovar Typhimurium, four Pi starvation-inducible uptake systems for organophosphates have been identified: Ugp and GlpT for glycerol 3-phosphate, UhpT for hexose 6-phosphates, and PgtP for phosphoenolpyruvate, 2-phosphoglycerate, and 3-phosphoglycerate (47). While the C. glutamicum genome contains homologs of the genes encoding the Ugp system, it lacks homologs of the genes encoding GlpT, UhpT, and PgtP (NCBI accession no. NC003450). However, expression of an operon encoding an ABC transport system which might be involved in the uptake of an as-yet-unidentified P compound increased after Pi starvation. Compared to Pi starvation in B. subtilis, Pi starvation in C. glutamicum led to different expression changes. The cell wall of C. glutamicum lacks teichuronic acid and teichoic acid (29), and the genome does not contain homologs of the B. subtilis tuaABCDEFGH operon for teichuronic acid biosynthesis or the teichoic acid biosynthesis operons tagAB and tagDEF (NCBI accession no. NC003450). In B. subtilis, Pi starvation also results in σB-dependent increases in expression of genes of the general stress response (4, 35). While no homologs of B. subtilis yjbC, ysnF, and yvgO were identified in the C. glutamicum genome, expression of homologs of B. subtilis yfhM (Cgl0297), csbD (Cgl0229), yheK (Cgl1407 and Cgl2853), and ykzA (Cgl2385) did not significantly increase during the C. glutamicum Pi starvation response.
Several genes obviously involved in P metabolism are not part of the Pi starvation stimulon of C. glutamicum. Expression of a gene homologous to E. coli pitA encoding a low-affinity Pi transporter (45) and expression of a gene encoding a predicted sodium-dependent phosphate transporter (Cgl2744) were not significantly changed in C. glutamicum upon Pi starvation. Whereas C. glutamicum accumulates up to 600 mM P units as polyphosphate (26), expression of the C. glutamicum homologs of the E. coli exopolyphosphatase gene ppx (2) (Cgl0408 and Cgl0977), expression of the homolog of the Pseudomonas aeruginosa polyphosphate kinase II gene ppk2 (49) (Cgl0917 and Cgl2714), and expression of the homolog of the M. tuberculosis polyphosphate glucokinase gene ppgK (21) (Cgl1910) were unchanged. Similarly, in E. coli the ppk-ppx operon is not part of the PhoB regulon (47), whereas phosphate-dependent regulation of genes involved in polyphosphate synthesis has been found in Saccharomyces cerevisiae (33) and Acinetobacter species (14, 15).
As a consequence of Pi starvation, expression of at least some ribosomal protein genes decreased. Significantly decreased expression of these genes was first observed 60 min after the shift from Pi-sufficient to Pi-limiting conditions. It is well known that in E. coli ribosome synthesis regulation involves growth rate-dependent control and stringent control (17). Apparently, the increases in expression of the genes and operons for uptake of Pi and glycerol 3-phosphate precede the decrease in ribosomal protein gene expression. Thus, the Pi starvation response is initiated before growth ceases and not as a consequence of the end of growth.
The transiently reduced expression of genes involved in iron metabolism and the transiently increased expression of genes involved in copper metabolism and in protocatechuate degradation after the shift to Pi-limiting conditions are not understood. In the shift experiment, changes in the concentrations of the medium components iron, copper, and protocatechuate, which was used as an iron chelator to facilitate iron uptake and optimal growth of C. glutamicum (30), might have influenced expression of genes involved in the transport and metabolism of these components. However, it is unlikely that a medium component was limiting before the shift as cells were growing exponentially and had only reached a low cell density (OD600, 5) compared to the density which is supported by the medium (OD600, 60). Furthermore, it is noteworthy that Pi seems to play a direct role in iron transport in Haemophilus influenzae (8), as well as in iron-dependent regulation in the related organism Corynebacterium diphtheriae (37).
Currently, the regulatory mechanism(s) governing the Pi starvation response of C. glutamicum is unknown. In E. coli and B. subtilis, the two-component regulatory systems PhoBR and PhoPR, respectively (4, 47), are involved in Pi-dependent regulation. In the case of B. subtilis, the Spo0 phosphorelay and the ResDE two-component system also play roles in the Pi starvation response (4). During the C. glutamicum Pi starvation response genes of two two-component regulatory systems showed transiently increased expression (subcluster 2) (Fig. 2). The genes of one system (ORFs 377 and 378, Cgl2964 and Cgl2965) are adjacent to genes for copper metabolism and showed up to sevenfold increases in expression. The genes of the other system (ORFs 2831 and 2832, Cgl2606 and Cgl2607) showed five- to eightfold-increased expression only 10 min after the shift to Pi starvation conditions. Future studies should reveal the roles of these systems for Pi-dependent control of gene expression.
Acknowledgments
This work was supported in part by the German Ministry of Education and Reseach (BMBF) within the framework of genome research on prokaryotic organisms (GenoMik).
We thank Martina Mickova for helpful discussions.
REFERENCES
- 1.Aguena, M., E. Yagil, and B. Spira. 2002. Transcriptional analysis of the pst operon of Escherichia coli. Mol. Genet. Genomics 268:518-524. [DOI] [PubMed] [Google Scholar]
- 2.Akiyama, M., E. Crooke, and A. Kornberg. 1993. An exopolyphosphatase of Escherichia coli. The enzyme and its ppx gene in a polyphosphate operon. J. Biol. Chem. 268:633-639. [PubMed] [Google Scholar]
- 3.Andersen, A. B., L. Ljungqvist, and M. Olsen. 1990. Evidence that protein antigen b of Mycobacterium tuberculosis is involved in phosphate metabolism. J. Gen Microbiol. 136:477-480. [DOI] [PubMed] [Google Scholar]
- 4.Antelmann, H., C. Scharf, and M. Hecker. 2000. Phosphate starvation-inducible proteins of Bacillus subtilis: proteomics and transcriptional analysis. J. Bacteriol. 182:4478-4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arfin, S. M., A. D. Long, E. T. Ito, L. Tolleri, M. M. Riehle, E. S. Paegle, and G. W. Hatfield. 2000. Global gene expression profiling in Escherichia coli K12. The effects of integration host factor. J. Biol. Chem. 275:29672-29684. [DOI] [PubMed] [Google Scholar]
- 6.Argast, M., and W. Boos. 1980. Co-regulation in Escherichia coli of a novel transport system for sn-glycerol-3-phosphate and outer membrane protein Ic (e, E) with alkaline phosphatase and phosphate-binding protein. J. Bacteriol. 143:142-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Birkey, S. M., W. Liu, X. Zhang, M. F. Duggan, and F. M. Hulett. 1998. Pho signal transduction network reveals direct transcriptional regulation of one two-component system by another two-component regulator: Bacillus subtilis PhoP directly regulates production of ResD. Mol. Microbiol. 30:943-953. [DOI] [PubMed] [Google Scholar]
- 8.Bruns, C. M., D. S. Anderson, K. G. Vaughan, P. A. Williams, A. J. Nowalk, D. E. McRee, and T. A. Mietzner. 2001. Crystallographic and biochemical analyses of the metal-free Haemophilus influenzae Fe3+-binding protein. Biochemistry 40:15631-15637. [DOI] [PubMed] [Google Scholar]
- 9.Brzoska, P., M. Rimmele, K. Brzostek, and W. Boos. 1994. The pho regulon-dependent Ugp uptake system for glycerol-3-phosphate in Escherichia coli is trans inhibited by Pi. J. Bacteriol. 176:15-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan, F. Y., and A. Torriani. 1996. PstB protein of the phosphate-specific transport system of Escherichia coli is an ATPase. J. Bacteriol. 178:3974-3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang, Z., A. Choudhary, R. Lathigra, and F. A. Quiocho. 1994. The immunodominant 38-kDa lipoprotein antigen of Mycobacterium tuberculosis is a phosphate-binding protein. J. Biol. Chem. 269:1956-1958. [PubMed] [Google Scholar]
- 12.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry 3rd, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, B. G. Barrell, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 13.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gavigan, J. A., L. M. Marshall, and A. D. Dobson. 1999. Regulation of polyphosphate kinase gene expression in Acinetobacter baumannii 252. Microbiology 145:2931-2937. [DOI] [PubMed] [Google Scholar]
- 15.Geissdorfer, W., A. Ratajczak, and W. Hillen. 1998. Transcription of ppk from Acinetobacter sp. strain ADP1, encoding a putative polyphosphate kinase, is induced by phosphate starvation. Appl. Environ. Microbiol. 64:896-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gornall, A. G., C. J. Bardawill, and M. M. David. 1949. Determination of serum proteins by means of biuret reaction. J. Biol. Chem. 177:751-766. [PubMed] [Google Scholar]
- 17.Gourse, R. L., T. Gaal, M. S. Bartlett, J. A. Appleman, and W. Ross. 1996. rRNA transcription and growth rate-dependent regulation of ribosome synthesis in Escherichia coli. Annu. Rev. Microbiol. 50:645-677. [DOI] [PubMed] [Google Scholar]
- 18.Hecker, M., and U. Volker. 1998. Non-specific, general and multiple stress resistance of growth-restricted Bacillus subtilis cells by the expression of the σB regulon. Mol. Microbiol. 29:1129-1136. [DOI] [PubMed] [Google Scholar]
- 19.Hoffer, S. M., P. Schoondermark, H. W. van Veen, and J. Tommassen. 2001. Activation by gene amplification of pitB, encoding a third phosphate transporter of Escherichia coli K-12. J. Bacteriol. 183:4659-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hommais, F., E. Krin, C. Laurent-Winter, O. Soutourina, A. Malpertuy, J. P. Le Caer, A. Danchin, and P. Bertin. 2001. Large-scale monitoring of pleiotropic regulation of gene expression by the prokaryotic nucleoid-associated protein, H-NS. Mol. Microbiol. 40:20-36. [DOI] [PubMed] [Google Scholar]
- 21.Hsieh, P. C., B. C. Shenoy, D. Samols, and N. F. Phillips. 1996. Cloning, expression, and characterization of polyphosphate glucokinase from Mycobacterium tuberculosis. J. Biol. Chem. 271:4909-4915. [DOI] [PubMed] [Google Scholar]
- 22.Hulett, F. M. 2002. The Pho regulon, p. 193-201. In A. L. Sonenshein, J. A. Hock, and R. M. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 23.Keilhauer, C., L. Eggeling, and H. Sahm. 1993. Isoleucine synthesis in Corynebacterium glutamicum: molecular analysis of the ilvB-ilvN-ilvC operon. J. Bacteriol. 175:5595-5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khodursky, A., J. A. Bernstein, B. J. Peter, V. Rhodius, V. F. Wendisch, and D. P. Zimmer. 2003. Escherichia coli spotted double strand DNA microarrays: RNA extraction, labeling, hybridization, quality control and data management, p. 61-78. In M. J. Brownstein and A. Khodursky (ed.), Methods in molecular biology—functional genomics: methods and protocols, vol. 224. Humana Press, Totowa, N.J. [DOI] [PubMed]
- 25.Lahooti, M., Z. Pragai, and C. R. Harwood. 2000. Phosphate regulation, p. 237-244. In W. Schumann, S. D. Ehrlich, and N. Ogasawara (ed.), Functional analysis of bacterial genes: a practical manual. Wiley, Chichester, United Kingdom.
- 26.Lambert, C., D. Weuster-Botz, R. Weichenhain, E. W. Kreutz, A. A. de Graaf, and S. M. Schoberth. 2002. Monitoring of inorganic polyphosphate dynamics in Corynebacterium glutamicum using a novel oxygen sparger for real time 31P in vivo NMR. Acta Biotechnol. 22:245-260. [Google Scholar]
- 27.Lefevre, P., M. Braibant, L. de Wit, M. Kalai, D. Roeper, J. Grotzinger, J. P. Delville, P. Peirs, J. Ooms, K. Huygen, and J. Content. 1997. Three different putative phosphate transport receptors are encoded by the Mycobacterium tuberculosis genome and are present at the surface of Mycobacterium bovis BCG. J. Bacteriol. 179:2900-2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lehnen, D., C. Blumer, T. Polen, B. Wackwitz, V. F. Wendisch, and G. Unden. 2002. LrhA as a new transcriptional key regulator of flagella, motility and chemotaxis genes in Escherichia coli. Mol. Microbiol. 45:521-532. [DOI] [PubMed] [Google Scholar]
- 29.Liebl, W., M. Ehrmann, W. Ludwig, and K. H. Schleifer. 1991. Transfer of Brevibacterium divaricatum DSM 20297T, “Brevibacterium flavum” DSM 20411, “Brevibacterium lactofermentum” DSM 20412 and DSM 1412, and Corynebacterium glutamicum and their distinction by rRNA gene restriction patterns. Int. J. Syst. Bacteriol. 41:255-260. [DOI] [PubMed] [Google Scholar]
- 30.Liebl, W., R. Klamer, and K. H. Schleifer. 1989. Requirement of chelating compounds for the growth of Corynebacterium glutamicum in synthetic media. Appl. Microbiol. Biotechnol. 32:205-210.
- 31.Menkel, E., G. Thierbach, L. Eggeling, and H. Sahm. 1989. Influence of increased aspartate availability on lysine formation by a recombinant strain of Corynebacterium glutamicum and utilization of fumarate. Appl. Environ. Microbiol. 55:684-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Metcalf, W. W., P. M. Steed, and B. L. Wanner. 1990. Identification of phosphate starvation-inducible genes in Escherichia coli K-12 by DNA sequence analysis of psi::lacZ(Mu d1) transcriptional fusions. J. Bacteriol. 172:3191-3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ogawa, N., J. DeRisi, and P. O. Brown. 2000. New components of a system for phosphate accumulation and polyphosphate metabolism in Saccharomyces cerevisiae revealed by genomic expression analysis. Mol. Biol. Cell 11:4309-4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Polen, T., D. Rittmann, V. F. Wendisch, and H. Sahm. 2003. DNA microarray analyses of the long-term adaptive response of Escherichia coli to acetate and propionate. Appl. Environ. Microbiol. 69:1759-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pragai, Z., and C. R. Harwood. 2002. Regulatory interactions between the Pho and sigma(B)-dependent general stress regulons of Bacillus subtilis. Microbiology 148:1593-1602. [DOI] [PubMed] [Google Scholar]
- 36.Price, C. W., P. Fawcett, H. Ceremonie, N. Su, C. K. Murphy, and P. Youngman. 2001. Genome-wide analysis of the general stress response in Bacillus subtilis. Mol. Microbiol. 41:757-774. [DOI] [PubMed] [Google Scholar]
- 37.Qiu, X., E. Pohl, R. K. Holmes, and W. G. Hol. 1996. High-resolution structure of the diphtheria toxin repressor complexed with cobalt and manganese reveals an SH3-like third domain and suggests a possible role of phosphate as co-corepressor. Biochemistry 35:12292-12302. [DOI] [PubMed] [Google Scholar]
- 38.Sahm, H., L. Eggeling, and A. A. de Graaf. 2000. Pathway analysis and metabolic engineering in Corynebacterium glutamicum. Biol. Chem. 381:899-910. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 40.Shaw, W. V. 1975. Chloramphenicol acetyltransferase from chloramphenicol-resistant bacteria. Methods Enzymol. 43:737-755. [DOI] [PubMed] [Google Scholar]
- 41.Steed, P. M., and B. L. Wanner. 1993. Use of the rep technique for allele replacement to construct mutants with deletions of the pstSCAB-phoU operon: evidence of a new role for the PhoU protein in the phosphate regulon. J. Bacteriol. 175:6797-6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun, G., S. M. Birkey, and F. M. Hulett. 1996. Three two-component signal-transduction systems interact for Pho regulation in Bacillus subtilis. Mol. Microbiol. 19:941-948. [DOI] [PubMed] [Google Scholar]
- 43.Torres, A., M. D. Juarez, R. Cervantes, and C. Espitia. 2001. Molecular analysis of Mycobacterium tuberculosis phosphate specific transport system in Mycobacterium smegmatis. Characterization of recombinant 38 kDa (PstS-1). Microb. Pathog. 30:289-297. [DOI] [PubMed] [Google Scholar]
- 44.VanBogelen, R. A., E. R. Olson, B. L. Wanner, and F. C. Neidhardt. 1996. Global analysis of proteins synthesized during phosphorus restriction in Escherichia coli. J. Bacteriol. 178:4344-4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Veen, H. W., T. Abee, G. J. Kortstee, W. N. Konings, and A. J. Zehnder. 1994. Translocation of metal phosphate via the phosphate inorganic transport system of Escherichia coli. Biochemistry 33:1766-1770. [DOI] [PubMed] [Google Scholar]
- 46.Vasicova, P., Z. Abrhamova, J. Nesvera, M. Patek, H. Sahm, and B. Eikmanns. 1998. Integrative and autonomously replicating vectors for analysis of promoters in Corynebacterium glutamicum. Biotechnol. Tech. 12:743-746. [Google Scholar]
- 47.Wanner, B. L. 1996. Phosphorus assimilation and control of the phosphate regulon, p. 1357-1381. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 48.Wendisch, V. F., D. P. Zimmer, A. Khodursky, B. Peter, N. Cozzarelli, and S. Kustu. 2001. Isolation of Escherichia coli mRNA and comparison of expression using mRNA and total RNA on DNA microarrays. Anal. Biochem. 290:205-213. [DOI] [PubMed] [Google Scholar]
- 49.Zhang, H., K. Ishige, and A. Kornberg. 2002. A polyphosphate kinase (PPK2) widely conserved in bacteria. Proc. Natl. Acad. Sci. USA 99:16678-16683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zimmer, D. P., E. Soupene, H. L. Lee, V. F. Wendisch, A. B. Khodursky, B. J. Peter, R. A. Bender, and S. Kustu. 2000. Nitrogen regulatory protein C-controlled genes of Escherichia coli: scavenging as a defense against nitrogen limitation. Proc. Natl. Acad. Sci. USA 97:14674-14679. [DOI] [PMC free article] [PubMed] [Google Scholar]