Abstract
The bacterium Shewanella frigidimarina can grow anaerobically by utilizing Fe(III) as a respiratory electron acceptor. This results in the synthesis of a number of periplasmic c-type cytochromes, which are absent when the organism is grown in the absence of added Fe(III). One cytochrome, IfcA, is synthesized when Fe(III) is present as the sole respiratory electron acceptor or when it is present in combination with oxygen, fumarate, or nitrate. The ifcA gene was thus selected for a study of iron-responsive gene regulation of respiratory proteins in S. frigidimarina. The monocistronic ifcA gene clusters with two other monocistronic genes, ifcO, encoding a putative outer membrane porin, and ifcR, encoding a putative transcriptional regulator of the LysR superfamily. Analysis of transcription of all three genes under a range of growth conditions in the wild type and an ifcR insertion mutant and analysis of a strain that constitutively expresses ifcR revealed that iron regulation is exerted at the level of ifcR transcription. In the presence of Fe(III) IfcR is synthesized and acts positively to regulate expression of ifcO and ifcA. Control of Fe(III) respiration by this novel regulatory system differs markedly from Fur-mediated regulation of iron assimilation, in which Fur serves as an Fe(II)-activated repressor.
Shewanella species are gram-negative, highly versatile, facultative, anaerobic bacteria that can use insoluble ferric iron and a variety of other substrates [e.g., manganese(IV), uranium(VI), nitrate, nitrite, fumarate, elemental sulfur, trimethylamine-N-oxide, and dimethyl sulfoxide] as terminal electron acceptors for anaerobic respiration. Fe(III) respiration has widespread environmental significance, influencing several biogeochemical cycles and affecting soil properties and plant growth (17). Fe(III)-respiring bacteria also have been exploited in the field of bioremediation (16), since oxidative degradation of aromatic hydrocarbons, such as benzene and toluene, to CO2 is coupled to Fe(III) reduction. Futhermore, Fe(III) respirers can reduce highly soluble U(VI) and Tc(VII) species, resulting in deposition of these metals as insoluble oxides, which effects their removal from contaminated waters (18).
At a neutral pH, Fe(III) oxides are found largely in crystalline form or as amorphous (oxy)hydroxide particles. Accordingly, Fe(III)-respiring bacteria localize a substantial proportion of their Fe(III) reductase activity in the outer membrane, although reductases able to reduce soluble Fe(III) chelates are also located in the periplasmic compartment (20). The generation of energy via respiratory reduction of insoluble Fe(III) raises a biochemical and bioenergetic problem. Electrons must move from the active site of primary dehydrogenases, associated with the cytoplasmic membrane, to the outer face of the outer membrane, where the insoluble Fe(III) species are reduced. In members of the Fe(III)-respiring genus Shewanella, it is emerging that electron transport to extracellular Fe(III) occurs via a network of low-potential multiheme c-type cytochromes located on the outer face of the cytoplasmic membrane, in the periplasmic compartment, and in the outer membrane. In Shewanella frigidimarina (formerly Shewanella putrefaciens NCIMB400) and Shewanella oneidensis MR-1 (formerly S. putrefaciens MR-1) some of these multiheme cytochromes have been purified and biochemically characterized (2, 7, 19, 20, 21, 24, 27). Spectroscopic and structural studies of a number of these multiheme cytochromes have suggested that the presence of multiple, low-redox-potential bis His-ligated hemes are a common feature of all of them (1, 14, 15, 30).
Analysis of the S. oneidensis genome sequence revealed the presence of genes for 39 c-type cytochromes, many of which are predicted to be multiheme in nature (12). The role of these cytochromes in electron transfer to metal oxides is currently the subject of intense study. The outer membrane cytochromes, including OmcA and OmcB, might be localized where they can make direct contact with extracellular metal oxides at the cell surface. The soluble periplasmic cytochromes could be involved in intermembrane electron transfer between the inner and outer membranes and also in the reduction of soluble Fe(III) chelates that cross the outer membrane (2, 5, 8).
To elucidate the functional role of the multiheme cytochromes of Shewanella species in more detail, it is important to complement biochemical and spectroscopic studies with an understanding of the genetic basis and mechanism(s) of regulation of the genes encoding these proteins. In this work, we studied regulation of the iron-induced ifcA gene in S. frigidimarina. The ifcA gene encodes a 63.9-kDa tetraheme flavocytochrome c. In this paper we provide the first characterization of Fe(III)-responsive gene regulation for the respiratory multiheme cytochromes of Shewanella species.
It has been shown previously that IfcA is synthesized in the periplasmic compartment of S. frigidimarina only when cells are grown with Fe(III) ions as the sole electron acceptor and not when fumarate or oxygen is used as the electron acceptor (5). IfcA was purified, and its crystal structure was determined (1, 5). The enzyme has a small (ca. 10-kDa) tetraheme cytochrome (STC) domain and a flavin domain. Although exhibiting a unidirectional fumarate reductase activity in vitro, the enzyme is not physiologically functional in fumarate reduction in vivo, since it is not synthesized during the respiration of fumarate. Although ifcA mutants retain the ability to grow anaerobically on Fe(III) citrate, it has been shown that such a mutant overproduced both a 35-kDa periplasmic c-type cytochrome and a 45-kDa outer membrane c-type cytochrome (5). It has been proposed that the STC domain of IfcA is involved in intermembrane electron transport and reduction of soluble Fe(III) (5).
Upstream of the ifcA gene and with opposite transcriptional polarity is a gene encoding a putative outer membrane β-barrel protein, designated ifcO. The function of the protein is not known, but it is conceivable that it transports soluble Fe(III) chelates into the periplasm, where they can be reduced by the flavocytochrome. The partial sequence of another open reading frame (ORF), encoding a putative LysR-type transcriptional regulator located directly downstream from ifcO, was also reported (5). We completed the sequence of this gene and designated the gene ifcR. In this study we examined the regulation of ifcR, ifcO, and ifcA expression under different respiratory conditions. We found that IfcR is a transcriptional activator essential for expression of ifcO and ifcA. We also found that ifcR transcription is dependent on the presence of Fe(III) in the external medium, even if the iron is not used extensively as a respiratory substrate. This is the first example of a specific Fe(III)-responsive system for regulating synthesis of respiratory multiheme cytochromes in Shewanella species. The features of this regulatory system are quite distinct from those of other well-characterized iron-responsive regulatory systems involved in regulating iron assimilation in many bacteria and fungi.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are described in Table 1. Escherichia coli strains were grown at 37°C on Luria-Bertani (LB) medium supplemented with antibiotics at the following concentrations: ampicillin, 100 μg/ml; chloramphenicol, 30 μg/ml; and tetracycline, 10 μg/ml. S. frigidimarina strains were grown on LB medium adjusted to pH 7.5 and supplemented with 50 mM dl-lactate. Antibiotics at the concentration indicated above were added when required. For microaerobic growth, S. frigidimarina strains were grown at 25°C by shaking 100-ml cultures in 250-ml conical flasks at 200 rpm. Anaerobic growth experiments were performed in completely filled 300-ml screw-cap bottles. For anaerobic growth, alternative soluble respiratory substrates were provided at the following concentrations: ferric citrate, 50 mM; sodium fumarate and potassium nitrate, 25 mM; and MnO2, 10 mM.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype | Reference or source |
|---|---|---|
| E. coli strains | ||
| DH5α | supE44 ΔlacU169 (φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | BRLa |
| S17-1 | thi pro hsdR hdsM+recA, chromosomal insertion of RP4-2(Tc::Mu Km::T7) | 29 |
| S. frigidimarina strains | ||
| NCIMB400R | Wild type | |
| PSD1 | NCBIMB400R ifcA::ΩCmr | 5 |
| PSD106 | NCBIMB400R ifcR::ΩCmr | This study |
| PSD201 | Complemented PSD106 carrying pPD1 Cmr, Tcr | This study |
| PSR33 | Complemented PSD106 carrying pFR33 Cmr, Tcr | This study |
| Plasmids | ||
| pBluescript II KS+ | Cloning vector, Ampr | Stratagene |
| pJQ200KS | Mobilizable suicide vector, Gmr | 26 |
| pRK415 | IncP broad-host-range cloning vector, Tcr | 13 |
| pHP45ΩCm | Cloning vector containing ΩCm, Ampr | 6 |
| pTRN1 | 2-kb XbaI-XhoI PCR fragment carrying ifcR cloned into pBluescript II KS+ | This study |
| pTRN2 | 4-kb EcoRI fragment containing ΩCm cloned into pTRN1 | This study |
| pTRN3 | 6-kb XbaI-ApaI fragment containing ifcR::ΩCmr cloned into pJQ200KS | This study |
| pTCM1 | 6-kb XbaI fragment carrying ifcR, ifcO, and ifcA::ΩCmr cloned into pBluescript II KS+ | This study |
| pPD1 | 1.3-kb XbaI-PstI fragment carrying ifcR plus 5′ and 3′ flanking sequences cloned into pRK415 | This study |
| pFR3 | 890-bp fragment carrying ifcR cloned into expression vector pT7-7 | This study |
| pFR33 | 932-bp fragment carrying a promotorless ifcR cloned into pRK415 | This study |
BRL, Bethesda Research Laboratories.
Preparation of periplasm and analysis of heme-stained SDS-PAGE gels.
To obtain the periplasmic fraction of S. frigidimarima, cells were harvested from microaerobic or anaerobic cultures by centrifugation, and each resulting pellet was resuspended in an appropriate volume of spheroplasting buffer (100 mM Tris-HCl [pH 8], 500 mM sucrose, 3 mM Na2EDTA). Lysozyme was added to a final concentration of 2 mg/ml, and the cells were then incubated for 20 min at 37°C. Periplasmic fractions were isolated by centrifugation at 12,000 × g for 30 min at 4°C. Periplasmic extracts were routinely analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) in either 10 or 15% (wt/vol) polyacrylamide gels. Samples were prepared for electrophoresis by incubating them with 3 M urea and 90 mM SDS at room temperature for 1 h. Gels were examined for the presence of c-type cytochromes by heme-linked peroxidase staining as previously described (31). The total protein concentration was determined by the bicinchoninic acid method (Sigma) with bovine serum albumin as the standard.
DNA manipulations.
Isolation of plasmid and genomic DNA and digestion, ligation, cloning, and subcloning of appropriate DNA fragments were performed by standard techniques by using the appropriate commercially available kits and enzymes. To obtain the complete DNA sequence of the ifcR gene, total genomic DNA from S. frigidimarina PSD1, which carries a chloramphenicol resistance cassette inserted into the ifcA gene (5), was isolated and digested with XbaI. The resulting fragments were ligated into the vector pBluescript II KS+, which had been previously digested with XbaI. The ligation mixture was transformed into E. coli DH5α, and colonies with chloramphenicol and tetracycline resistance were selected. DNA isolation from one of these colonies resulted in a plasmid (pTCM1) with a 6-kb XbaI DNA fragment including the full-length ifcR gene. The entry in the database has been updated to include the complete sequence of the ifcR gene (GenBank accession number AJ236923).
Primer extension analysis.
Total RNA was isolated from an S. frigidimarina wild-type strain or S. frigidimarina PSD106 (ΔifcR) after either microaerobic or anaerobic growth on LB medium supplemented with the appropriate respiratory substrate by using a Qiagen RNeasy kit under the conditions recommended by the manufacturer. In order to determine the transcription initiation sites of the ifcR, ifcO, and ifcA genes, 2-pmol portions of primers P1 (5′GAAATTTGGCCGCAGGTATGAATT3′), P2 (5′GTAGACAATAATGCTATGGAAATTAGG3′), and P3 (5′GCACAACCAATGCCATCGCTGATACC3′), which are complementary to the regions immediately upstream of the initiation codons of ifcR, ifcO, and ifcA, respectively, were 5′ end labeled with [γ-32P]ATP and T4 polynucleotide kinase. Labeled primers (0.1 pmol) were mixed with 25 μg of total RNA in hybridization buffer (100 mM KCl, 50 mM Tris-HCl; pH 8.3) and incubated for 30 s at 95°C and then for 20 min at 55°C. Primer extension reactions were performed at 41°C for 90 min in a reaction buffer containing 6.5 U of avian myeloblastosis virus reverse transcriptase (Pharmacia). After the reaction was stopped with gel-loading buffer, samples were boiled for 3 min and loaded onto a denaturing 6% (wt/vol) polyacrylamide gel. To determine the sizes of the extension products, manual DNA sequencing of plasmid pTCM1 primed with labeled oligonucleotide P1, P2, or P3 was performed.
Construction of S. frigidimarina ifcR mutant PSD106.
To construct S. frigidimarina ifcR insertion mutant PSD106, a DNA fragment containing the ifcR gene interrupted by an EcoRI cassette encoding chloramphenicol resistance was generated. To do this, a PCR was performed to amplify DNA from wild-type S. frigidimarina with primers TRN1 (5′GAATCTAGATATAACGTCAGTGGC3′) and TRN2 (5′GTAGAATTCCAATGTTTAAACAAC3′), yielding PCR-1. A second PCR product, PCR-2, was obtained by using primers TRN3 (5′TTGGAATTCTACGTGATAAAACATAG3′) and TRN4 (5′CCTCTCGAGGTGTTGGACAAATC3′). The PCR-1 and PCR-2 extension products were digested with XbaI plus EcoRI and EcoRI plus XhoI, respectively, and cloned into pBluescript II KS+ that had previously been digested with XbaI and XhoI, yielding plasmid pTRN1. Next, an Ω-cassette encoding resistance to chloramphenicol, excised from pHP45 on an EcoRI fragment, was ligated into pTRN1 that had previously been digested with the same enzyme. Digestion of the resulting plasmid, pTRN2, with XbaI and ApaI released a 6-kb DNA fragment containing the interrupted ifcR gene plus 1 kb of upstream and downstream DNA sequence. This fragment was subcloned into the suicide vector pJQ200KS digested with XbaI and ApaI, yielding plasmid pTRN3. Plasmid pTRN3 was transformed into E. coli S17-1 and used in subsequent matings with S. frigidimarina NCIMB400R at 25°C. Selection for mutants was performed on agar plates containing 50 mg of sucrose per ml and chloramphenicol. Gene replacement was confirmed by Southern blot analysis.
The broad-host-range plasmid pRK415 was used for complementation studies of S. frigidimarina PSD106.
A 1.3-kb XbaI-PstI DNA fragment which encompassed the region from 209 bp downstream of the stop codon of ifcR to 213 bp upstream of the start codon of ifcR, including the entire ifcR gene, was obtained by performing PCR with genomic DNA and primers PA (5′AGCTGCAGAGCGGTATAAGGTTTTCCATG3′) and PB (5′AAATCTAGAGACTACAGGCTCGATAAC3′). The PCR product was digested with XbaI and PstI and cloned into pRK415 digested with the same restriction enzymes, yielding plasmid pPD1. To obtain strain PSD201, E. coli S17-1 was transformed with pPD1 and then used to transfer this plasmid into PSD106 by conjugation at 25°C. Recombinant colonies were selected on LB agar plates supplemented with lactate, tetracycline, and chloramphenicol.
To obtain a promotorless ifcR DNA fragment, an 890-bp DNA fragment was amplified from genomic DNA by using primers P5 (5′CCGGCATATGCGAATAACCTTAAAGC3′) and P6 (5′GAATTAAGCTTTTCCATTTAGC3′). After digestion with NdeI and HindIII, the fragment was cloned into the expression vector pT7-7, yielding plasmid pFR3. Plasmid pFR33 was generated from pFR3 by digesting pFR3 with HindIII, creating a blunt end with mung bean nuclease, and digesting the preparation with XbaI. The corresponding 932-bp DNA fragment carrying the ifcR gene was subsequently cloned into pRK415, which previously had been digested with EcoRI, blunt ended with mung bean nuclease, and digested with XbaI. To obtain the complemented strain PSR33, E. coli S17-1 was transformed with pFR33 and then used to transfer the plasmid into PSD106 by conjugation. Recombinant colonies were selected on LB agar plates supplemented with lactate, tetracycline, and chloramphenicol.
RESULTS AND DISCUSSION
Fe(III)-dependent synthesis of c-type cytochromes and identification of ifcA as a target for studies of Fe(III)-dependent gene expression.
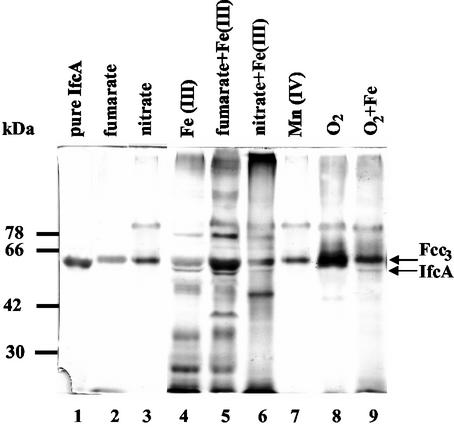
We initiated our studies of regulation of expression of c-type cytochromes in response to Fe(III) by examining the synthesis of c-type cytochromes in periplasmic fractions prepared from cultures of S. frigidimarina grown either microaerobically or anaerobically with various respiratory electron acceptors and with lactate as the electron donor. The c-type cytochromes synthesized were visualized by heme staining following resolution of the periplasmic proteins by SDS-PAGE (Fig. 1). A major heme-stained band at approximately 64-kDa was detected under all growth conditions investigated. This heme-stained band corresponds to the Fcc3 protein, the soluble, periplasmic, tetraheme flavocytochrome that functions as a physiological respiratory fumarate reductase in Shewanella (8, 9). Fcc3 was the dominant heme-stained band during microaerobic growth or anaerobic growth with fumarate or nitrate as the sole electron acceptor. However, synthesis of a large number of additional cytochromes was observed in the periplasmic fractions prepared from cultures that were grown anaerobically with ferric citrate as the electron acceptor (Fig. 1, lanes 4, 5, and 6).
FIG. 1.
Heme-linked peroxidase staining of periplasmic fractions of wild-type S. frigidimarina. Aliquots (40 μg) of periplasmic fractions were loaded onto an SDS-10% (wt/vol) PAGE gel. Lane 1, purified sample of IfcA (1 μg); lane 2, periplasm from cells grown anaerobically with fumarate; lane 3, periplasm from cells grown anaerobically with nitrate; lane 4, periplasm from cells grown anaerobically with ferric citrate; lane 5, periplasm from cells grown anaerobically with ferric citrate and fumarate; lane 6, periplasm from cells grown anaerobically with ferric citrate and nitrate; lane 7, periplasm from cells grown anaerobically with manganese; lane 8, periplasm from cells grown microaerobically; lane 9 periplasm from cells grown microaerobically with ferric citrate.
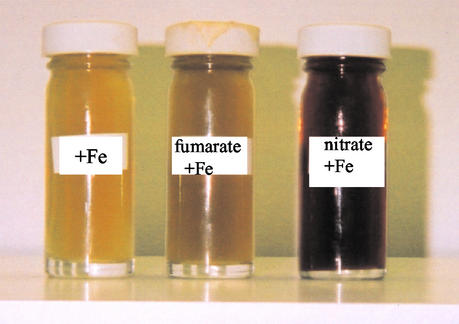
A negative effect of NO3− on Fe(III) reduction by S. putrefaciens 200 has been reported previously (3). S. putrefaciens cells grown with nitrate as the electron acceptor did not translocate protons when Fe(III) was provided, whereas fumarate-grown cells did translocate protons (22). Fe(III) reduction can be readily measured qualitatively by examining the color of the medium after growth, which changes from brown to yellow when Fe(III) is reduced. As Fig. 2 shows, S. frigidimarina extensively reduced Fe(III) to Fe(II) when it was grown with just Fe(III) or with fumarate and Fe(III). However, Fe(III) was not extensively reduced when S. frigidimarina was grown with nitrate and Fe(III). Thus, the apparent decreases in the amounts of periplasmic cytochromes after growth of S. frigidimarina with nitrate and iron compared to the amounts of the cytochromes present when iron or fumarate plus iron were used to grow cells might contribute to the lower extent of iron reduction during growth.
FIG. 2.
Fe(III) reduction by wild-type S. frigidimarina. A color change from black to yellow in the medium is an indication of Fe(III) reduction, as observed when S. frigidimarina cells were grown with ferric citrate or with fumarate and ferric citrate as electron acceptors but not when S. frigidimarina cells were grown with nitrate and ferric citrate as respiratory substrates.
Mn(IV) is an alternative metal ion respiratory substrate that can be used during anaerobic growth of Shewanella species (4, 23). Thus, it was of interest to analyze the cytochrome composition of the periplasm when cells were grown anaerobically with MnO2 as a terminal electron acceptor. It was notable that anaerobic growth on Mn(IV) resulted in the synthesis of many fewer periplasmic cytochromes compared to the number observed after growth with Fe(III) (Fig. 1, lane 7).
Taken together, these results indicate that S. frigidimarina responds to the presence of extracellular iron by increasing the number of c-type cytochromes that are allocated in the periplasmic compartment under anoxic conditions. This synthesis is more pronounced when Fe(III) is reduced efficiently. Moreover, this response is specific to iron since it does not occur if alternative respiratory substrates, including the insoluble metal MnO2, are used as terminal electron acceptors (23).
One of the heme-stained polypeptides synthesized exclusively during growth with Fe(III) migrated just below the Fcc3 protein and corresponded to the IfcA protein (Fig. 1). This 63.9-kDa iron-induced flavocytochrome protein has previously been purified from anaerobic cultures of S. frigidimarina with soluble iron(III) citrate as the sole electron acceptor, and it has been shown that it displays ferric reductase activity (5). IfcA was not synthesized when fumarate, nitrate, manganese, or oxygen was present as the sole respiratory substrate (Fig. 1). However, unlike some of the other cytochromes that were detected only during respiratory growth with Fe(III) (e.g., the 35-kDa band which might correspond to the S. oneidensis periplasmic decaheme protein MtrA [Fig. 1]), IfcA was synthesized in all the cultures in which iron was present as a respiratory substrate, regardless of whether iron was used as the main terminal electron acceptor (Fig. 1 and 2). Thus, IfcA was still produced after anaerobic growth with both ferric citrate and nitrate and after microaerobic growth on LB medium supplemented with ferric citrate. For this reason regulation of the ifcA gene was selected as a good system with which to begin studying the iron-dependent regulation of expression of c-type cytochrome genes in Shewanella species.
Identification of IfcR as a positive regulator of ifcA.
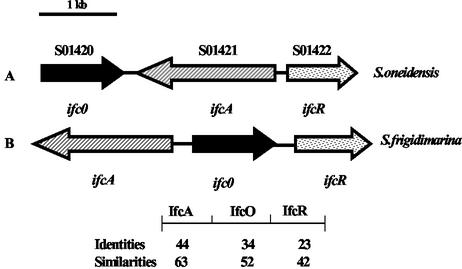
The ifcA gene has previously been identified on a 4,275-bp SacI DNA fragment (5). Sequence analysis of this fragment revealed the presence of one gene, ifcO, and one partial ORF, both of which are upstream of ifcA and have the opposite transcriptional orientation. The ifcO gene lies 271 bp upstream of ifcA (Fig. 3). Analysis of the IfcO primary sequence and database searches suggested that ifcO might encode an outer membrane β-barrel porin. The incomplete ORF was located 213 bp downstream of the ifcO stop codon and had the same transcriptional orientation. Completion of the nucleotide sequence of this ORF showed that this gene, designated ifcR, encodes a 293-amino-acid protein with a high level of amino acid similarity to members of the LysR family of transcriptional regulators. LysR-type transcriptional regulators probably comprise the largest family of prokaryotic regulatory proteins. So far, all of these proteins are DNA-binding proteins that positively regulate transcription of target genes, and most of them also regulate their own expression. The common family features are the similar sizes of the proteins (between 300 and 350 amino acids), the presence of a helix-turn-helix DNA-binding motif in the N-terminal region, and the requirement for a small molecule that acts as a coinducer (28). Genome sequence analysis of S. oneidensis indicated that the same three genes are also present in a cluster (SO1420 to SO1422); however, the genome organizations of this region in the two Shewanella species are different. The organizations of these three genes in the two species and the identities and similarities between the proteins are shown in Fig. 3.
FIG. 3.
Schematic representation of the organization of the ifcR, ifcO, and ifcA genes in S. frigidimarina and S. oneidensis. The arrows indicate directions of transcription. Annotation and preliminary genomic sequence data were provided by The Institute for Genomic Research (http://www.tigr.org). The numbers indicate the levels of amino acid identity and similarity (expressed as percentages) between the corresponding proteins from the two species.
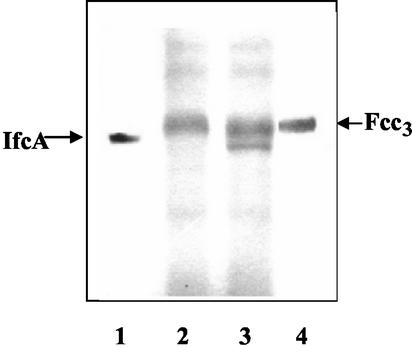
To determine whether the LysR homologue is involved in ifcA expression, mutant strain PSD106 of S. frigidimarina carrying a disruption of the gene encoding IfcR was constructed. The mutation did not noticeably affect the ability of the strain to grow and reduce various electron acceptors in batch cultures under anaerobic respiratory conditions (data not show). However, inspection of heme-stained SDS-PAGE gels of periplasmic fractions revealed that PSD106 grown anaerobically in the presence of Fe(III) did not produce the IfcA protein (Fig. 4, lane 2). Complementation of S. frigidimarina PSD106 with plasmid pPD1, a derivative of the replicative plasmid pRK415, which carries the complete ifcR gene plus 213 bp of upstream sequence, restored synthesis of IfcA (Fig. 4, lane 3). These results indicate that IfcR is a transcriptional activator required for IfcA production.
FIG. 4.
Heme-linked peroxidase staining of periplasmic fractions from S. frigidimarina PSD106 (ifcR) and PSD201 (ifcR+) after anaerobic growth with ferric citrate. Aliquots (40 μg) of periplasmic fractions were loaded onto an SDS-10% (wt/vol) PAGE gel. Lane 1, pure IfcA protein (1 μg); lane 2, periplasm from S. frigidimarina PSD106 (ifcR); lane 3, periplasm from S. frigidimarina PSD201 (PSD106 complemented with ifcR+); lane 4, purified Fcc3 protein (1 μg).
Fe(III)-dependent transcription of ifcA and ifcO is mediated by IfcR.
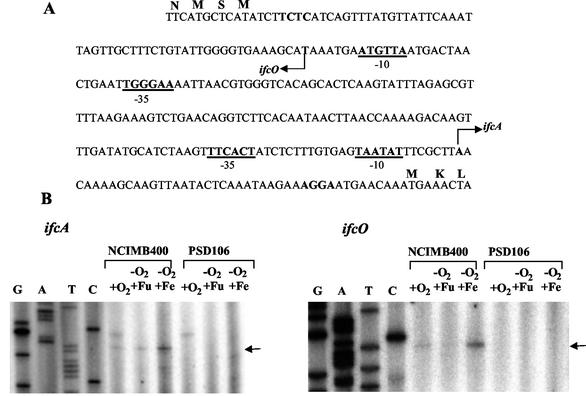
In order to investigate the regulatory role of IfcR in ifcA and ifcO expression, transcription of the ifcO and ifcA genes in both the wild type and the ifcR mutant was examined by primer extension analysis. Transcription initiation sites were identified 43 and 60 bp upstream from the translational start codons of the ifcA and ifcO genes, respectively (Fig. 5). In both cases, analysis of the DNA regions upstream of the ifcO and ifcA transcription initiation sites revealed the presence of −10 hexamers with only two mismatches compared with the −10 element from E. coli. Similarly, the −35 regions matched the E. coli consensus sequence at four of six positions (Fig. 5A). For both genes, the maximal levels of transcripts were observed after anaerobic growth with Fe citrate as the electron acceptor. Transcripts were less prominent following microaerobic or anaerobic growth with fumarate as the electron acceptor (Fig. 5B). Notably, no transcript of either gene was detected with total RNA isolated from ifcR mutant strain PSD106 (Fig. 5). These results show that both ifcA and ifcO are regulated in an iron-dependent manner that is dependent on IfcR. Thus, the requirement for IfcR for IfcA synthesis revealed by the analysis of heme-stained SDS-PAGE gels (Fig. 4) is due to transcriptional control.
FIG. 5.
Regulation of ifcO and ifcA transcription. (A) Intergenic region of ifcO and ifcA. The arrows indicate the transcription start site of each gene. Putative ribosome-binding sites are indicated by boldface type, and the −35 and −10 RNA polymerase-binding sites of each gene are indicated by boldface type and underlining. (B) Primer extension analysis of ifcA and ifcO performed with total RNA isolated from wild-type cells of S. frigidimarina and cells of ifcR mutant strain PSD106. +O2, RNA from cells grown microaerobically on LB medium; −O2 +Fu, RNA from cells grown anaerobically with fumarate as the sole electron acceptor; −O2+Fe, RNA from cells grown anaerobically with iron as the electron acceptor. Primer extension reactions were performed by using oligonucleotides P2 and P3 (see Materials and Methods), which hybridized with ifcO and ifcA, respectively. Sequencing reactions were performed with the oligonucleotides that were used in the primer extension reactions.
IfcR does not mediate Fe(III)-dependent transcription of the ifcR gene.
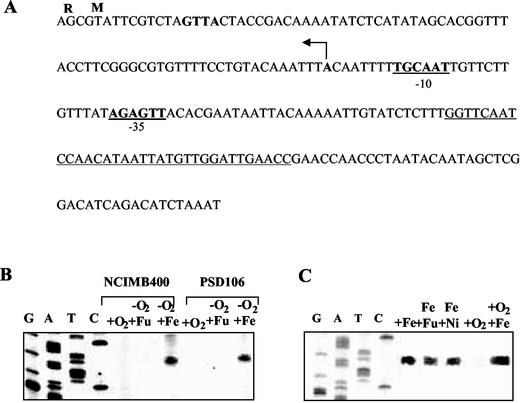
The transcription initiation site of ifcR was determined with RNA prepared from the wild-type strain after anaerobic growth with Fe(III) citrate. The transcription initiation site was 76 bp upstream of the translation initiation codon of the ifcR gene (Fig. 6). Putative sequences for the −10 and −35 elements of RNA polymerase binding sites could be identified (Fig. 6A). There was also a 34-bp palindromic sequence (bases 173 to 140) upstream of ifcR, but the significance of this sequence with regard to Fe(III)-dependent regulation has not been investigated yet. No transcript could be identified from the results of primer extension reactions performed with RNA prepared from cells incubated anaerobically with fumarate or microaerobically (Fig. 6B).
FIG. 6.
Regulation of ifcR expression. (A) Regulatory region of the ifcR gene. The arrow indicates the transcription start site of the ifcR gene. The proposed ribosome-binding site is indicated by boldface type, and the putative −35 and −10 RNA polymerase recognition sequences are indicated by boldface type and underlining. A perfect palindromic sequence is underlined. (B) Primer extension analysis of ifcR performed with total RNA isolated from wild-type cells of S. frigidimarina and cells of ifcR mutant strain PSD106. +O2, RNA from cells grown microaerobically on LB medium; −O2+Fu, RNA from cells grown anaerobically with fumarate as the sole electron acceptor; −O2+Fe, RNA from cells grown anaerobically with iron as the sole electron acceptor. Primer extension reactions were performed by using labeled oligonucleotide P1 (see Materials and Methods). Sequencing reactions were performed with the oligonucleotide that was used in the primer extension reactions. (C) Primer extension analysis of ifcR performed with total RNA isolated from wild-type cells of S. frigidimarina. +Fe, RNA from cells grown anaerobically with iron as the sole electron acceptor; Fe+Fu, RNA from cells grown anaerobically with fumarate and iron as electrons acceptors; Fe+Ni, RNA from cells grown anaerobically with iron and nitrate as electrons acceptors; +O2, RNA from cells grown microaerobically on LB medium; +O2 +Fe, RNA from cells grown microaerobically with ferric citrate.
Notably, a transcript was also observed in reactions carried out with RNA from the ifcR mutant. This transcript was only seen with RNA isolated from cells grown anaerobically with iron as the electron acceptor (Fig. 6B). These results indicate that transcription of the ifcR gene is regulated in response to Fe(III). The fact that the ifcR gene is transcribed even in the absence of IfcR indicates that Fe(III)-dependent transcription of ifcR is not mediated by the IfcR protein and points to the existence of a distinct Fe(III)-responsive regulator, which is required for expression of ifcR.
As mentioned above and shown in Fig. 2, S. frigidimarina completely reduces Fe(III) to Fe(II) in cultures supplemented with fumarate, whereas it does not reduce Fe(III) when cultures are supplemented with nitrate. To investigate whether ifcR expression requires Fe(III) per se or rather the utilization of Fe(III) as a terminal electron acceptor for respiration, transcription was examined in wild-type cells grown anaerobically with Fe(III) together with either nitrate or fumarate as an alternative respiratory substrate. Transcripts were also analyzed in cells grown microaerobically on LB medium supplemented with iron citrate, in which oxygen was the principal electron acceptor (Fig. 6C). A transcript was observed with RNA from both anaerobically and microaerobically grown cells as long as iron was provided in the external medium, regardless of whether the iron citrate was used as a respiratory substrate. Moreover, no significant differences in the amounts of transcript were apparent under the different respiratory conditions. These results strongly suggest that in S. frigidimarina the presence of Fe(III) rather than Fe(III) respiratory activity appears to be the trigger for ifcR expression and that anoxia is not a requirement.
Constitutive expression of ifcR in S. frigidimarina eliminates the requirement for extracellular Fe(III) for IfcA synthesis.
Transcript analyses indicated that expression of ifcR occurs when iron is supplied to the medium. In order to determine whether iron is specifically required for ifcR transcription or whether it is necessary for IfcR to be functionally active, a strain expressing ifcR constitutively was constructed. We placed the ifcR gene under control of the lac promoter, which previously has been shown to be functional in S. putrefaciens MR-1 (2). A DNA fragment lacking the entire promoter region of ifcR was prepared and cloned into pRK415. The resulting plasmid, pFR33, was introduced by conjugation into ifcR mutant strain PSD106, yielding strain PSR33. Plasmid pFR33 complemented the ifcR mutation, and synthesis of IfcA was restored in the periplasmic compartment during Fe(III) respiration of Shewanella strain PSR33 (data not shown).
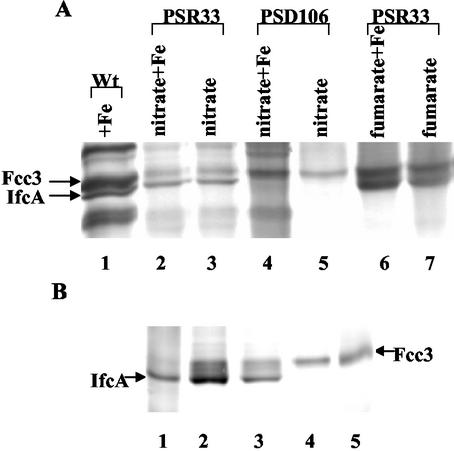
The ability of PSR33 to synthesize IfcA during anaerobic growth with various combinations of electron acceptors was studied. We also included in our experiments ifcR mutant strain PSD106 as a negative control. As expected, strain PSD106 did not produce IfcA (Fig. 7A). In agreement with the results obtained with periplasmic fractions from the wild-type strain (Fig. 1), IfcA was synthesized in periplasmic fractions from PSR33 cells that were grown with nitrate or fumarate in addition to Fe(III) citrate (Fig. 7A, lanes 2 and 6). Interestingly, and in contrast to what occurred with the wild type, the complemented strain produced IfcA when it was grown with nitrate or fumarate as the sole respiratory substrate, even if extracellular iron was not provided (Fig. 7A, lanes 3 and 7). IfcA was also synthesized in S. frigidimarina PSR33 when it was grown microaerobically in the absence of iron (Fig. 7B, compare lanes 2 and 3 with lane 4). It should also be noted that under all growth conditions IfcA was synthesized regardless of whether isopropyl-β-d-thiogalactopyranoside (IPTG) was added, suggesting that in S. frigidimarina sufficient expression of ifcR from the lac promoter occurs even without IPTG addition (Fig. 7B, compares lane 2 and 3). Taken together, these results indicate that extracellular iron is not required for IfcR to be able to activate ifcA transcription; rather, the requirement for iron for synthesis of IfcA is imposed at the level of transcription of ifcR.
FIG. 7.
Heme-linked peroxidase staining of periplasmic fractions from S. frigidimarina PSR33 (ifcR+). (A) Aliquots (40 μg) of periplasmic fractions were loaded onto an SDS-10% PAGE gel. Lane 1, periplasm from wild-type cells grown anaerobically with ferric citrate (positive control); lane 2, periplasm from PSR33 cells grown anaerobically with nitrate and ferric citrate; lane 3, periplasm from PSR33 cells grown anaerobically with nitrate; lane 4, periplasm from PSD106 cells grown anaerobically with nitrate and ferric citrate; lane 5, periplasm from PSD106 cells grown anaerobically with nitrate; lane 6, periplasm from PSR33 cells grown anaerobically with fumarate and ferric citrate; lane 7, periplasm from PSR33 cells grown anaerobically with fumarate. (B) Lane 1, purified IfcA (1 μg); lane 2, periplasm from PSR33 cells grown microaerobically on LB medium with 1 mM IPTG; lane 3, periplasm from PSR33 cells grown microaerobically on LB medium without IPTG; lane 4, periplasm from wild-type cells carrying plasmid pRK415 grown microaerobically on LB medium with 1 mM IPTG; lane 5, purified Fcc3 (1 μg).
Conclusions.
The results described here revealed that there is an Fe(III)-responsive regulatory system that has features which are clearly distinct from those of the well-characterized transcription factors that regulate iron assimilation in both bacteria and fungi. For example, the Fur protein negatively regulates transcription of many genes involved in iron assimilation in a phylogenetically diverse range of bacteria in response to Fe(II) (10). Similarly, in fungi such as Aspergillus nidulans, Neurospora crassa, or Ustilago maydis GATA family proteins serve as transcriptional repressors by binding to GATA motifs in the promoters of genes involved in siderophore biosynthesis (11, 25). The recent annotation of the genome sequence of S. oneidensis MR-1 revealed the presence of a Fur homolog with 73 and 79% amino acid identity to the homologs in E. coli and Vibrio cholerae, respectively (12). Preliminary data from partial genome microarrays of a fur knockout strain of S. oneidensis agree with the well-established model in which Fur is a negative regulator of siderophore-receptor-mediated iron acquisition but do not suggest involvement in regulation of Fe(III) respiration (32).
The Fe(III)-dependent regulatory system which we identified here is clearly distinct from Fur. Elucidation of the nature of this Fe(III)-responsive regulator is important for establishing how Shewanella coordinates assimilatory and respiratory iron metabolism.
Acknowledgments
This work was supported by BBSRC grant 83/P11894 to D.J.R. and P.D. and by Wellcome Trust grant 059531/Z/99/Z to D.J.R. Work in the laboratory of G.S. was supported by a competitive strategic grant from the BBSRC to the John Innes Centre.
We are grateful to Ann Reilly for technical assistance.
REFERENCES
- 1.Bamford, V., P. S. Dobbin, D. J. Richardson, and A. M. Hemmings. 1999. Open conformation of a flavocytochrome c3 fumarate reductase. Nat. Struct. Biol. 6:1104-1107. [DOI] [PubMed] [Google Scholar]
- 2.Beliaev, A. S., D. A. Saffarini, J. L. McLaughlin, and D. Hunnicutt. 2001. MtrC, an outer membrane decahaem c cytochrome required for metal reduction in Shewanella putrefaciens MR-1. Mol. Microbiol. 39:722-730. [DOI] [PubMed] [Google Scholar]
- 3.DiChristina, T. J. 1992. Effects of nitrate and nitrite on dissimilatory iron reduction by Shewanella putrefaciens 200. J. Bacteriol. 174:1891-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DiChristina, T. J., and E. F. DeLong. 1994. Isolation of anaerobic respiratory mutants of Shewannella putrefaciens and genetic analysis of mutants deficient in anaerobic growth on Fe3+. J. Bacteriol. 176:1468-1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dobbin, P. S., J. N. Butt, A. K. Powell, G. A. Reid, and D. J. Richardson. 1999. Characterization of a flavocytochrome that is induced during the anaerobic respiration of Fe3+ by Shewanella frigidimarina NCIMB400. Biochem. J. 342:439-448. [PMC free article] [PubMed] [Google Scholar]
- 6.Fellay, R., J. Frey, and H. Krisch. 1987. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of gram-negative bacteria. Gene 52:147-154. [DOI] [PubMed] [Google Scholar]
- 7.Field, S. J., P. S. Dobbin, M. R. Cheesman, N. J. Watmough, A. J. Thomson, and D. J. Richardson. 2000. Purification and magneto-optical spectroscopic characterization of cytoplasmic membrane and outer membrane multiheme c-type cytochromes from Shewanella frigidimarina NCIMB400. J. Biol. Chem. 275:8515-8522. [DOI] [PubMed] [Google Scholar]
- 8.Gordon, E. H., S. L. Pealing, S. K. Chapman, F. B. Ward, and G. A. Reid. 1998. Physiological function and regulation of flavocytochrome c3, the soluble fumarate reductase from Shewanella putrefaciens NCIMB 400. Microbiology 144:937-945. [DOI] [PubMed] [Google Scholar]
- 9.Gordon, E. H., A. D. Pike, A. E. Hill, P. M. Cuthbertson, S. K. Chapman, and G. A. Reid. 2000. Identification and characterization of a novel cytochrome c(3) from Shewanella frigidimarina that is involved in Fe(III) respiration. Biochem. J. 349:153-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guerinot, M. L. 1994. Microbial iron transport. Annu. Rev. Microbiol. 48:743-772. [DOI] [PubMed] [Google Scholar]
- 11.Haas, H., I. Zadra, G. Stoffler, and K. Angermayr. 1999. The Aspergillus nidulans GATA factor SREA is involved in regulation of siderophore biosynthesis and control of iron uptake. J. Biol. Chem. 274:4613-4619. [DOI] [PubMed] [Google Scholar]
- 12.Heidelberg, J. F., I. T. Paulsen, K. E. Nelson, E. J. Gaidos, W. C. Nelson, T. D. Read, J. A. Eisen, R. Seshadri, N. Ward, B. Methe, R. A. Clayton, T. Meyer, A. Tsapin, J. Scott, M. Beanan, L. Brinkac, S. Daugherty, R. T. DeBoy, R. J. Dodson, A. S. Durkin, D. H. Haft, J. F. Kolonay, R. Madupu, J. D. Peterson, L. A. Umayam, O. White, A. M. Wolf, J. Vamathevan, J. Weidman, M. Impraim, K. Lee, K. Berry, C. Lee, J. Mueller, H. Khouri, J. Gill, T. R. Utterback, L. A. McDonald, T. V. Feldblyum, H. O. Smith, J. C. Venter, K. H. Nealson, and C. M. Fraser. 2002. Genome sequence of the dissimilatory metal ion-reducing bacterium Shewanella oneidensis. Nat. Biotechnol. 20:1118-1123. [DOI] [PubMed] [Google Scholar]
- 13.Keen, N. T., S. Tamaki, D. Koayashi, and D. Trolliger. 1988. Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria. Gene 70:191-197. [DOI] [PubMed] [Google Scholar]
- 14.Leys, D., T. E. Meyer, A. S. Tsapin, K. H. Nealson, M. A. Cusanovich, and J. J. Van Beeumen. 2002. Crystal structures at atomic resolution reveal the novel concept of ′electron-harvesting' as a role for the small tetraheme cytochrome c. J. Biol. Chem. 277:35703-35711. [DOI] [PubMed] [Google Scholar]
- 15.Leys, D., A. S. Tsapin, K. H. Nealson, T. E. Meyer, M. A. Cusanovich, and J. J. Van Beeumen. 1999. Structure and mechanism of the flavocytochrome c fumarate reductase of Shewanella putrefaciens MR-1. Nat. Struct. Biol. 6:1113-1117. [DOI] [PubMed] [Google Scholar]
- 16.Lovely, D. R. 1995. Bioremediation of organic and metal contaminants with dissimilatory metal reduction. J. Ind. Microbiol. Biotechnol. 14:85-93. [DOI] [PubMed] [Google Scholar]
- 17.Lovely, D. R. 1991. Dissimilatory Fe(III) and Mn(IV) reduction. Microbiol. Rev. 55:259-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lovely, D. R., and J. D. Coates. 1997. Bioremediation of metal contamination. Curr. Opin. Biotechnol. 8:285-289. [DOI] [PubMed] [Google Scholar]
- 19.Myers, C. R., and J. M. Myers. 1997. Cloning and sequence of cymA, a gene encoding a tetraheme cytochrome c required for reduction of iron(III), fumarate, and nitrate by Shewanella putrefaciens MR-1. J. Bacteriol. 179:1143-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Myers, C. R., and J. M. Myers. 1993. Ferric reductase is associated with the membranes of anaerobically grown Shewanella putrefaciens MR-1. FEMS Microbiol. Lett. 108:15-22. [Google Scholar]
- 21.Myers, C. R., and J. M. Myers. 1997. Outer membrane cytochromes of Shewanella putrefaciens MR-1: spectral analysis, and purification of the 83-kDa c-type cytochrome. Biochim. Biophys. Acta 1326:307-318. [DOI] [PubMed] [Google Scholar]
- 22.Myers, C. R., and K. H. Nealson. 1990. Respiration-linked proton translocation coupled to anaerobic reduction of manganese(IV) and iron(III) in Shewanella putrefaciens MR-1. J. Bacteriol. 172:6232-6238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Myers, J. M., and C. R. Myers. 2001. Role for outer membrane cytochromes OmcA and OmcB of Shewanella putrefaciens MR-1 in reduction of manganese dioxide. Appl. Environ. Microbiol. 67:260-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Myers, J. M., and C. R. Myers. 2000. Role of the tetraheme cytochrome CymA in anaerobic electron transport in cells of Shewanella putrefaciens MR-1 with normal levels of menaquinone. J. Bacteriol. 182:67-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oberegger, H., M. Schoeser, I. Zadra, B. Abt, and H. Haas. 2001. SREA is involved in regulation of siderophore biosynthesis, utilization and uptake in Aspergillus nidulans. Mol. Microbiol. 41:1077-1089. [DOI] [PubMed] [Google Scholar]
- 26.Quandt, J., and M. F. Hynes. 1993. Versatile suicide vectors which allow direct selection for gene replacement in Gram negative bacteria. Gene 127:15-21. [DOI] [PubMed] [Google Scholar]
- 27.Roldan, M. D., H. J. Sears, M. R. Cheesman, S. J. Ferguson, A. J. Thomson, B. C. Berks, and D. J. Richardson. 1998. Spectroscopic characterization of a novel multiheme c-type cytochrome widely implicated in bacterial electron transport. J. Biol. Chem. 273:28785-28790. [DOI] [PubMed] [Google Scholar]
- 28.Schell, M. A. 1993. Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47:597-626. [DOI] [PubMed] [Google Scholar]
- 29.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host-range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 30.Taylor, P., S. L. Pealing, G. A. Reid, S. K. Chapman, and M. D. Walkinshaw. 1999. Structural and mechanistic mapping of a unique fumarate reductase. Nat. Struct. Biol. 6:1108-1112. [DOI] [PubMed] [Google Scholar]
- 31.Thomas, P. E., D. Ryan, and W. Levin. 1976. An improved staining procedure for the detection of the peroxidase activity of cytochrome P-450 on sodium dodecyl sulfate polyacrylamide gels. Anal. Biochem. 75:168-176. [DOI] [PubMed] [Google Scholar]
- 32.Thompson, D. K., A. S. Beliaev, C. S. Giometti, S. L. Tollaksen, T. Khare, D. P. Lies, K. H. Nealson, H. Lim, J. Yates 3rd, C. C. Brandt, J. M. Tiedje, and J. Zhou. 2002. Transcriptional and proteomic analysis of a ferric uptake regulator (Fur) mutant of Shewanella oneidensis: possible involvement of Fur in energy metabolism, transcriptional regulation, and oxidative stress. Appl. Environ. Microbiol. 68:881-892. [DOI] [PMC free article] [PubMed] [Google Scholar]