Abstract
Comparison of the proteomes of the wild-type and Fe-only hydrogenase mutant strains of Desulfovibrio vulgaris Hildenborough, grown in lactate-sulfate (LS) medium, indicated the near absence of open reading frame 2977 (ORF2977)-coded alcohol dehydrogenase in the hyd mutant. Hybridization of labeled cDNA to a macroarray of 145 PCR-amplified D. vulgaris genes encoding proteins active in energy metabolism indicated that the adh gene was among the most highly expressed in wild-type cells grown in LS medium. Relative to the wild type, expression of the adh gene was strongly downregulated in the hyd mutant, in agreement with the proteomic data. Expression was upregulated in ethanol-grown wild-type cells. An adh mutant was constructed and found to be incapable of growth in media in which ethanol was both the carbon source and electron donor for sulfate reduction or was only the carbon source, with hydrogen serving as electron donor. The hyd mutant also grew poorly on ethanol, in agreement with its low level of adh gene expression. The adh mutant grew to a lower final cell density on LS medium than the wild type. These results, as well as the high level of expression of adh in wild-type cells on media in which lactate, pyruvate, formate, or hydrogen served as the sole electron donor for sulfate reduction, indicate that ORF2977 Adh contributes to the energy metabolism of D. vulgaris under a wide variety of metabolic conditions. A hydrogen cycling mechanism is proposed in which protons and electrons originating from cytoplasmic ethanol oxidation by ORF2977 Adh are converted to hydrogen or hydrogen equivalents, possibly by a putative H2-heterodisulfide oxidoreductase complex, which is then oxidized by periplasmic Fe-only hydrogenase to generate a proton gradient.
The role of the cytoplasmic membrane in the conservation of chemiosmotic energy is fairly well understood for aerobic bacteria, because of similarities to the mitochondrial inner membrane paradigm. However, this role is much less clear for anaerobes. For instance, chemiosmotic energy conservation is crucial for sulfate-reducing bacteria (SRB), when growing with lactate as the electron donor, because the ATP yield of substrate-level phosphorylation exactly balances what is required for sulfate activation (19, 21, 33). Odom and Peck proposed that SRB of the genus Desulfovibrio conserve chemiosmotic energy by hydrogen cycling (17). In their model, hydrogen generated from cytoplasmic lactate oxidation was proposed to diffuse across the cytoplasmic membrane, where it is oxidized by periplasmic hydrogenase to form a proton gradient with the electrons being conducted back to the cytoplasmic sulfate reduction pathway. Analysis of the genome sequence for Desulfovibrio vulgaris Hildenborough (http://www.tigr.org) has indicated the presence of two cytoplasm-facing, membrane-bound hydrogenases, four periplasmic hydrogenases, and a variety of transmembrane electron-transporting complexes. Hence, all components required for a hydrogen cycling mechanism of energy conservation are present in D. vulgaris. However, detailed physiological analysis of mutants lacking either the hyd genes for periplasmic Fe-only hydrogenase or the hmc genes for the transmembrane, electron-transporting high-molecular-weight cytochrome (Hmc) complex has revealed a more complicated picture (5, 18, 30). Relative to the wild-type strain, the hyd mutant appeared to produce significant amounts of CO from pyruvate. CO was formed transiently in cultures containing lactate or pyruvate and excess sulfate (30), indicating that it was both produced and consumed. The genome sequence of D. vulgaris also indicates the presence of a cytoplasmic pyruvate-formate lyase and of multiple periplasmic formate dehydrogenases. Hence, cycling of metabolites other than hydrogen (i.e., CO and formate) may contribute to chemiosmotic energy conservation during sulfate reduction with lactate (30).
Because the D. vulgaris genome sequence is known, the question of which proteins are involved in chemiosmotic energy conservation can now be pursued by proteomic or functional genomic approaches. In view of the significant physiological differences between the hyd mutant and wild-type strain (30), we focused initial studies on a comparison of these two strains.
MATERIALS AND METHODS
Materials.
Mixed gases (85% [vol/vol] N2, 10% [vol/vol] CO2, and 5% [vol/vol] H2; as well as 90% [vol/vol] N2 and 10% [vol/vol] CO2) were obtained from Praxair Products, Inc., Edmonton, Alberta, Canada. Restriction enzymes, Taq polymerase, DNA ligase, and Hybond-N membranes were obtained from Amersham Pharmacia Biotech, Baie d'Urfe, Quebec, Canada, whereas Superscript II RNase H− reverse transcriptase was obtained from Invitrogen, Burlington, Ontario, Canada. [α-32P]dCTP (10 mCi/ml [3,000 Ci/mmol]) was purchased from ICN Biomedicals, Inc., Irvine, Calif. Reagent-grade chemicals were obtained from BDH, Fisher, or Sigma. RNeasy kits, RNAprotect reagent, and RNase-free DNase were purchased from Qiagen, Mississauga, Ontario, Canada. Deoxyoligonucleotide primers were obtained from University Core DNA Services of the University of Calgary.
Bacterial strains, plasmids, and primers.
The strains and plasmids used in this study are listed in Table 1. The primers listed in Table 1 were used for construction of the D. vulgaris adh mutant. Sequences of 290 forward and reverse primers used to PCR amplify 145 genes involved in energy metabolism of D. vulgaris chosen from a preliminary annotation of the genome were designed with Primer 3 (http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi) to have a Tm of 60°C and were synthesized by University Core DNA Services with a 96-well MerMade IV system from Bioautomation (Plano, Tex.). The sequences used have been listed elsewhere (http://www.ucalgary.ca/SC/BI/divisions/cmmb/voordouw2.html).
TABLE 1.
Bacterial strains, primers, vectors, and plasmids used
| Strain, plasmid, or primer | Genotype, sequence (position), and/or comment(s) | Source or reference |
|---|---|---|
| Strains | ||
| D. vulgaris subsp. vulgaris Hildenborough | ||
| Wild type | NCIMB 8303; isolated from clay soil near Hildenborough, United Kingdom | 19 |
| Hyd100 | ΔhydA,B; Cmr | 18 |
| ADH100 | Δadh; Cmr | This study |
| ADH8 | pNotΔAdhCmMob integrated into the chromosome; Sucs Cmr | This study |
| E. coli S17-1 | thi pro hsdR hsdM+ recA RP4-2 (Tc::Mu, Km::Tn7) | 25 |
| Plasmids | ||
| pUC19Cm | pUC19 containing the cat gene | 7 |
| pNOT19 | Cloning vector pUC19; NdeI site replaced by a NotI site | 24 |
| pMOB2 | Contains oriT of plasmid RP4 and Bacillus subtilis sacBR genes on a 4.5-kb NotI fragment; Kmr Cmr | 24 |
| pNotAdh, pNotΔAdh, pNotΔAdhBam, pNotΔAdhCmMob | This study (see text) | |
| Primers | ||
| P206-f | 5′-cccggtaCCATGCTTCAGGGGCTGAAGCCC (−581 to −560)a | |
| P207-r | 5′-ACAACTGGAGCAGGACAGGC (1627 to 1647)a |
Positions relative to the adh coding region (bp 1 to 1179). Lowercase letters in P206-f are bases added to create a KpnI cleavage site.
Growth of bacteria.
For construction of the adh mutant, Escherichia coli and D. vulgaris strains were cultured in media described elsewhere (7, 18). For proteomic and gene array analyses, D. vulgaris wild-type or mutant strains were grown in Widdel Pfennig (WP) medium formulated according to Widdel and Bak (34) under an anoxic atmosphere of 90% N2-10% CO2 (vol/vol). WP-LS medium contained 38 mM lactate and 28 mM sulfate. Pyruvate (30 mM), formate (70 mM), and ethanol (40 mM) were used as alternative organic electron donors. Cultures in WP medium with hydrogen as the electron donor were in a mixed gas atmosphere (85% [vol/vol] N2, 10% [vol/vol] CO2, 5% [vol/vol] H2) with either CO2 and 3 mM acetate or CO2 and 3 mM ethanol as the carbon source. The optical density of cultures at 600 nm (OD600) was determined with a Shimadzu UV-visible spectrophotometer. Sulfate and sulfide concentrations were determined as described elsewhere (15).
2DE.
Cultures (50 to 250 ml) in WP-LS medium were harvested in the mid-exponential phase (OD600 = 0.3). Following harvesting of cells by centrifugation (10,000 × g, 20 min, 4°C), Tris-EDTA (TE) extracts were prepared by resuspending the pellets in 0.1 M Tris-0.1 M EDTA (pH 9) equivalent to 0.01 V × OD600, where V is the volume (milliliters) and OD600 is the OD of the culture at the time of harvesting and processing as described elsewhere (6). Aliquots (50 μl) of TE extracts were frozen in liquid nitrogen and stored at −80°C. TE extracts had a protein content of 3.1 to 5.4 mg/ml, as determined by the method of Bradford (2). Isoelectric focusing was carried out with the IPGphor system using 18-cm strips with immobilized pH gradients (IPGs) of 3 to 10 nonlinear (NL), 4 to 7, and 6 to 9 (Amersham Biosciences, Freiburg, Germany). For silver and colloidal Coomassie brilliant blue (cCBB) staining, 20 to 40 μg and 50 to 200 μg of protein were loaded into the IPG strip holders, respectively. The appropriate volume of TE extract was diluted with rehydration buffer (8). Conditions for rehydration, sample entry, and isoelectric focusing were as described by Rabus et al. (20). After isoelectric focusing, the strips were incubated in equilibration buffer containing dithiothreitol (DTT) and then in equilibration buffer containing iodoacetamide (8, 20). Proteins were separated by two-dimensional gel electrophoresis (2DE) using an electrophoresis system from Genomic Solutions, Inc. (Ann Arbor, Mich.), as recently described (8, 20). Following electrophoresis, gels were fixed and stained with silver or cCBB (16). Data acquisition of stained gels was performed with the IMAGE scanner (Amersham Biosciences).
Amino acid and mass spectrometric analysis.
Protein spots stained with cCBB were excised manually. Tryptic digest of proteins and separation of peptide fragments by high-performance liquid chromatography was carried out as described before (12). Determination of amino acid sequences by Edman-degradation (11) and of peptide masses by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) (4) was carried out by TopLab (Martinsried, Germany). Protein identification and genome analysis were based on a preliminary list of annotated genes from the genomic sequence of D. vulgaris. Peptide mass fingerprints were mapped to the coding genes by using the MS-Digest program (3). For matching of experimentally determined peptide masses with theoretical values, differences of about 50 ppm were allowed as thresholds.
Design and production of a D. vulgaris macroarray.
A set of 177 open reading frames (ORFs) encoding proteins involved in energy metabolism (for a complete list, see http://www.ucalgary.ca/SC/BI/divisions/cmmb/voordouw2.html) was identified from a preliminary annotation of the D. vulgaris genome. Primers for PCR amplification of this set covered as much of each ORF as possible. PCR products ranged in length from 0.1 to 2.9 kb. In some cases, two or more ORFs in the same operon were amplified together. PCRs (50 μl) included 5 μl of 10× PCR buffer (500 mM KCl, 15 mM MgCl2, 100 mM Tris-HCl [pH 9]), 1 μl of deoxynucleoside triphosphates (dNTPs [2.5 mM each]), 1 μl of each primer (20 pmol/μl), 0.2 μl of Taq polymerase (5 U/μl), and either a large (100 ng) or small (1 ng) amount of D. vulgaris chromosomal DNA. Amplification was in a Perkin-Elmer GeneAmp 2400 PCR system by incubation for 5 min at 94°C, followed by 30 cycles of 30 s at 94°C; 30 s at 55°C; and 90, 120, or 180 s at 72°C, depending on the size of the expected product (up to 1 kb, 1 to 2 kb, or 2 to 3 kb, respectively). When using 100 ng of chromosomal DNA as the template, the product of the first PCR was diluted 500-fold, and 1 μl was used as the template in a second PCR. When using 1 ng of DNA, the product of the first PCR was used directly. PCR products were visualized on 0.7% (wt/vol) agarose gels stained with ethidium bromide. Those used for array construction (145 PCR products covering 159 of the 177 ORFs identified initially) consisted of single bands of the expected size. PCR products (25 to 100 ng/μl, as determined fluorimetrically) were incubated in a boiling water bath (3 min) and then in an ice bath (3 min). Aliquots (1 μl) of denatured PCR products were spotted 0.6 cm apart on Hybond-N membranes with a PB600 repeating dispenser (Hamilton, Co., Reno, Nevada), fitted with a 50-μl no. 710 Hamilton syringe. A PCR-negative control reaction mixture with 1 ng of chromosomal DNA but no primers was spotted as a negative control (0.02 ng of chromosomal DNA per spot). Bacteriophage λ DNA was also spotted as a negative control, whereas denatured D. vulgaris chromosomal DNA (12.5 to 1.6 ng per spot) was spotted as positive controls. After drying, DNAs were covalently linked to the arrays by UV irradiation. Macroarrays were stored at room temperature.
Hybridization with labeled cDNA.
RNA was prepared from 50-ml cultures grown to half-maximal density in WP medium. One volume of culture was added to 2 volumes of Qiagen RNAprotect reagent, and RNA was extracted and purified with the Qiagen RNeasy kit with on-column DNase treatment. Samples of purified, DNA-free RNA in RNase-free water were stored at −80°C. Reverse transcription reaction mixtures (50 μl) contained 5 μg of RNA, 1 μl of random hexamers (0.25 μg/μl), 2.5 μl of dNTP mix (20 mM each dATP, dGTP, and dTTP), 200 U of reverse transcriptase, 100 μCi of [α-32P]dCTP (3,000 Ci/mmol), 10 μl of 5× First Strand buffer (250 mM Tris-Cl [pH 8.3], 375 mM KCl, 15 mM MgCl2), and 5 μl of 0.1 M DTT and were carried out according to the manufacturer's instructions. The RNA substrate was degraded by adding 50 μl of 0.27 M NaOH-20 mM EDTA and incubating this mixture at 65°C for 30 min. Reactions were neutralized by adding 50 μl of 1 M Tris-Cl (pH 7.4). RNA-free, chromosomal DNA of wild-type D. vulgaris (14) was labeled by using random hexamers (26, 32). Labeled cDNA or genomic DNA probes were hybridized to the arrays at 68°C by a high-stringency procedure (22, 31). After washing and drying, the arrays were exposed to BAS-IIIS type imaging plates for 4 h. These were scanned with a BAS1000 bio-imaging analyzer (Fuji). MacBas 2.2 software gave hybridization intensities in units of photostimulable luminescence (PSL) for all spots. Net values (ΔPSL) were obtained by subtracting negative control values, and the sum, ΣΔPSL, of all 145 ΔPSL values was calculated. ΔPSLS values were calculated as (ΔPSL/ΣΔPSL) · 100. On average, ΔPSLS values differed by 4.5% from the mean in duplicate experiments in which a labeled cDNA from the same mRNA pool was split and hybridized to two macroarrays. ΔPSLS values obtained for labeled cDNAs from independently prepared mRNA pools differed on average by 16.9% from the mean. Average ΔPSLS values were calculated for four array hybridizations by using labeled cDNAs from two independently prepared RNA samples hybridized in duplicate. Average values for ΔPSLS were divided by average values obtained with the genomic DNA probe to obtain net relative hybridization intensities (IR), corrected for different concentrations of the immobilized target gene, as well as for differences in signal intensity due to the different lengths of the PCR products.
Construction of the adh mutant.
The gene for ORF2977 Adh, flanked by 500-bp upstream and downstream regions, was amplified with primers P206-f and P207-r (Table 1), cut with KpnI and PstI, and cloned in pNOT19 to give pNotAdh. Cleavage with SalI and ligation gave pNotΔAdh, in which bp 387 to 502 of the 1,179-bp adh coding region were deleted. Plasmids pNotΔAdhBam, pNotΔAdhCm, and pNotΔAdhCmMob were created by sequential insertion of (i) a SalI-BamHI adapter (5′-TCGAGGGATCCC), (ii) the 1.4-kb BamHI fragment from pUC19Cm containing the cat gene, and (iii) the 4.2-kb NotI fragment from pMOB2 containing oriT and sacB. The suicide plasmid was transferred to D. vulgaris by conjugation with E. coli S17-1 (pNotΔAdhCmMob). A single-crossover integrant, D. vulgaris ADH11, in which pNotΔAdhCmMob had integrated through the upstream homologous regions, was selected. Computer analysis indicated that Southern blotting of SalI-digested chromosomal DNAs, using the labeled wild-type P206-f and P207-r amplicon as the probe, would give distinctly sized hybridizing fragments for the wild type (1.7 and 2.3 kb), the integrant (1.3, 2.3, and 8.3 kb), and the double-crossover deletion mutant (5.4 kb). Growth of D. vulgaris ADH11 in the presence of sucrose and chloramphenicol followed by colony purification gave D. vulgaris ADH100 with this expected Southern blot pattern (results not shown).
RESULTS
2DE.
The protein compositions of TE extracts of the wild-type and hyd mutant D. vulgaris strains were compared by using IPG strips with IPGs of 3 to 10, 4 to 7, or 6 to 9 in the first dimension. The largest number of distinct protein spots was resolved with a pH gradient of 4 to 7 (Fig. 1). Twenty-nine prominent proteins were excised from cCBB-stained 2DE gels and identified by MALDI-TOF analysis of trypsin digests (Table 2). Their positions in silver-stained gels are indicated in Fig. 1 and 2. Their theoretical molecular masses agreed well with their positions on the gel, except for spots 3 to 7, 12, and 14. Spots 4 and 5 were identified as a 94.1-kDa membrane-protein that would not be expected in the TE extract, unless the observed mass (32 kDa) represents a smaller, soluble fragment. As reported before (6), the TE extract contains both periplasmic and cytoplasmic proteins, even though this extraction method was thought to specifically extract periplasmic proteins (29). Although periplasmic proteins were present (Table 2), most of the identified proteins were cytoplasmic. This included EF-Ts and EF-Tu (Fig. 1, spots 13 and 20), elongation factors in protein synthesis that are among the most highly expressed proteins in growing bacterial cells. A protein typical for anaerobic energy metabolism was acetate kinase (Fig. 1, spot 25), whereas an enzyme typical for sulfate reducers, sulfate adenylyltransferase (ATP sulfurylase), was represented by spots 16 and 17, indicating posttranslational processing. Nigerythrin, which has an oxygen defense function, was also identified (Fig. 1, spot 11). The TE extract thus contains a wide variety of soluble cytoplasmic and periplasmic proteins. Comparison of more than 10 2DE gels indicated that TE extraction represents an easy and reproducible way to characterize the D. vulgaris proteome.
FIG. 1.
Silver-stained 2DE gels of TE extracts from D. vulgaris wild-type (WT) and hyd mutant (Hyd100) strains. Proteins were separated by isoelectric focusing with IPGs of 4 to 7, as indicated. All numerically marked spots were identified by MALDI-TOF-MS (Table 2). Protein 1 is at the right edge of this pH gradient.
TABLE 2.
Mass spectrometric identification of proteins from D. vulgaris separated by 2DE
| Spot no.a | IDb | Sequence coverage (%)c | Significanced | pIe | Mrf | Locationg | Possible function (gene)h |
|---|---|---|---|---|---|---|---|
| 1a | ORF02977 | 32 | 5.8 e-22 | 6.8 | 41.8 | C | Alcohol dehydrogenase |
| 1b | ORF02977 | 44 | 5.7 e-27 | 6.8 | 41.8 | C | Alcohol dehydrogenase |
| 2 | ORF03570 | 26 | 3.0 e-06 | 6.0 | 28.5 | P | Conserved hypothetical protein (periplasmic) |
| 3 | ORF04254 | 13 | 1.5 e-04 | 6.7 | 28.4* | C | Ubiquinone/menaquinone biosynthesis methyltransferase (ubiE) |
| 4 | ORF02836 | 68 | 6.5 e-50 | 5.1 | 94.1* | M | Cation-transporting ATPase, E1-E2 family |
| 5 | ORF02836 | 58 | 2.0 e-37 | 5.1 | 94.1* | M | Cation-transporting ATPase, E1-E2 family |
| 6 | ORF00572 | 40 | 1.9 e-31 | 4.7 | 87.1* | C | PTS system, fructose-specific EIIA/HPr/EI components (ptsI) |
| 7 | ORF00572 | 41 | 7.7 e-39 | 4.7 | 87.1* | C | PTS system, fructose-specific EIIA/HPr/EI components (ptsI) |
| 8 | ORF00988 | 63 | 1.4 e-29 | 5.8 | 26.8 | P | Arginine ABC transporter, periplasmic arginine-binding protein, putative |
| 9 | ORF02865 | 52 | 1.3 e-19 | 7.0 | 29.7 | P | Amino acid ABC transporter, amino acid-binding protein, putative |
| 10 | ORF00176 | 32 | 2.1 e-14 | 7.1 | 30.4 | P | Amino acid ABC transporter, amino acid-binding protein, putative |
| 11 | ORF04661 | 42 | 1.1 e-19 | 4.9 | 20.2 | C | Nigerythrin |
| 12 | ORF04056 | 9 | 2.2 e-04 | 5.7 | 63.5* | C | Sensory box/GGDEF family protein |
| 13 | ORF00383 | 56 | 4.6 e-29 | 5.0 | 30.5 | C | Translation elongation factor Ts (tsf) |
| 14 | ORF00347 | 54 | 5.6 e-18 | 4.9 | 20.2* | C | Hypothetical protein |
| 15 | ORF00590 | 57 | 1.4 e-23 | 5.4 | 21.2 | C | ThiJ/PfpI family superfamily |
| 16 | ORF01081 | 45 | 2.8 e-37 | 6.0 | 47.5 | C | Sulfate adenylyltransferase (sat) |
| 17 | ORF01081 | 52 | 1.7 e-41 | 6.0 | 47.5 | C | Sulfate adenylyltransferase (sat) |
| 18 | ORF03883 | 69 | 3.4 e-17 | 5.4 | 17.0 | C | Nitrogen assimilation regulatory protein (Klebsiella, putative) |
| 19 | ORF02097 | 42 | 2.4 e-13 | 4.6 | 17.7 | C | Purine-binding chemotaxis protein CheW (cheW-1) |
| 20 | ORF03811 | 43 | 5.9 e-25 | 4.8 | 43.4 | C | Translation elongation factor Tu (tuf) |
| 21 | ORF03178 | 60 | 1.6 e-36 | 5.1 | 41.4 | C | Phosphoglycerate kinases |
| 22 | ORF03040 | 54 | 4.3 e-37 | 5.3 | 42.4 | C | S-Adenosylmethionine synthetase (metK) |
| 23 | ORF01685 | 46 | 6.5 e-45 | 5.2 | 42.7 | C | Aminotransferase, class I (aspC4) |
| 24 | ORF04028 | 42 | 3.6 e-30 | 5.2 | 38.3 | C | Aspartate-semialdehyde dehydrogenase (asd) |
| 25 | ORF03996 | 54 | 1.3 e-37 | 5.8 | 36.7 | C | Acetate kinase (ackA) |
| 26 | ORF05210 | 26 | 5.1 e-20 | 4.9 | 28.9 | C | 3-Deoxy-d-manno-octulosonate cytidylyltransferase (kdsB) |
| 27 | ORF04793 | 58 | 2.3 e-35 | 5.5 | 38.9 | P | Periplasmic spermidine/putrescine-binding protein (potD-1) |
| 28 | ORF00568 | 52 | 3.8 e-42 | 5.2 | 38.1 | C | Dihydroxyacetone kinase family protein (b1200) |
Identification by mass spectrometry in a database of coding genes (http://www.tigr.org). Spots 1a and 1b were also identified by N-terminal sequence analysis.
Extent to which the combined peptide masses cover the indicated sequence. Low coverage of proteins 3 and 12 was due to problems with mass spectrometry calibration and N-terminal truncation, respectively.
Significance of the match between experimental and theoretical peptide masses as expressed by the probability that the match is due to random chance.
pI is the average theoretical value calculated with programs found at http://www.embl-heidelberg.de/cgi/pi-wrapper.pl and http://www.biologie.uni-freiburg.de/data/PcIP.html.
Mr is the theoretical molecular mass derived from the genome sequence. *, disagreement with Mr observed in Fig. 3.
Most likely location—periplasmic (P), cytoplasmic (C), or in the membrane (M)—based on evaluation of the amino acid sequence.
Annotated function.
FIG. 2.
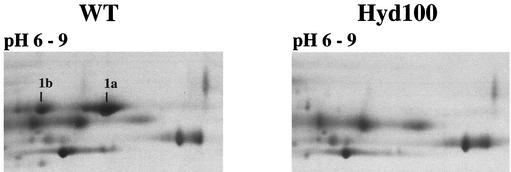
Silver-stained 2DE gels of TE extracts from D. vulgaris wild-type (WT) and hyd mutant (Hyd100) strains. Proteins were separated by isoelectric focusing with IPGs of 6 to 9. Proteins 1a and 1b were identified by MALDI-TOF-MS and Edman degradation. Only a portion of each gel is shown.
Despite the significant physiological differences between wild-type and hyd mutant D. vulgaris (30), it appeared that the proteomes of WP-LS-grown cells were quite similar (Fig. 1). Significant differences were only seen in spot 1, resolved in spots 1a and 1b when a pH gradient of 6 to 9 was used (Fig. 2). These were seen in the wild type but not in the hyd mutant, whereas spots 2 and 3 were more prominent in the hyd mutant than in the wild-type proteome. Because of its similar molecular mass, spot 1 was thought to represent the 46-kDa α-subunit of Fe-only hydrogenase (HydA), which is absent from the TE extract of the hyd mutant as judged by immunoblotting (18). However, MALDI-TOF analysis of trypsin digests indicated that spots 1a and 1b both represent ORF2977 Adh, not HydA. This was confirmed by Edman degradation analysis, which indicated AVQEQVYGFFIPR as the N terminus for both spots, matching that predicted for ORF2977 Adh. The presence of two spots with the same Mr, but distinct pIs, indicates posttranslational modification of ORF2977 Adh. HydA was not identified either on 2DE gels with different IPGs or with protein blotting of a wide-pH-range gel, indicating its loss during isoelectric focusing. Proteins 2 and 3, which appeared more abundant in the hyd mutant than in the wild type, did not have a function associated with hydrogen metabolism (Table 2). Both were more abundant in the wild-type strain during the stationary phase (not shown). The more sluggish growth of the hyd mutant on WP-LS medium (30) may have caused higher abundance of these proteins to occur earlier. In contrast, the absence of spots 1 in the hyd mutant was a constant feature. Thus the main difference in the two proteomes is a near absence of ORF2977 Adh in the hyd mutant, as compared to the wild type.
Gene arrays.
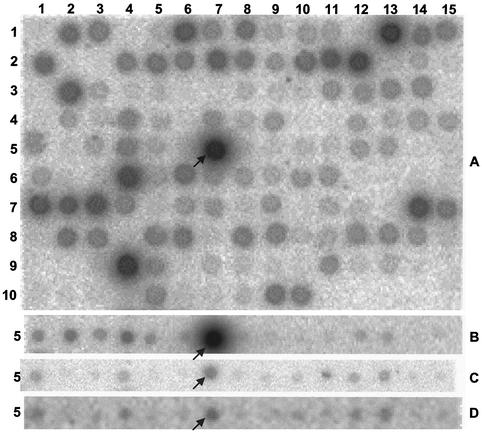
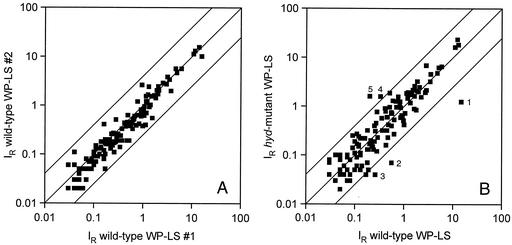
The pattern resulting from hybridization of labeled cDNA prepared from WP-LS grown wild-type D. vulgaris RNA to a macroarray is shown in Fig. 3A. IR values for all array results are listed at http://www.ucalgary.ca/SC/BI/divisions/cmmb/voordouw2.html. The reproducibility of macroarray hybridizations for RNA samples extracted from two separate cultures of the wild type in WP-LS is shown in Fig. 4A. All data points fall within a corridor that represents a fourfold change in RNA expression. Hence, gene expression changes within this fourfold corridor may not be significant. Under these conditions, the most highly expressed genes encode proteins involved in sulfate reduction (sat, apsBA, dsrABD, dsrC, and ppaC), lactate oxidation (por and ack), ethanol oxidation (adh), electron transport in the periplasm (cyc) and across the cytoplasmic membrane (thcE), ATP synthesis (atpA and atpD), and oxygen resistance (sor and rub) (Table 3). Interestingly, ORF2977 adh was the most highly expressed of the set of 145 genes represented on the macroarray. Expression of ORF2977 adh was further increased in wild-type cells grown with ethanol, but significantly decreased in the hyd mutant (Fig. 3B and C). A comparison of array data for the hyd mutant and wild type is shown in Fig. 4B. Spots outside the fourfold corridor were ORF2977 adh, ORF2976 and ORF2967, putative H2-heterodisulfide reductase complex subunits (Fig. 4B, spots 1 to 3, downregulated), as well as ORF2128-2130 hynAB NiFe hydrogenase and an ORF3201-encoded hybrid cluster protein of unknown function (Fig. 4B, spots 4 and 5, upregulated). The ORF2976 and ORF2967 putative H2-Hdr complex was also downregulated in the adh mutant in addition to ORF3423 flavodoxin, whereas ORF2437 carbon monoxide dehydrogenase was upregulated (data not shown). In the hyd mutant, hydAB gave no significant hybridization signal, in agreement with the fact that the hydAB genes were completely deleted (18). There was a residual adh message signal in the adh mutant (Fig. 3D). Because only a portion of the adh gene was deleted, this likely represented the 5′ end of the adh gene. 2DE analysis of the adh mutant showed the absence of ORF2977 Adh as in Fig. 2 (data not shown).
FIG. 3.
Hybridization of D. vulgaris macroarrays with labeled cDNA synthesized in vitro. (A) RNA extracted from wild-type cells grown in WP-LS medium. The entire macroarray is shown (145 spots, 15 columns, 10 rows). (B) RNA extracted from wild-type cells grown in WP medium with ethanol as the sole electron donor. Only row 5 of the array is shown. (C) RNA extracted from hyd mutant cells grown in WP-LS medium. Only row 5 of the array is shown. (D) RNA extracted from adh mutant cells grown in WP-LS medium. Only row 5 of the array is shown. The location of ORF2977 adh is indicated by an arrow. The array positions of other highly expressed genes are indicated in Table 3.
FIG. 4.
Plots of relative hybridization intensities (IR) for macroarray hybridizations of wild-type and hyd mutant D. vulgaris strains. The upper and lower lines represent a fourfold change in gene expression from the central line. (A) Comparison of RNA extracted from two separate cultures of the wild type in WP-LS medium. The data are means of two array hybridizations. (B) Comparison of gene expression for the wild type and hyd mutant grown in WP-LS. The data are means of four array hybridizations. Genes outside the fourfold expression corridor are as follows: 1, ORF2977 adh; 2, ORF2976 putative H2:Hdr reductase complex subunit gene; 3, ORF2967 putative H2:Hdr complex subunit gene; 4, ORF2128-2130 NiFe hydrogenase genes; and 5, ORF3201 hybrid cluster protein gene.
TABLE 3.
Relative expression level (IR) of the most highly expressed genes in a set of 145 represented on the macroarraya
| ORFb | Gene | Description | IRc | Positiond |
|---|---|---|---|---|
| 2977 | adh | Alcohol dehydrogenase | 14.8 ± 0.9 | 5, 7 |
| 3581 | dsrC | Dissimilatory sulfite reductase | 12.9 ± 2.8 | 6, 4 |
| 4250 | cyc | Cytochrome c3 | 12.3 ± 0.8 | 7, 14 |
| 336, 338 | apsBA | APS reductase | 11.0 ± 0.4 | 1, 13 |
| 5313, 5314, 5316 | dsrABD | Dissimilatory sulfite reductase | 5.9 ± 0.5 | 9, 4 |
| 219 | atpD | ATP synthase F1 β-subunit | 5.1 ± 0.5 | 1, 6 |
| 1081 | sat | Sulfate adenylyltransferase | 4.8 ± 0.2 | 2, 12 |
| 3986 | por | Pyruvate:ferredoxin oxidoreductase | 3.5 ± 0.3 | 7, 1 |
| 5316 | dsrD | Dissimilatory sulfite reductase | 3.4 ± 0.2 | 9, 5 |
| 4273 | rub | Rubredoxin | 2.9 ± 0.1 | 8, 1 |
| 4271 | sor | Superoxide reductase | 2.8 ± 0.9 | 7, 15 |
| 1642 | ppaC | Pyrophosphatase | 2.7 ± 0.2 | 3, 2 |
| 3996 | ack | Acetate kinase | 2.2 ± 0.2 | 7, 3 |
| 223 | atpA | ATP Synthase F1 α-subunit | 2.1 ± 0.1 | 1, 8 |
| 1072 | thcE | Triheme cytochrome c operon membrane protein | 2.0 ± 0.1 | 2, 11 |
Macroarrays were hybridized with cDNA reverse transcribed from RNA extracted from wild-type D. vulgaris grown in WP-LS medium.
ORF numbers in a preliminary annotation of the genome sequence (http://www.tigr.org).
Calculated as indicated in the text. The mean of four determinations and the average deviation from the mean are shown.
Position (row number, column number) of gene on the macroarray depicted in Fig. 3A.
Expression of the adh gene relative to that of the wild type grown in WP-LS is summarized in Table 4. Expression was found to be even higher for the wild type grown with ethanol (see also Fig. 3B) and to be low in the hyd mutant (Fig. 3C and Table 4). Although in wild-type cells, expression of adh depended somewhat on the choice of electron donor, it was one of the most highly expressed genes irrespective of whether lactate, pyruvate, formate, ethanol, or hydrogen was used (Table 4). The array data thus confirm the results of proteomic analysis in that adh expression is low in the hyd mutant.
TABLE 4.
Relative expression level (IR) of ORF2977 adh as determined by macroarray hybridization
| Strain | Electron donor | IR of adha |
|---|---|---|
| Wild type | Lactate | 14.8 ± 0.9 |
| Pyruvate | 14.9 ± 5.0 | |
| Ethanol | 44.4 ± 5.5 | |
| Formate | 30.5 ± 5.2 | |
| Hydrogen | 34.6 ± 1.2 | |
| hyd mutant | Lactate | 1.24 ± 0.3 |
Calculated as indicated in the text. The mean of four determinations and the average deviation from the mean are shown.
Physiological analysis of the adh mutant.
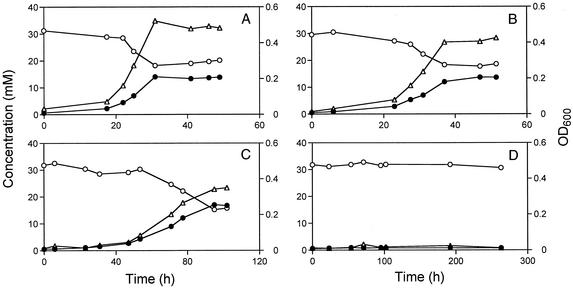
Despite the high level of expression of the adh gene, it was relatively easy to make a mutant, indicating that ORF2977 Adh is not essential to D. vulgaris when cells are grown with either lactate or hydrogen as the electron donor for sulfate reduction. Growth in WP-LS medium (lactate as the electron donor) is compared in Fig. 5A and B. Although the adh mutant appeared to grow well in this medium, it reached a lower final cell density: OD600 = 0.420 ± 0.015 and 0.475 ± 0.019 for the adh mutant and wild-type strains, respectively (mean of five determinations). This indicates that, although not essential, ORF2977 Adh contributes to the bioenergetics of D. vulgaris during growth on lactate-containing medium. The hyd mutant also grew to lower final cell density compared to the wild type (OD600 = 0.435 ± 0.032) in WP-LS medium.
FIG. 5.
Comparison of the growth physiologies of D. vulgaris wild-type and adh mutant strains. The cell density (▵; OD600, right ordinate), millimolar sulfate concentration (○; left ordinate), and millimolar sulfide concentration (•; left ordinate) are shown as a function of time. (A to D) Strains and electron donors. (A) Wild type with lactate. (B) adh mutant with lactate. (C) Wild type with ethanol. (D) adh mutant with ethanol.
When using ethanol as a carbon source and electron donor, no sulfate reduction and associated growth was seen for the adh mutant, whereas the wild type reached a final cell density (OD600) of 0.335 ± 0.012, as shown in Fig. 5C and D. The same result was obtained for cultures with 5% (vol/vol) H2 as the electron donor and ethanol (and CO2) as the carbon source (results not shown). Hence, despite the fact that the genome sequence indicates the presence of multiple ORFs that are annotated as “alcohol dehydrogenase,” ORF2977 Adh appears to be the only enzyme catalyzing ethanol oxidation. The hyd mutant reached a final cell density (OD600) of 0.248 ± 0.013 with ethanol as the carbon source and electron donor. Cultures of the hyd mutant transferred from WP-LS medium to WP medium with ethanol as the carbon source and electron donor had a lag phase of 200 to 250 h. Ethanol-grown cultures of the hyd mutant transferred to ethanol-containing media started to grow within 24 h.
DISCUSSION
The power of proteomics, functional genomics, or other molecular biology techniques for identification of proteins that function in energy metabolism of anaerobes has already been demonstrated. Schut et al. (23) identified genes involved in hydrogen metabolism and sulfur reduction from hybridization to a limited Pyrococcus furiosus microarray. Tersteegen and Hedderich (27) have shown by reverse transcription-PCR that two membrane-bound, energy-converting hydrogenases are induced in Methanobacterium thermoautotrophicum under hydrogen limitation, suggesting a role in energy conservation by some form of hydrogen cycling.
Our experiments point to an important role of ORF2977 Adh in the energy metabolism of D. vulgaris, which we wish to understand. Hensgens et al. (10) purified a close homolog of ORF2977 Adh from D. gigas cells grown on media containing ethanol and sulfate. The enzyme had a molecular mass of 43 kDa by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (similar to ORF2977 Adh), assembled into decamers as judged by electron microscopy, and had a very similar N terminus, AVREQVYGFFIPSVTLIGIGASKEI, compared to that for ORF2977 Adh, AVQEQVYGFFIPRVTLIGIGASKAI. Hensgens et al. (10) found this enzyme to be active towards ethanol, isopropanol, and a variety of other alcohols at lower activity. Because it was isolated from cells grown on ethanol, the authors concluded that catabolic oxidation of ethanol must be its primary function in D. gigas. We have confirmed here by mutation that ORF2977 Adh is the sole enzyme catalyzing ethanol oxidation in D. vulgaris (Fig. 5). Interestingly, it also functions during growth with lactate, pyruvate, formate, or hydrogen as an electron donor for sulfate reduction, because it is prominently expressed under all of these conditions (Table 4). The enzyme likely reoxidizes ethanol formed during metabolism of these different electron donors. For instance, D. vulgaris incompletely oxidizes lactate to acetate and CO2 (21), and some of the acetate is converted to ethanol (28). The enzyme or enzymes forming the ethanol are currently unknown. With excess sulfate, ORF2977 Adh likely reoxidizes the ethanol formed later in the growth curve. Growth of D. vulgaris with hydrogen as the sole electron donor requires the presence of acetate as a carbon source. The need for ORF2977 Adh under these conditions can be understood when assuming that some of the acetate is transiently converted into ethanol.
Since ORF2977 Adh is a cytoplasmic enzyme, growth on ethanol as the sole electron donor for sulfate reduction requires some form of cycling of hydrogen (H2) or hydrogen equivalents ([H]). The need for this is even more pressing than for growth on lactate (30), because oxidation of ethanol to acetate, the expected end product in D. vulgaris, does not yield ATP by substrate-level phosphorylation. Oxidation of 2 ethanol to 2 acetate per sulfate reduced, through the action of ORF2977 Adh and aldehyde dehydrogenase, yields 8 [H]. The proton gradient resulting from oxidation of 8 [H] by periplasmic hydrogenases can give sufficient ATP for the activation of sulfate prior to reduction (2 ATP per sulfate) and other metabolic processes (maximally 0.7 ATP per sulfate, assuming formation of 2.7 ATP per 8 H+ by ATP synthase). The proteomic and functional genomic data collected in the present study shed some light on the mechanism by which the [H] liberated in ethanol oxidation are converted into a proton gradient.
Proteomic analysis indicated that deletion of the hydAB genes, encoding Fe-only hydrogenase (HydAB), significantly lowers the cellular content of ORF2977 Adh (Fig. 2). This result was confirmed by macroarray hybridizations in which transcription patterns of wild-type and hyd mutant D. vulgaris strains were compared (Fig. 4B, spot 1). Figure 4B also indicated downregulation of ORF2976 and ORF2967 putative H2:Hdr complex (Fig. 4B, spots 2 and 3) and upregulation of ORF2128-2130 hynAB (Fig. 4B, spot 4) encoding NiFe-hydrogenase, possibly to compensate for the loss of HydAB. The ORF2976 and ORF2967 genes belong to a six-gene operon located 230 bp downstream of ORF2977 adh. The adh gene is not believed to be part of this operon, because the intergenic region is rather large and contains a putative hairpin loop. The operon consists of ORFs 2976, 2975, 2972, 2971, 2969, and 2967, encoding proteins predicted to be cytoplasmic. A BLAST search (1) showed that the closest homologs to ORFs 2976, 2975, and 2972 are subunits HdrC, HdrB, and HdrA of heterodisulfide reductase from Carboxydothermus hydrogenovorans or M. thermoautotrophicum (9). The closest homologs to ORF2971, -2969, and -2967 are the δ-, α-, and γ-subunits of (sulf)hydrogenase from Pyrococcus. The operon may thus encode a cytoplasmic heterodisulfide reductase-hydrogenase. Corresponding enzymes in methanogens are referred to as H2-heterodisulfide oxidoreductase (H2:Hdr). Macroarray analysis of the adh mutant also indicated downregulation of the genes encoding the ORF2976-2967 putative H2:Hdr complex (data not shown). The close physical relationship and coexpression of ORF2977 adh and the operon for putative H2:Hdr complex strongly suggest that [H] originating from ethanol oxidation are processed by H2:Hdr.
In methanogens, a heterodisulfide of coenzymes M and B (CoM-S-S-CoB) and methane are formed in the final step of methanogenesis. Membrane-bound H2:Hdr regenerates CoM-SH and CoB-SH by reduction of the heterodisulfide with hydrogen (9). This reaction is energy conserving and contributes to the proton motive force. The H2:Hdr from M. thermoautotrophicum can be dissociated into a hydrogenase and a heterodisulfide reductase subcomplex, each consisting of three subunits (9). In addition to methanogens, heterodisulfide reductase homologs are found in the sulfate-reducing bacteria D. vulgaris and D. desulfuricans, in the sulfate-reducing archaeon Archaeoglobus fulgidus (13), in the metal-reducing bacterium Geobacter metallireducens, in the green sulfur bacterium Chlorobium tepidum, and in the CO-reducing bacterium C. hydrogenoformans, among others. The role of H2:Hdr in nonmethanogens is unclear. In D. vulgaris, the ORF2976-2967 putative H2:Hdr complex likely has a role in ethanol oxidation. A possible mechanism is that ORF2977 Adh transfers [H] from ethanol oxidation to a disulfide in H2:Hdr, forming reduced enzyme (HS-E-SH), which is then converted to the oxidized form and H2. In the hydrogen cycling hypothesis, this cytoplasmically generated H2 diffuses to the periplasm, where it is oxidized by periplasmic hydrogenases (17). The latter model may be too simplistic, because it fails to explain why of several periplasmic hydrogenases, HydAB is specifically required for ethanol oxidation. If H2 were to diffuse freely, it could equally well be oxidized by any of three periplasmic NiFe-hydrogenases present in D. vulgaris. The specific requirement for HydAB suggests that the [H] formed in cytoplasmic ethanol oxidation by ORF2977 Adh are somehow transmitted through a specific chain involving cytoplasmic ORF2976-2967 H2:Hdr and as yet unknown transmembrane components. Chemically the chain may consist of a number of linked H2-disulfide oxidation-reduction events that eventually delivers 2 [H] as H2 in the active site of periplasmic HydAB for final oxidation to protons. Oxidation of [H] originating from ethanol oxidation is not the only function of HydAB, as shown by physiological studies (18, 30) and by the fact that the HydAB content in the adh mutant is similar to that in the wild type as determined by immunoblotting (results not shown).
In summary, the proteomics, functional genomics, and gene mutation results obtained here further illustrate the complex transmembrane cycling mechanisms that conserve the free energy liberated when organic compounds are oxidized with sulfate in the sulfate-reducing bacteria.
Acknowledgments
This work was supported by a Research Grant from the Natural Science and Engineering Research Council of Canada to G.V. and by the Max-Planck Society of Germany. G.V. acknowledges the support of a fellowship from the Hanse Wissenschaftskolleg in Bremen, Germany, and of a visiting professorship of the CNRS in Marseille, France. A database of the D. vulgaris Hildenborough genome was searched at the web site of The Institute for Genomic Research at http://www.tigr.org. Sequencing of this genome is financially supported by the U.S. Department of Energy.
We thank Richard Pon for synthesizing the oligonucleotides, Janine Wildschut and Daniela Lange for technical assistance, Thomas Halder for MS analysis, and Fritz Widdel for his interest in this work and many discussions.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipmann. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 3.Clauser, K. R., P. Baker, and A. L. Burlingame. 1999. Role of accurate mass measurement (±10 ppm) in protein identification strategies employing MS or MS/MS and database searching. Anal. Chem. 71:2871-2882. [DOI] [PubMed] [Google Scholar]
- 4.Corthals, G. L., S. P. Gygi, R. Aebersold, and S. D. Patterson. 2000. Identification of proteins by mass spectrometry, p. 197-231. In T. Rabilloud (ed.), Proteome research: two-dimensional gel electrophoresis and identification methods. Springer, Berlin, Germany.
- 5.Dolla, A., B. K. J. Pohorelic, J. K. Voordouw, and G. Voordouw. 2000. Deletion of the hmc-operon of Desulfovibrio vulgaris subsp. vulgaris Hildenborough hampers hydrogen metabolism and low-redox potential niche establishment. Arch. Microbiol. 174:143-151. [DOI] [PubMed] [Google Scholar]
- 6.Fournier, M., Y. Zhang, J. D. Wildschut, A. Dolla, J. K. Voordouw, D. C. Schriemer, and G. Voordouw. 2003. Function of oxygen resistance proteins in the anaerobic sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough. J. Bacteriol. 185:71-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fu, R., and G. Voordouw. 1997. Targeted gene replacement mutagenesis of dcrA, encoding an oxygen sensor of the sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough. Microbiology 143:1815-1826. [DOI] [PubMed] [Google Scholar]
- 8.Görg, A., C. Obermaier, G. Boguth, A. Harder, B. Scheibe, R. Wildgruber, and W. Weiss. 2000. The current state of two-dimensional electrophoresis with immobilized pH gradients. Electrophoresis 21:1037-1053. [DOI] [PubMed] [Google Scholar]
- 9.Hedderich, R., J. Koch, D. Linder, and R. K. Thauer. 1994. The heterodisulfide reductase from Methanobacterium thermoautotrophicum contains sequence motifs characteristic of pyridine-nucleotide-dependent thoredoxin reductases. Eur. J. Biochem. 225:253-261. [DOI] [PubMed] [Google Scholar]
- 10.Hensgens, C. M. H., J. Vonck, J. van Beeumen, E. F. J. van Bruggen, and T. A. Hansen. 1993. Purification and characterization of an oxygen-labile NAD-dependent alcohol dehydrogenase from Desulfovibrio gigas. J. Bacteriol. 175:2859-2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hunkapillar, M. W., R. M. Hewick, J. M. Dreyer, and L. E. Hood. 1983. High-sensitivity sequencing with a gas-phase sequenator. Methods Enzymol. 91:399-413. [DOI] [PubMed] [Google Scholar]
- 12.Jenö, P., T. Mini, S. Moes, E. Hintermann, and M. Horst. 1995. Internal sequences from proteins digested in polyacrylamide gels. Anal. Biochem. 224:75-82. [DOI] [PubMed] [Google Scholar]
- 13.Mander, G. J., E. C. Duin, D. Linder, K. O. Stetter, and R. Hedderich. 2002. Purification and characterization of a membrane-bound enzyme complex from the sulfate-reducing archaeon Archaeoglobus fulgidus related to heterodisulfide reductase from methanogenic archaea. Eur. J. Biochem. 269:1895-1904. [DOI] [PubMed] [Google Scholar]
- 14.Marmur, J. 1961. A procedure for the isolation of deoxyribonucleic acid from micro-organisms. J. Mol. Biol. 3:208-218. [Google Scholar]
- 15.Nemati, M., T. J. Mazutinec, G. E. Jenneman, and G. Voordouw. 2001. Control of biogenic H2S production with nitrite and molybdate. J. Ind. Microbiol. Biotechnol. 26:350-355. [DOI] [PubMed] [Google Scholar]
- 16.Neuhoff, V., R. Stamm, I. Pardowitz, N. Arold, W. Ehrhardt, and D. Taube. 1990. Essential problems in quantification of proteins following colloidal staining with Coomassie brilliant blue dyes in polyacrylamide gels, and their solution. Electrophoresis 11:101-117. [DOI] [PubMed] [Google Scholar]
- 17.Odom, J. M., and H. D. Peck, Jr. 1981. Hydrogen cycling as a general mechanism for energy coupling in the sulfate-reducing bacteria Desulfovibrio sp. FEMS Microbiol. Lett. 12:47-50. [Google Scholar]
- 18.Pohorelic, B. K. J., J. K. Voordouw, E. Lojou, A. Dolla, J. Harder, and G. Voordouw. 2002. Effects of deletion of genes encoding Fe-only hydrogenase of Desulfovibrio vulgaris Hildenborough on hydrogen and lactate metabolism. J. Bacteriol. 184:679-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Postgate, J. R. 1984. The sulphate-reducing bacteria, 2nd ed. Cambridge University Press, Cambridge, United Kingdom.
- 20.Rabus, R., D. Gade, R. Helbig, M. Bauer, F. O. Glöckner, M. Kube, H. Schlesner, R. Reinhardt, and R. Amann. 2002. Analysis of N-acetylglucosamine metabolism in the marine bacterium Pirellula sp. strain 1 by a proteomic approach. Proteomics 2:649-655. [DOI] [PubMed] [Google Scholar]
- 21.Rabus, R., T. Hansen, and F. Widdel. 2001. Dissimilatory sulfate-and sulfur-reducing prokaryotes. In M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes: an evolving electronic resource for the microbiological community. [Online.] Springer Science Online, Heidelberg, Germany. http://www.tigr.org.
- 22.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 23.Schut, G. J., J. Zhou, and M. W. W. Adams. 2001. DNA microarray analysis of the hyperthermophilic archaeon Pyrococcus furiosus: evidence for a new type of sulfur-reducing enzyme complex. J. Bacteriol. 183:7027-7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schweizer, H. P. 1992. Allelic exchange in Pseudomonas aeruginosa using novel ColE1-type vectors and a family of cassettes containing a portable oriT and the counter-selectable Bacillus subtilis sacB marker. Mol. Microbiol. 6:1195-1204. [DOI] [PubMed] [Google Scholar]
- 25.Simon, R., U. Priefer, and A. Pühler. 1983. A broad-host-range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 26.Telang, A. J., S. Ebert, J. M. Foght, D. W. S. Westlake, G. E. Jenneman, D. Gevertz, and G. Voordouw. 1997. Effect of nitrate injection on the microbial community in an oil field as monitored by reverse sample genome probing. Appl. Environ. Microbiol. 63:1785-1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tersteegen, A., and R. Hedderich. 1999. Methanobacterium thermoautotrophicum encodes two multisubunit membrane-bound [NiFe] hydrogenases. Eur. J. Biochem. 264:930-943. [DOI] [PubMed] [Google Scholar]
- 28.Traore, A. S., C. E. Hatchikian, J. P. Belaich, and J. Le Gall. 1981. Microcalorimetric studies of the growth of sulfate-reducing bacteria: energetics of Desulfovibrio vulgaris growth. J. Bacteriol. 145:191-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van der Westen, H. M., S. G. Mayhhew, and C. Veeger. 1978. Separation of hydrogenase from intact cells of Desulfovibrio vulgaris. FEBS Lett. 86:122-126. [DOI] [PubMed] [Google Scholar]
- 30.Voordouw, G. 2002. Carbon monoxide cycling by Desulfovibrio vulgaris Hildenborough. J. Bacteriol. 184:5903-5911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Voordouw, G., J. D. Strang, and F. R. Wilson. 1989. Organization of genes encoding [Fe] hydrogenase in Desulfovibrio vulgaris subsp. oxamicus Monticello. J. Bacteriol. 171:3881-3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Voordouw, G., J. K. Voordouw, T. R. Jack, J. Foght, P. M. Fedorak, and D. W. S. Westlake. 1992. Identification of distinct communities of sulfate-reducing bacteria in oil fields by reverse sample genome probing. Appl. Environ. Microbiol. 58:3542-3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.White, D. 2000. The physiology and biochemistry of prokaryotes, 2nd ed., p. 318. Oxford University Press, New York, N.Y.
- 34.Widdel, F., and F. Bak. 1992. Gram-negative mesophilic sulfate-reducing bacteria, p. 3352-3378. In A. Balows, H. G. Truper, M. Dworkin, W. Harder, and K. H. Schleifer (ed.), The prokaryotes, 2nd ed., vol. 4. Springer-Verlag, New York, N.Y.