Abstract
The mre genes of Escherichia coli and Bacillus subtilis are cell shape determination genes. Mutants affected in mre function are spheres instead of the normal rods. Although the mre determinants are not required for viability in E. coli, the mreB determinant is an essential gene in B. subtilis. Conflicting results have been reported as to whether the two membrane-associated proteins MreC and MreD are essential proteins. Furthermore, although the MreB protein has been studied in some detail, the roles of the MreC and MreD proteins in cell shape determination are unknown. We constructed a strain of B. subtilis in which expression of the mreC determinant is dependent upon the addition of isopropyl-β-d-thiogalactopyranoside to the culture medium. Utilizing this conditional strain, it was shown that mreC is an essential gene in B. subtilis. Furthermore, it was shown that cells lacking sufficient quantities of MreC undergo morphological changes, namely, swelling and twisting of the cells, which is followed by cell lysis. Electron microscopy was utilized to demonstrate that a polymeric material accumulated at one side of the division septum of the cells and that the presence of this material correlated with the bending of the cell. The best explanation for the results is that the MreC protein is involved in the control of septal versus long-axis peptidoglycan synthesis, that cells lacking MreC perform aberrant septal peptidoglycan synthesis, and that lysis results from a deficiency in long-axis peptidoglycan synthesis.
Bacteria have characteristic shapes, which are important taxonomic features. Most bacteria possess a fixed shape, although members of the genus Arthrobacter transition from rods to cocci depending on growth conditions (12). The bacterial cell wall is thought to be the major determinant of cell shape, as the purified murein sacculus retains the characteristic morphology of the cell, and specific changes in the chemical composition of the peptidoglycan have been noted in shape-altered mutants (25). However, shape determination is a complicated process, which involves not only the topology of peptidoglycan biosynthesis but also contributions of other components of the cell envelope as well as involvement of the cell division machinery. A mutation in the tagF (rodC) determinant of Bacillus subtilis gives rise to a shift from rod-shaped to spherical cells, indicating a contribution by teichoic acid to the determination of cell morphology (4, 16, 23).
A model for bacterial shape regulation, the two-competing-sites model, which relates cell shape to a balance between cell wall synthesis along the long axis of a rod-shaped cell and septum-specific peptidoglycan synthesis, has been proposed (21, 24). This model proposes that shape determination in bacterial rods depends on the activity of two peptidoglycan synthesis reactions (sites) which are mutually exclusive. Consequently, the lateral wall is not extended during the septum formation step of cell division, and septal peptidoglycan synthesis does not occur during lateral wall elongation. The shape of the cell is determined by the balance between the competing reactions. The normal balance produces rods, an abnormal prevalence for lateral wall elongation yields filaments, and a prevalence for the site for septum formation leads to the formation of coccobacilli or cocci. Bacteria which have the characteristic coccal shape fall into two categories. Some possess only the site for septum formation and can only exist as cocci, whereas others carry both peptidoglycan biosynthetic sites and can transition to rods when the septum-specific site is blocked by mutation or antibiotic treatment (21).
Mutations in the genome of Escherichia coli which result in altered cell morphology have been identified, but the precise biochemical functions of the affected genes have not been determined. The mre operon of E. coli was identified as a locus associated with cell shape determination and sensitivity to amdinocillin (11, 27-30). The operon consists of five genes, mreBCD, orfE, and cafA. A mutation in mreB or deletion of mreBCD produces a morphological shift in the cell population, resulting in spherical cells. The MreB protein shows sequence similarity to members of the Hsp70 superfamily, as well as to the FtsA cell division protein (6, 14). Inactivation of mreB is associated with an increased activity of the FtsI (penicillin binding protein 3) protein, which is responsible for septal peptidoglycan synthesis (30). It was proposed that this increase leads to hyperseptation activity and spherical cell formation. Introduction of the mre genes on a multicopy plasmid led to a reduction in FtsI activity and an associated filamentation phenotype, presumably due to an increased production of MreB. On the basis of these observations, it was postulated that MreB is a negative regulator of FtsI (30). The roles of the MreC, MreD, and OrfE proteins in shape determination remain undefined.
The MreB homolog of B. subtilis is contained within the divIVB minicell operon (20, 26). This operon contains the mreBCD determinants as well as the minCD minicell-associated genes. A mutant allele of mreD, rodB1, induces the formation of spherical cells. Thus, the B. subtilis mre genes are also associated with cell shape determination. However, it was reported that the B. subtilis mre genes appear to be essential for cell viability, unlike their E. coli counterparts (20, 26). Two additional determinants whose products have substantial sequence identity to MreB have been identified on the B. subtilis chromosome. The mbl determinant appears to play a role in cell morphology, but inactivation of this nonessential determinant does not result in spherical cell formation (1). The mreBH determinant, which maps near kinC on the B. subtilis chromosome, has not been characterized (GenBank accession number D37799). The B. subtilis Mre proteins are even less well studied than their E. coli counterparts. Their sequence similarity to the E. coli Mre proteins, and the phenotype of the rodB1 allele, suggests that their function also involves an interaction with the septum-specific peptidoglycan biosynthetic enzyme, namely, Pbp2B (20, 26, 32).
Introduction of the mreC and mreD genes from Bacillus stearothermophilus into B. subtilis resulted in an overexpression of protease (18). This effect on protease expression required the presence of both mre determinants; no effect was seen with either determinant introduced alone into this host. It has not been determined whether this effect on protease expression is a direct or indirect effect of the production of the B. stearothermophilus MreCD proteins in B. subtilis. The MreC protein has been shown to localize to the division septum in dividing cells of B. subtilis (19).
Jones and coworkers indicated that the MreB and Mbl proteins of B. subtilis are filamentous helical structures which may have a cytoskeleton-like role in bacterial shape determination (17). They proposed a model in which the MreB protein is concerned with cell width control, whereas Mbl is more important in maintaining the linearity of the longitudinal axis of the cell. Their studies confirmed earlier suggestions that the mreB determinant is essential for cell viability but that the mreC and mreD determinants could be inactivated with no loss of viability and only “mild phenotypic consequences.” The Streptomyces coelicolor mreB determinant was also shown to be essential for viability, although the mreC gene when inactivated by gene disruption resulted in mutants which were viable but showed significant growth retardation in comparison to the wild type (7). To better understand the biological role of the MreC protein in B. subtilis and to reconcile the conflicting conclusions regarding the essential nature of this determinant, we created a strain of B. subtilis which conditionally expresses the MreC protein and examined the effect of depletion of this protein on growth and morphology of the cell.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Media and growth conditions for E. coli and B. subtilis were as described previously (8, 26). Phleomycin (zeocin) was utilized at 10 and 7 μg/ml for selection with E. coli and B. subtilis, respectively; kanamycin was utilized at 20 and 5 μg/ml for selection with E. coli and B. subtilis, respectively; spectinomycin was utilized at a concentration of 100 μg/ml, and chloramphenicol was utilized at 10 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Source or reference |
|---|---|---|
| B. subtilis 168 strains | ||
| KSS1001 | trpC2 | K. Bott |
| KSS1570 | amyE::pdivIVB mreD-minCD | This study |
| KSS1571 | Ω pLEE100, amyE::pdivIVB mreD-minCD | This study |
| KSS1572 | trpC2, Ω pLEE100 | This study |
| KSS1573 | KSS1571 (pLEE504) | This study |
| MO1099 | amyE::erm | 9 |
| E. coli strains | ||
| DH5α | endA1 gyrA96 hsdR17 ΔlacU169 (φ80lacZΔM15) recA1 supE44 thi-1 | BRLc |
| M15 | StrrlacZΔM15 (pRep4 Kmr) | Qiagen |
| Plasmids | ||
| pCHA2489 | mreC ORFa PCR product cloned into pT7Blue | This study |
| pCHA2506 | NotI (in mreC, blunted)-SalI (vector site) mreC DNA fragment from pCHA2489 cloned into EcoICRI-SalI site of pQE30 | This study |
| pCR2.1 | PCR fragment cloning vector, Apr Kmr | Invitrogen |
| pDG364 | amyE locus integration vector | BGSCd (9) |
| pDG3053 | Shuttle expression plasmid utilizing the S. aureus lactose operon promoter; Apr Kanr | This study |
| pLEE50 | pUS19 with the lacI spac EcoRI-BamHI insert from pSI-1 | This study |
| pLEE100 | MreC-inducible expression integration vector | This study |
| pLEE200 | pDG364 containing pdivIVB mreC-minCD | This study |
| pLEE504 | pDG3053 containing truncated mreCb | This study |
| pQE30 | Six-histidine tag vector, Apr | Qiagen |
| pSI-1 | IPTG-inducible expression shuttle plasmid | D. Henner (2, 33) |
| pT7Blue | PCR product cloning vector, Apr | Invitrogen |
| pUS19 | pUC19 with Spr expressed in B. subtilis | 3 |
ORF, open reading frame.
Truncated mreC (contains bp 1 to 540).
BRL, Bethesda Research Laboratories.
BGSC, Bacillus Genetic Stock Center, Ohio State University.
To grow B. subtilis strain KSS1571, tryptic soy broth (TSB) (Difco) and tryptic soy agar (TSA) were used. The culture media were supplemented with spectinomycin and 2 mM isopropyl-β-d-thiogalactopyranoside (IPTG). After the cells were grown in broth overnight, they were harvested by centrifugation, washed with an equal volume of phosphate-buffered saline, diluted 100-fold in fresh broth, and grown for 6 h with 100 μg of spectinomycin per ml and various amounts of IPTG. All incubations were performed at 37°C with shaking at 200 rpm, and cell density was monitored spectrophotometrically at 500 nm. At the end of the 6-h cultivation period, absorbance values were determined. The cells were then harvested by centrifugation, and the cells were resuspended to identical cell densities. One-milliliter samples were collected for electron microscopy observation or methylene blue staining and conventional microscopy. The remainder of the culture was utilized for Western blot analyses and muramic acid assays.
To induce a switch from teichoic acid to teichuronic acid synthesis, a low-phosphate medium was used as described previously (31).
General methods.
Competent E. coli and B. subtilis cells were prepared and transformed by the methods of Dagert and Ehrlich (10) and Erickson and Copeland (13), respectively. Plasmid DNA from E. coli was prepared by the alkaline lysate procedure of Birnboim and Doly (5). Isolation of chromosomal and plasmid DNAs from B. subtilis was performed as described previously (8, 26).
Construction of an MreC-inducible strain.
The HincII-HindIII sequence of the multiple cloning site on pUS19 (3) was removed, and the DNA fragment containing the spac promoter and the lacI coding region was excised from plasmid pSI-1 (2, 33) with EcoRI and BamHI and then ligated into the modified pUS19 plasmid cleaved with the same enzymes, generating plasmid pLEE50. A 500-bp fragment encoding the 3′ end of mreB was generated by PCR and positioned as a HindIII-SphI fragment downstream of the IPTG-inducible spac promoter of the plasmid pLEE50 to create pLEE100 (Fig. 1). This plasmid cannot replicate in B. subtilis cells, but it contains a spectinomycin resistance determinant which can be expressed in the gram-positive host. Integration of the plasmid into the B. subtilis chromosome at the mreB locus results in mreB expression from the divIVB operon promoter, but expression of mreCD is directed by spac. The distal determinants minC and minD would be driven by both the spac promoter and a weak internal promoter in the divIVB operon (26). The 3′ terminus of mreB was positioned downstream of spac, instead of the 5′ terminus of mreC. The reason for this was to avoid a duplication of part of the mreC determinant upon integration of the plasmid. It was preferable to have a 5′ untranslated leader on the mreC-containing transcript than to have an expressed truncated mreC, which would include its transmembrane domain (19).
FIG. 1.
(A) Integration of pLEE100 into the B. subtilis chromosome. As a result of the plasmid integration, the mreB determinant of the divIVA operon remains under the control of its natural promoter (pdivIVA), whereas the mreB-distal genes in the operon are now under the control of the IPTG-inducible spac promoter (pspac). (B) The mreD-minC-minD determinants, expressed from the divIVA operon promoter, were positioned within the amyE locus (9). Abbreviations: Apr, ampicillin resistance determinant; Spr, spectinomycin resistance determinant; pspac, spac promoter; pdivIVB, divIVB operon promoter.
To eliminate possible polar effects on the mreC-distal genes of the divIVB operon, a merodiploid strain of B. subtilis was constructed in which an extra copy of mreD minCD, controlled by the divIVB operon promoter, was present. The divIVB promoter was amplified by PCR (forward primer, 5′ CGGAATTCATCTATTGTCCACCCGCG 3′; reverse primer, 5′ GCTCTAGAGCAAAAATACCCTAAAGGG 3′) and cloned using the EcoRI and XbaI sites incorporated into the 5′ ends of the primers. The promoter fragment was positioned in front of the mreD-minC-minD determinants (amplified using 5′ GCTCTAGAACAGAGGAGGAAGGATCG 3′ and 5′ CGGGATCCATTAAGATCTTACTCCG 3′ as PCR primers, introducing XbaI and BamHI sites, respectively) and this was inserted into the plasmid pDG364 (9) as an EcoRI-BamHI fragment. All cloned PCR products were sequenced to ensure that the constructs were constructed correctly. The resulting plasmid, pLEE200, was transformed into B. subtilis strain MO1099 to create strain KSS1570, which is merodiploid for the mreD, minC, and minD genes. The second copy of these determinants is positioned within the amyE locus (9) (Fig. 1B).
To express a truncated form of MreC, the sequence corresponding to the ribosome binding site and the first 504 bp of the mreC open reading frame was amplified by PCR (forward primer, 5′ GAGCTCGAGTTCAATAGAAGAG 3′; reverse primer, 5′ GCATGCCAGACGTAAAATTGTTAA 3′). The PCR product was cloned as a SacI-SphI fragment immediately downstream of the Staphylococcus aureus lactose operon promoter in plasmid pDG3053 to create plasmid pLEE504. This plasmid is a shuttle plasmid with gram-positive replication functions provided by the staphylococcal plasmid pUB110 and ampicillin resistance and replication functions for E. coli provided by pUC19. Expression from this promoter is constitutive because the plasmid lacks a functional lacR repressor-encoding determinant.
Scanning electron microscopy.
B. subtilis cells were cultured to the mid-logarithmic growth phase, and 1 ml of this culture was fixed overnight at 4°C in 20 volumes of modified Karnowsky's fixative (2% paraformaldehyde, 2.5% glutaraldehyde, 1.7 mM CaCl2 in 0.1 M cacodylate buffer [pH 7.4]). After the cells were fixed, they were washed twice in 0.1 M cacodylate buffer (pH 7.4) and postfixed in 1% osmium tetroxide in 0.1 M cacodylate buffer overnight at 4°C. After the cells were washed twice in double-distilled water, they were resuspended in double-distilled water, mounted on coverslips coated with poly-l-lysine, and dehydrated in a series of alcohol solutions (30, 50, 70, 95, and 100%). Dehydrated samples were treated for 5 min with hexamethyldisilazane (Electron Microscopy Supplies, Ft. Washington, Pa.), dried, mounted, and sputter coated with gold (S150A sputter coater). Samples were viewed in a Hitachi H-300 electron microscope with a 3010 scanning image accessory.
Transmission electron microscopy.
B. subtilis cells were cultured to the mid-logarithmic growth phase. The cells were pelleted and fixed in modified Karnowsky's fixative, washed, and postfixed in 1% osmium tetroxide as described above. After three 5-min washes in cold double-distilled water, the cells were immobilized in 2% TSA and dehydrated in a series of ethanol solutions (50, 70, 70, 95, and 100%). Dehydrated specimens were stained for 60 min en bloc with 5% uranyl acetate in 70% ethanol, and the dehydration was completed by two 20-min rinses in 100% acetone, which was used as the transitional solvent. The cell blocks were then infiltrated with three increasing concentrations of resin in acetone, ending in 100% resin for periods of 2 to 12 h or 8 to 12 h. All samples were then embedded in Epon LX112 embedding medium for 18 to 20 h at 42°C and then for 24 h at 60°C. Embedded samples were trimmed and sectioned on an Ultracut E-Reichert-Jung ultramicrotome (C. Reichert Optische Werke AG, Vienna, Austria). Thin sections (approximately 80 μm thick) sputter coated with silver and gold were retrieved with copper grids and stained with uranyl acetate and lead citrate. Grids were examined and viewed with a Hitachi H-300 electron microscope.
Muramic acid assay.
The peptidoglycan content of the bacterial cells was determined by the method of Hadzija (15). B. subtilis cells were cultured in TSB to the late-logarithmic growth phase, harvested by centrifugation, and washed in TE buffer (10 mM Tris-HCl, 1 mM EDTA [pH 8.0]). The cell amounts were then adjusted to the same culture densities (A500 = 1.8). The cells were then lysed using a mini beadbeater-8 (Biospec Products, Bartlesville, Okla.). Following cell disruption, the samples were incubated for 15 min at 75°C. Samples were kept at −80°C until assayed. The cell wall samples were resuspended with 20 ml of chloroform-saturated TE buffer. DNase (0.5 ml of a 1-mg/ml solution) and RNase (1.0 ml of a 5-mg/ml solution) were added and incubated at 37°C for 30 min. Trypsin (100 μg) was added and incubated at 37°C for 6 h. The cell wall fraction was recovered by centrifugation (12,000 × g, 45 min) and washed one time with TE buffer.
Aliquots of the lysed cells were brought to a 0.5-ml volume with 1.0 M NaOH and incubated at 38°C for 30 min. Then 0.5 ml of 0.5 M H2SO4 and 5 ml of concentrated H2SO4 were added, and the samples were placed in a boiling water bath for 5 min. The samples were cooled, and then 0.05 ml of 4% (wt/vol) CuSO4 · 5H2O and 0.1 ml of 1.5% (wt/vol) p-hydroxydiphenyl (in 95% ethanol) were added. The samples were then incubated at 30°C for 30 min, and the absorbance was determined at 560 nm. A standard curve consisting of 0 to 20 μg of muramic acid (Sigma Chemical Co., St. Louis, Mo.) was utilized in the assay.
Preparation of polyclonal antiserum against MreC.
Histidine-tagged MreC protein was purified, and anti-MreC polyclonal antiserum was raised in rabbits as previously described (19).
Western blot analysis.
B. subtilis cells were cultured in TSB with or without IPTG to the mid-logarithmic growth phase. The cultures densities were determined spectrophotometrically at 500 nm, and the culture volumes were adjusted to give equivalent cell densities. Equivalent amounts of cells from the cultures were harvested by centrifugation and resuspended in 1 ml of TE buffer. The cells were lysed using a mini beadbeater-8, and the lysate was loaded onto sodium dodecyl sulfate-4 to 20% polyacrylamide gradient gels (Ready Gels; Bio-Rad). The electrophoretically resolved proteins were transferred to a nitrocellulose filter and probed with the anti-MreC antiserum. Goat anti-rabbit immunoglobulin G conjugated to alkaline phosphatase (Sigma Chemical Company) was used as the secondary antibody, and the immunoreactive proteins were detected using nitroblue tetrazolium and 5-bromo-4-chloro-3-indolylphosphate as substrates.
RESULTS
Construction of an MreC-inducible strain.
Although the MreB protein has been proposed to be a helical cytoskeletal element involved in cell length determination in B. subtilis (17), the functions of the mreC and mreD determinants have not been defined. To examine the effect of a loss of MreC production on B. subtilis, we created a strain with conditional expression of the MreC protein. This construct is pLEE100 and is depicted in Fig. 1. To eliminate possible polar effects on the mreC-distal genes of the divIVB operon, a merodiploid strain of B. subtilis, KSS1571, was constructed. In this strain, an extra copy of mreD minCD, controlled by the divIVB operon promoter, was inserted into the amyE locus in the chromosome of B. subtilis. Western blot analysis utilizing a rabbit polyclonal anti-MinC antiserum indicated that the MinC was produced at normal levels in an IPTG-independent fashion, indicating that the determinants at the amyE locus were expressed (data not shown).
B. subtilis strain KSS1571 fails to grown on TSA plates lacking the inducer of MreC expression. Only residual growth in the initial area of the streak on the plate is obtained, presumably due to intracellular carryover of IPTG from the overnight broth culture used to inoculate the plates. Therefore, this strain is a conditional mutant which is dependent on the presence of IPTG in the culture medium for growth. This result suggests that MreC is essential for B. subtilis viability.
Effect of MreC concentration on cell growth.
Because mreC appeared to be required for growth, B. subtilis KSS1571 was cultured in TSB containing different concentrations of IPTG (0, 10, 50, and 250 μM) to investigate the growth yields of this B. subtilis strain as a function of IPTG concentration. In addition, Western blots of cellular proteins were prepared, utilizing a rabbit polyclonal anti-MreC antibody. The amount of growth achieved with each concentration of IPTG correlated with the amount of MreC protein produced, as indicated by Western blot analysis (Fig. 2). Low levels of MreC observed in the absence of IPTG induction may result from either leaky uninduced expression from the spac promoter or carryover of IPTG from the inoculum culture. The growth level, and the amount of MreC present in cells, is higher in the wild-type strain of B. subtilis than that achieved with strain KSS1571 in the presence of 250 μM IPTG. Increasing the IPTG concentration above 250 μM did not noticeably improve the level of growth or MreC production, and it did not eliminate the changes in cell morphology described below (data not shown). The dependence of cell growth on MreC production supports the conclusion that the MreC protein is an essential protein in B. subtilis. Integration of pLEE100 into B. subtilis lacking the extra copies of the mreD minCD determinants (KSS1572) gives the same growth profile as for KSS1571, indicating that any increased expression of the divIVB-distal determinants were not responsible for the observed growth deficit.
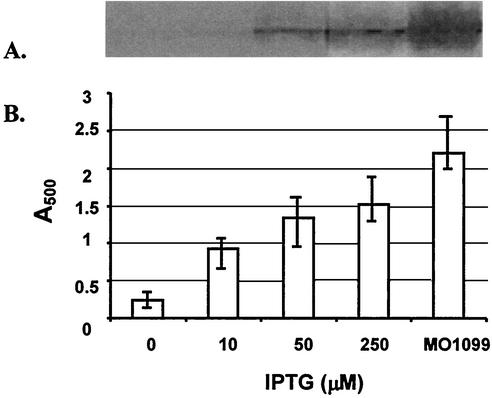
FIG. 2.
Growth and MreC expression by the MreC-inducible strain. B. subtilis KSS1571 was cultured in TSB containing 2 mM IPTG overnight at 37°C, diluted 1:100 in fresh medium supplemented with different concentrations of IPTG, and then incubated for 6 h. MO1099, the wild-type MreC control strain, was cultured in TSB without IPTG. Growth was determined by absorbance at 500 nm (B). Then the cells were harvested and lysed, and the cell lysates were electrophoresed and Western blotted. The blots were probed with rabbit anti-MreC antibody (A). In panel B, the mean values from three determinations are shown, and the range of values is indicated.
To determine the effects of reduced mreC expression levels on cell morphology, the phenotypic changes of B. subtilis KSS1571 were examined by light microscopy under two different IPTG conditions, 250 and 0 μM, and compared to the “wild-type” MO1099 (Fig. 3). MO1099 had the normal rod shape (Fig. 3A), whereas altered cell morphologies were observed with the KSS1571 cultures. At 250 μM IPTG, the cells were shorter, and there was evidence for curvature of the cells, especially at the ends (Fig. 3B). In the absence of IPTG induction, swollen cells were observed, cells had become more spherical, and the presence of debris and cell ghosts indicated that lysis had occurred (Fig. 3C).
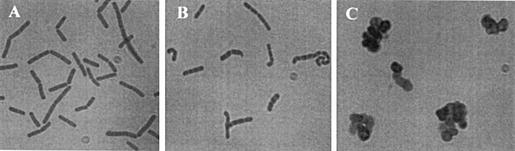
FIG. 3.
Morphology of B. subtilis cells. Smears of MO1099 (wild-type parent) (A), KSS1571 cultured in the presence of 250 μM IPTG (B), and KSS1571 cultured in the absence of IPTG (C) were stained with crystal violet and examined by microscopy. All images are at the same magnification.
Effect of MreC on the cell morphology of B. subtilis.
To better define the morphological changes obtained at different IPTG concentrations, cells were prepared for scanning electron microscopy (Fig. 4). Reduction of cellular MreC levels resulted in a shift from the normal rod shape to that of a bent rod, with the bend usually occurring near the end of the cell, giving a “shepherd's crook” appearance (Fig. 4B and C). At 250 μM IPTG, a hook was often observed at one end of cell, with the rest of cell having a normal appearance (Fig. 4B). As the concentration of IPTG was reduced, the bent rods begin to swell, as indicated by an increased cell diameter (Fig. 4C), and twist into irregular shapes (Fig. 4C and D). Further reduction in IPTG concentrations produced cells with an irregular spherical shape with a reduction in cell length and an increased diameter relative to the control strain MO1099. Culture in the absence of IPTG results in little growth. Under these conditions, the B. subtilis cells were predominantly spherical, residual rods that tended to remain associated as chains, and cell lysis was evident with the production of ghost cells (Fig. 4E). MreC-depleted B. subtilis appear to be vulnerable to osmotic pressure-induced lysis due to weakened cell wall structure. To minimize cell lysis by osmotic imbalance, 0.3 M sucrose was incorporated into the growth medium containing 0 μM IPTG. Under this condition, the majority of the cells assumed a spherical shape (Fig. 4F). The observed morphological shifts and the stabilization of the cells in an iso-osmotic environment suggest that a reduction in MreC levels may be associated with a deficiency in the cell wall structure in these bacteria.
FIG. 4.
Scanning electron microscopy of MreC-depleted B. subtilis cells. Images are of strain MO1099 (A) and KSS1571 (B to F). The MreC-inducible strain was cultured in the presence of 250 μM (B), 50 μM (C), and 10 μM (D) IPTG and in the absence of IPTG (E and F). (F) The MreC-inducible strain was cultured in the presence of 0.3 M sucrose. Bars in the micrographs are the indicated sizes in micrometers.
Transmission electron microscopy was used to examine the ultrastructure of the MreC-deficient strain (Fig. 5). At an IPTG concentration of 250 μM, the rod-shaped cells showed evidence of bending. The bending always appeared to be centered at a site of septum formation (Fig. 5B). Examination of the septum site indicated that an accumulation of a polymeric material appeared on one side of the cell at the site of septation. Furthermore, this accumulated material was always present on the concave side of the bent cell. On the basis of its location in the cell envelope and its appearance under the electron microscope, it is likely that this accumulated material is peptidoglycan.
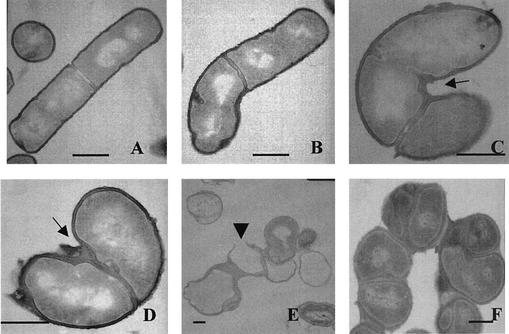
FIG. 5.
Transmission electron microscopy of MreC-depleted B. subtilis cells. Images are of strain MO1099 (A) and KSS1571 (B to F). The MreC-inducible strain was cultured in the presence of 250 μM (B), 50 μM (C), and 10 μM (D) IPTG and in the absence of IPTG (E and F). (F) The MreC-inducible strain was cultured in the presence of 0.3 M sucrose. The arrows in panels C and D indicate the areas of polymer accumulation. The arrowhead in panel E indicates a region where the cell wall had become thinner and ruptured. Bars, 1 μm.
At lower IPTG concentrations, the accumulation of polymeric material increased. The cells began to show evidence of swelling and rounding, and twisting of rod-shaped cells occurred (Fig. 5C and D). Removal of the IPTG inducer from B. subtilis KSS1571 resulted in cells with irregular-appearing septa, giving the appearance of different-sized daughter cells (Fig. 5E). However, this may be the result of sectioning through cells which had become twisted. The frequency of septation may also have been affected, producing different-sized progeny cells. The micrographs suggest that mislocalization or misdirection of septum formation may occur in the MreC-depleted cells. Cell lysis was a typical observation in the absence of IPTG. The cell walls from the resulting ghosts did not have a uniform thickness (Fig. 5E). The apparent rupture point of the cell wall was thinner than the rest of the wall (Fig. 5E). Sucrose supplementation of the growth medium reduced the apparent amount of cell lysis (Fig. 5F). Growth under these conditions allowed progression of the cellular defect to proceed for a longer period of time. With the longer time, the accumulation of the polymeric material progressed from the septum site to spread around the surface of the spherical cells (Fig. 5F).
Muramic acid content in MreC-depleted B. subtilis.
MreC depletion resulted in morphological changes and lysis of B. subtilis, suggesting that a defect in cell wall biosynthesis may be involved. Peptidoglycan is the major structural component of the gram-positive cell wall and comprises 40 to 90% of the cell wall. To determine whether there was a deviation in peptidoglycan content in the cells that conditionally express MreC when they were grown in medium containing various amounts of IPTG, a muramic acid assay was used. Muramic acid assays in this study suffer from certain limitations. At the lower concentrations of IPTG, cell death and lysis occur in the cultures, which affect both the amount of muramic acid recovered and the dry weight of the harvested cells. Despite these limitations, the muramic acid content remained constant at the different IPTG concentrations and did not differ from the wild-type level (Table 2).
TABLE 2.
Muramic acid content of MreC-deficient cells
| Strain | IPTG concn (μM) | Muramic acid contenta (μg/mg of cells [dry wt]) |
|---|---|---|
| KSS1001 | 0 | 1.9 ± 0.1 |
| KSS1571 | 10 | 1.8 ± 0.1 |
| KSS1571 | 50 | 2.0 ± 0.2 |
Values are the means ± standard deviations for three determinations.
The conclusion of this analysis is that the polymeric material detected by electron microscopy represents a substance other than peptidoglycan or the material is peptidoglycan and the distribution, but not the total amount of this cell wall material, is altered in the MreC-deficient strain.
To evaluate the possibility that it is teichoic acid rather than peptidoglycan that is the accumulated polymer, B. subtilis KSS1571 was cultured in a reduced phosphate medium. Under these conditions, the teichoic acid is replaced with teichuronic acid. The morphological changes associated with a defective teichoic acid resulting from a tagF (rodC) mutation were eliminated by the replacement of teichoic acid by teichuronic acid under these growth conditions (31). When KSS1571 was cultured under phosphate-limiting and phosphate-replete growth conditions, there were no differences seen in MreC-dependent growth, morphology, or polymeric material accumulation in the cells (data not shown). Therefore, a switch in synthesis from teichoic acid to teichuronic acid did not override the MreC-associated defect, and it appears the accumulated material is not teichoic acid.
Lack of complementation of MreC activity with a truncated mreC mutant.
The mreC mutant described by Jones et al. was created by integrating a plasmid containing an internal fragment of mreC (a 242-bp fragment initiating at the unique HindIII site) into the B. subtilis chromosome (17). The integration event would generate a truncated expressed copy of mreC with positions 1 to 504 of the open reading frame, resulting in the loss of 122 amino acid residues from the protein. We have been unable to obtain viable transformants of B. subtilis utilizing this procedure.
To evaluate the biological activity of the truncated MreC protein, the truncated allele of mreC (contains bp 1 to 504) was positioned downstream of a constitutive S. aureus lactose operon promoter (22) in plasmid pDG3053 to create pLEE504 (Fig. 6A). This plasmid was introduced into the mreC expression strain KSS1571 to produce KSS1573. This isolate exhibits only limited growth and ultimately lyses without the inclusion of IPTG in the culture medium, indicating that the truncated MreC protein lacks sufficient biological activity to support growth (Fig. 6B). Western blot analysis indicated that the truncated MreC protein was expressed in this cell, so a lack of expression was not the explanation for the failure to complement the MreC deficiency (Fig. 6C). In fact, the presence of the plasmid resulted in a higher level of expression of wild-type MreC in the cell. This results from the presence of the pUC plasmid-derived lactose operon operator site in the plasmid, which is able to bind the LacI repressor protein and thus titrate the repressor from the spac promoter operator. Despite this increased production of wild-type MreC, the culture still grew poorly (Fig. 6B) and also displayed the characteristic cell swelling and twisting of MreC-depleted cells (Fig. 6D). The strain expressing the truncated MreC protein grew more poorly than KSS1571 in the absence and presence of IPTG. Thus, the truncated MreC protein is not able to complement the phenotype associated with the loss of MreC protein in the cells, but it appears to be able to compete with the wild-type protein. This gives the cell the appearance of a more severe MreC-deficient phenotype than warranted by the level of full-length MreC in the cell.
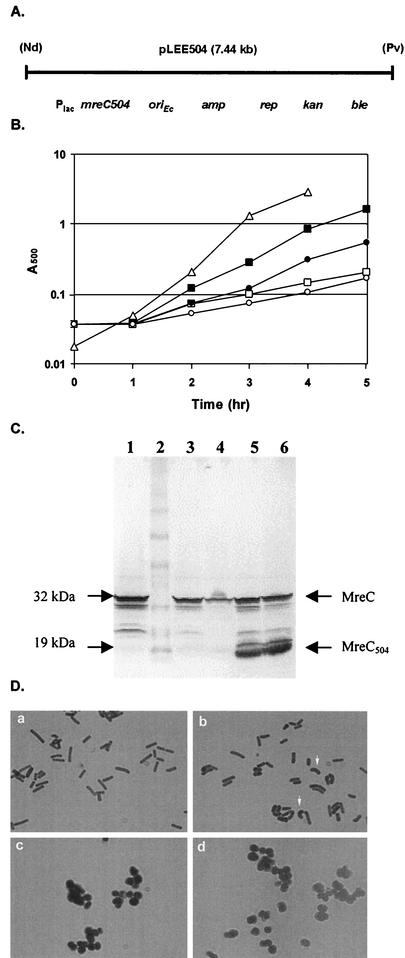
FIG. 6.
(A) Map of pLEE504, a shuttle plasmid which expresses the 168-residue truncated form of MreC from the S. aureus lac promoter. The ampicillin resistance determinant (amp), replication protein gene (rep), kanamycin resistance determinant (kan), phleomycin (zeocin) resistance determinant (ble), and NdeI (Nd) and PvuII (Pv) restriction sites are shown. (B) Growth of B. subtilis strains in TSB. Symbols: ▵, KSS1001 (wild type); ▪, KSS1571 cultured in the presence of 1 mM IPTG; □, KSS1571 cultured in the absence of IPTG; •, KSS1573 cultured in the presence of 1 mM IPTG; ○, KSS1573 cultured in the absence of IPTG. (C) Western blot of B. subtilis cell lysates probed with rabbit anti-MreC antiserum. Lanes: 1, MO1099 (wild-type control); 2, molecular size markers; 3, KSS1571 grown in the presence of 250 μM IPTG; 4, KSS1571 grown in the absence of IPTG; 5, KSS1573 grown in the presence of 250 μM IPTG; 6, KSS1573 grown in the absence of IPTG. The positions of full-length MreC (32 kDa) and the truncated form of MreC (19 kDa) are indicated. The amount of cell lysate loaded per well was eight times that used in the blot shown in Fig. 2A. (D) Photomicrographs of methylene blue-stained cells. B. subtilis KSS1571 (a and c) and KSS1573 (b and d) were cultured in the presence of 1 mM IPTG (a and b) or in the absence of IPTG (c and d). The arrows in panel b indicate cells that appear swollen and bent.
DISCUSSION
The two-competing-sites model for peptidoglycan assembly predicts that peptidoglycan assembly along the long axis of the cell and septal peptidoglycan synthesis during division are mutually exclusive processes and that control of these two activities influence cell morphology (21, 24). Hyperexpression of the septal peptidoglycan biosynthetic apparatus leads to a spherical shape, whereas hyposeptation activity results in filamentation. Wachi and Matsuhashi had concluded that the MreB protein may play a regulatory role in septal synthesis by regulating expression of FtsI (30). Our results extend the findings of Wachi and Matsuhashi by suggesting that the MreC transmembrane protein may function as part of the control network which determines which peptidoglycan biosynthesis process is active. When MreC production is turned off, the cells begin to synthesize cell wall material at the division site in an aberrant manner. Overproduction of the wall polymer occurs on one side of the cell, the consequence of which is a bending of the cell, with the concave side being the side of overproduction of this material. Continued synthesis of the cell wall leads to a progressive spread of this material from one side of the septum until it eventually covers the cell. However, during this process, the overall cell wall appears to weaken. This leads to an influx of water into the cell. The swelling of the cell exaggerates the twisting of the cell. Eventually, the cell wall ruptures, and the cell lyses. The loss of wall integrity may be due to the loss of a regulated switch from septum synthesis to lateral wall extension or to the abnormal septal wall synthesis pattern. Prevention of cell lysis by culturing the cells in an iso-osmotic environment leads to a complete progression of the thickened wall around the perimeter of the cell. The cells take on a more spherical shape, consistent with a loss of lateral wall extension capability.
The septation process in B. subtilis involves the formation of the FtsZ ring, a structure which then recruits other division-associated proteins (2). This division ring forms the leading edge of the constriction which progresses during the division process. Up to now, there has been no evidence for a sidedness to this cytokinesis ring. The electron microscopic observations reported herein indicate that the septal ring associated with cytokinesis in B. subtilis does indeed have a sidedness to it. The overexpression of the wall polymeric material occurs on one side of the cell. The differences in the cell envelope, or in the ring formed by FtsZ and other division proteins, that accounts for the septal ring asymmetry is unknown.
The mreC determinant is an essential determinant in B. subtilis and appears to function in the septation process. MreC has been shown to localize to the division septum of B. subtilis (19). Truncation of this protein destroys its proper functioning in the cell, although the 168-amino-acid polypeptide does appear to be able to compete with the full-length version of the protein. The truncated protein may be able to compete with the wild-type protein for interaction with another component(s) of the septation apparatus, or if MreC is a multimeric protein, it may be able to be included in the MreC complex, and this inclusion would result in a loss of biological activity.
Acknowledgments
We thank David George for technical assistance and operation of the DNA sequencer, Lloyd Willard for the electron microscopy, and Patricia W. Stewart at DVM for assistance with the production of the anti-MreC antiserum.
This work was supported in part by grant GM57049 from the National Institutes of Health.
Footnotes
Contribution 03-298-J from the Kansas Agricultural Experiment Station.
REFERENCES
- 1.Abhayawardhane, Y. K., and G. C. Stewart. 1995. Bacillus subtilis possesses a second determinant with extensive sequence similarity to the Escherichia coli mreB morphogene. J. Bacteriol. 177:765-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beall, B., and J. Lutkenhaus. 1991. FtsZ in Bacillus subtilis is required for vegetative septation and for asymmetric septation during sporulation. Genes Dev. 5:447-455. [DOI] [PubMed] [Google Scholar]
- 3.Benson, A. K., and W. G. Haldenwang. 1993. Regulation of σB levels and activity in Bacillus subtilis. J. Bacteriol. 175:2347-2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhavsar, A. P., T. J. Beveridge, and E. D. Brown. 2001. Precise deletion of tagD and controlled depletion of its product, glycerol 3-phosphate cytidylyltransferase, leads to irregular morphology and lysis of Bacillus subtilis grown at physiological temperature. J. Bacteriol. 183:6688-6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bork, P., C. Sander, and A. Valencia. 1992. An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and Hsp70 shock proteins. Proc. Natl. Acad. Sci. USA 89:7290-7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burger, A., K. Sichler, G. Kelemen, M. Buttner, and W. Wohlleben. 2000. Identification and characterization of the mre gene region of Streptomyces coelicolor A3(2). Mol. Gen. Genet. 263:1053-1060. [DOI] [PubMed] [Google Scholar]
- 8.Cha, J.-H., and G. C. Stewart. 1997. The divIVA minicell locus of Bacillus subtilis. J. Bacteriol. 179:1671-1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cutting, S. M., and P. B. Vander Horn. 1990. Genetic analysis, p. 27-74. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley & Sons, Chichester, United Kingdom.
- 10.Dagert, M., and S. D. Ehrlich. 1979. Prolonged incubation in calcium chloride improves the competence of Escherichia coli cells. Gene 6:23-28. [DOI] [PubMed] [Google Scholar]
- 11.Doi, M., M. Wachi, F. Ishino, S. Tomioka, M. Ito, Y. Sakagami, A. Suzuki, and M. Matsuhashi. 1988. Determinations of the DNA sequence of the mreB gene and of the gene products of the mre region that function in formation of the rod shape of Escherichia coli cells. J. Bacteriol. 170:4619-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ensign, J. C., and R. S. Wolfe. 1964. Nutritional control of morphogenesis in Arthrobacter crystallopoietes. J. Bacteriol. 47:137-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Erickson, R. J., and J. C. Copeland. 1972. Structure and replication of chromosomes in competent cells of Bacillus subtilis. J. Bacteriol. 109:1075-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gupta, R. S., and B. Singh. 1992. Cloning of the Hsp70 gene from Halobacterium marismortui: relatedness of archaebacterial Hsp70 to its eubacterial homologs and a model for the evolution of the Hsp70 gene. J. Bacteriol. 174:4594-4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hadzija, O. 1974. A simple method for the quantitative determination of muramic acid. Anal. Biochem. 60:512-517. [DOI] [PubMed] [Google Scholar]
- 16.Honeyman, A. L., and G. C. Stewart. 1989. The nucleotide sequence of the rodC operon of Bacillus subtilis. Mol. Microbiol. 3:1257-1268. [DOI] [PubMed] [Google Scholar]
- 17.Jones, L. J. F., R. Carballido-Lopez, and J. Errington. 2001. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell 104:913-922. [DOI] [PubMed] [Google Scholar]
- 18.Kubo, M., D. J. Pierro, Y. Mochizuki, T. Kojima, T. Yamazaki, S. Satoh, N. Takizawa, and H. Kiyohara. 1996. Bacillus stearothermophilus cell shape determinant genes, mreC and mreD, and their stimulation of protease production in Bacillus subtilis. Biosci. Biotechnol. Biochem. 60:271-276. [DOI] [PubMed] [Google Scholar]
- 19.Lee, J.-C., J.-H. Cha, D. B. Zerby, and G. C. Stewart. Heterospecific expression of the Bacillus subtilis cell shape determination genes mreBCD in Escherichia coli. Curr. Microbiol., in press. [DOI] [PubMed]
- 20.Levin, P. A., P. S. Margolis, P. Setlow, R. Losick, and D. Sun. 1992. Identification of Bacillus subtilis genes for septum placement and shape determination. J. Bacteriol. 174:6717-6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lleo, M. M., P. Canepari, and G. Satta. 1990. Bacterial cell shape regulation: testing of additional predictions unique to the two-competing-sites model for peptidoglycan assembly and isolation of conditional rod-shaped mutants from some wild-type cocci. J. Bacteriol. 172:3758-3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oskouian, B., and G. C. Stewart. 1990. Repression and catabolite repression of the lactose operon of Staphylococcus aureus. J. Bacteriol. 172:3804-3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pooley, H. M., F.-X. Abellan, and D. Karamata. 1992. CDP-glycerol:poly(glycerophosphate) glycerophosphotransferase, which is involved in the synthesis of the major wall teichoic acid in Bacillus subtilis 168, is encoded by tagF (rodC). J. Bacteriol. 174:646-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Satta, G., P. Canepari, and R. Fontana. 1983. A novel hypothesis to explain regulation of the murein sacculus shape, p. 135-140. In R. Hakenbeck, J. V. Holtje, and H. Labischinski (ed.), The target of penicillin. Walter de Gruyter, Berlin, Germany.
- 25.Signoretto, C., F. Di Stefano, and P. Canepari. 1996. Modified peptidoglycan chemical composition in shape-altered Escherichia coli. Microbiology 142:1919-1926. [DOI] [PubMed] [Google Scholar]
- 26.Varley, A. W., and G. C. Stewart. 1992. The divIVB region of the Bacillus subtilis chromosome encodes homologs of Escherichia coli septum placement (MinCD) and cell shape determinants (MreBCD). J. Bacteriol. 174:6729-6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wachi, M., M. Doi, Y. Okada, and M. Matsuhashi. 1989. New mre genes mreC and mreD, responsible for formation of the rod shape of Escherichia coli cells. J. Bacteriol. 171:6511-6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wachi, M., M. Doi, S. Tamaki, W. Park, S. Nakajima-Iijima, and M. Matsuhashi. 1987. Mutant isolation and molecular cloning of mre genes which determine cell shape, sensitivity to mecillinam, and amount of penicillin-binding proteins in Escherichia coli. J. Bacteriol. 169:4935-4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wachi, M., M. Doi, T. Ueda, M. Ueki, K. Tsuritani, K. Nagai, and M. Matsuhashi. 1991. Sequence of the downstream flanking region of the shape-determining genes mreBCD of Escherichia coli. Gene 106:135-136. [DOI] [PubMed] [Google Scholar]
- 30.Wachi, M., and M. Matsuhashi. 1989. Negative control of cell division by mreB, a gene that functions in determining the rod shape of Escherichia coli cells. J. Bacteriol. 171:3123-3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wagner, P. M., and G. C. Stewart. 1991. Role and expression of the rodC operon of Bacillus subtilis. J. Bacteriol. 173:4341-4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yanouri, A., R. A. Daniel, J. Errington, and C. E. Buchanan. 1993. Cloning and sequencing of the cell division gene pbpB, which encodes penicillin-binding protein 2B in Bacillus subtilis. J. Bacteriol. 175:7604-7616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yansura, D. G., and D. J. Henner. 1984. Use of the Escherichia coli lac repressor and operator to control gene expression in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 81:439-443. [DOI] [PMC free article] [PubMed] [Google Scholar]