Abstract
The bacterial peptidoglycan consists of glycan chains of repeating β-1,4-linked N-acetylglucosaminyl-N-acetylmuramyl units cross-linked through short peptide chains. The polymerization of the glycans, or glycosyltransfer (GT), and transpeptidation (TP) are catalyzed by bifunctional penicillin-binding proteins (PBPs). The β-lactam antibiotics inhibit the TP reaction, but their widespread use led to the development of drug resistance in pathogenic bacteria. In this context, the GT catalytic domain represents a potential target in the antibacterial fight. In this work, the in vitro polymerization of glycan chains by the extracellular region of recombinant Streptococcus pneumoniae PBP2a, namely, PBP2a* (the asterisk indicates the soluble form of the protein) is presented. Dansylated lipid II was used as the substrate, and the kinetic parameters Km and kcat/Km were measured at 40.6 μM (± 15.5) and 1 × 10−3 M−1 s−1, respectively. The GT reaction catalyzed by PBP2a* was inhibited by moenomycin and vancomycin. Furthermore, the sequence between Lys 78 and Ser 156 is required for enzymatic activity, whereas it is dispensable for lipid II binding. In addition, we confirmed that this region of the protein is also involved in membrane interaction, independently of the transmembrane anchor. The characterization of the catalytically active GT domain of S. pneumoniae PBP2a may contribute to the development of new inhibitors, which are urgently needed to renew the antibiotic arsenal.
The development of antibiotic resistance is an unavoidable phenomenon, since it is associated with the natural evolution of the highly flexible bacterial genome. Consequently, physicians are deprived of solutions to combat infections caused by highly resistant pathogenic strains. β-Lactam molecules represent 60% of the total consumption of antibiotics in human therapy today. Fifty percent of the clinical strains isolated in some European countries, and more than 90% isolated in Asia, are resistant to β-lactams (1), while resistance levels of Streptococcus pneumoniae have increased by over 100-fold compared to that of the reference strain isolated before the use of antibiotics.
β-Lactam action relies on the inhibition of a crucial step in the biosynthesis of the peptidoglycan, leading to cell lysis. The peptidoglycan is a strong netlike polymer responsible for maintaining the shape and size of the bacterial cell and for resisting the high intracellular osmotic pressure. Peptidoglycan consists of glycan chains of repeating β-1,4-linked N-acetylglucosaminyl-N-acetylmuramyl units cross-linked through short peptide chains. The final steps of peptidoglycan biosynthesis occur extracellularly, associated with the plasma membrane, and are catalyzed by penicillin-binding proteins (PBPs) (6). In addition to harboring a short cytoplasmic region and a transmembrane helix, class A PBPs bear two enzymatic functions, namely, glycosyltransfer (GT) and transpeptidation (TP). The former is responsible for the elongation of the glycan strands, using as a substrate the disaccharide (N-acetylglucosaminyl-N-acetylmuramyl) pentapeptide anchored to the membrane by a 55-carbon undecaprenyl chain (lipid II), while the latter cross-links the peptide chains attached to the glycan strands. In addition to bifunctional PBPs, all bacterial species contain class B PBPs which harbor only the TP activity, as well as N- and C-terminal domains of unknown function (7). Penicillin and other β-lactam antibiotics are specific inhibitors of the transpeptidase activity catalyzed by PBPs. β-Lactams form a covalent complex with the active serine of the TP domain, preventing the cross-linking of stem peptides and weakening the peptidoglycan structure (9, 14).
PBPs have been validated as therapeutic targets through the efficiency of β-lactams. It is then clear that the enzymatic GT domain, harbored by these same PBPs but insensitive to β-lactams, represents a very promising therapeutic target. Deletion of class A PBPs genes in Escherichia coli, S. pneumoniae, and Bacillus subtilis showed that some combinations are lethal or induced dramatic morphological alterations, indicating that bifunctional PBPs have important roles in cell division and cell growth (2, 10, 16-18, 26). Furthermore, GT domains are located on the outer surface of the bacterial membrane and are consequently accessible to drug molecules. The unique competitive antibiotic directed against the GT activity is moenomycin, a molecule with a structure analogous to that of the lipid II substrate (12). Addition of moenomycin in E. coli cultures induced morphological modifications leading to cell lysis (23). In spite of this lethal effect, moenomycin is not used in human therapy due to poor absorption and long half-life (8).
Several points can be raised to understand why a new GT inhibitor is not yet available. Extensive screening of chemical libraries (of natural and synthesized molecules) did not succeed in the identification of lead molecules. Consequently, development of an inhibitor of GT activity requires the rational design of such molecules. This approach cannot be undertaken without the three-dimensional structure of a GT domain, complexed or not with ligands (lipid II and/or analogues and moenomycin and/or analogues). However, a major limitation is the limited availability of GT ligands (13). van Heijenoort and coworkers, as well as two industrial groups, have recently succeeded in purifying lipid II, allowing significant progress in the functional characterization of the GT activity of E. coli PBPs (19-22, 24). In addition, the isolation of the bifunctional proteins themselves, in soluble and stable form, poses a challenge which is undoubtedly linked to a membrane-recognizing region present in the GT region (5, 15, 25).
In this work, the polymerization of glycan chains by the extracellular region of S. pneumoniae PBP2a, namely, PBP2a* (the asterisk indicates the soluble form of the protein), is demonstrated for the first time. Dansylated lipid II was used as the substrate, and kinetic parameters were measured. We observed that the sequence between Lys 78 and Ser 156 is required for enzymatic activity, whereas it is dispensable for lipid II binding. In addition, we confirmed that this region of the protein is also involved in membrane interaction independently of the transmembrane anchor. The characterization of the catalytically active GT domain of S. pneumoniae PBP2a may contribute to the development of new inhibitors, which are urgently needed to renew the antibiotherapeutic arsenal.
MATERIALS AND METHODS
Materials.
Glutathione, phenylmethylsulfonyl fluoride (PMSF), and vancomycin were purchased from Sigma (Saint Quentin Fallavier, France). Isopropyl- β-d-thiogalactopyranoside (IPTG), ampicillin, and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) were purchased from Euromedex (Mundolsheim, France). Complete protease inhibitor cocktail was obtained from Roche. Dansylated lipid II was generously provided by Eli Lilly (Indianapolis, Ind.). Moenomycin was a gift from Aventis (Frankfurt am Main, Germany). Restriction and modification DNA enzymes were from Promega (Charbonnières, France). DNA sequencing was performed by Genome Express (Grenoble, France).
Construction of the periplasmic form of pbp2aSGG from S. pneumoniae in pGEX-4T1 vector.
The unencapsulated R6 strain of S. pneumoniae was anaerobically propagated in glucose-buffered broth (Diagnostics Pasteur) for 16 h at 30°C. Genomic DNA was extracted from 5 ml of culture with the High Pure PCR template preparation kit (Roche Molecular Biochemicals), following the manufacturer's instructions; 100 ng of genomic DNA was recovered in 200 μl of water. Genomic DNA from the R6 strain of S. pneumoniae was used as template (2 ng) to amplify the sequence of the pbp2aSGG gene (for which SGG represents the first three amino acids of the deleted protein [Fig. 1B]) by using Vent polymerase (New England Biolabs) with the upstream primer 5′ CCCATATGGAGGAGAAACTTGTGCCGACCATC, which contains the NdeI site (in boldface), and the downstream primer 5′ CCGCGGCCGCACCTCCTTCCATCAAAAACTTTTT, which contains the NotI site (in boldface). The PCR product was cloned into the pCR-Script vector (Stratagene) by following the manufacturer's instructions. After transformation into Epicurian Coli XL1-Blue supercompetent cells, clones containing the pbp2aSGG gene were selected as white colonies on IPTG X-Gal-ampicillin-containing agar plates. The pbp2aSGG gene was subcloned into a pGEX-4T1 vector (Amersham Biosciences). The resulting construct, pDG2aSGG, encodes a PBP2a variant, the so-called PBP2a*SGG, encompassing amino acids S156 to R678. PBP2a*, encoded by construct pJAH144 (5), and PBP2a*SGG are both fused to the glutathione S-transferase (GST) fused at the GT domain.
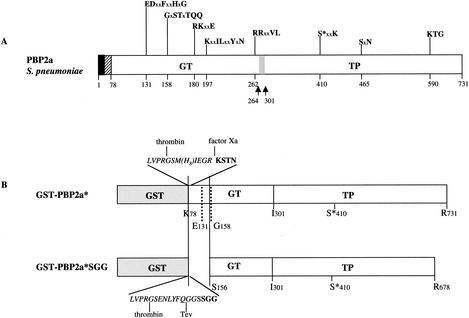
FIG. 1.
Schematic diagrams of S. pneumoniae PBP2a. (A) Topology of the native PBP2a protein. The solid and hatched boxes indicate the N-terminal cytoplasmic region and the membrane anchor, respectively. GT and TP domains are represented together with their conserved motifs; active site serine 410 is indicated by an asterisk. x represents any amino acid. The gray box and the arrows indicate the location of the identified GT and TP junction (5). (B) Schematic diagrams of the PBP2a-derived constructs. The peptide at the GST-GT junction of the PBP2a* construct includes sequences specific for thrombin, Tev protease, and factor Xa (italic characters) and N-terminal amino acids of GT (bold characters).
Purification of GST-fused PBP2a* variants.
Overnight cultures of E. coli MC1061 strains transformed with plasmids pJAH144 and pDG2aSGG were used to inoculate (1:50) 1 liter of Luria-Bertani medium supplemented with 100 μg of ampicillin/ml. On achieving a culture with an optical density at 600 nm of 1 at 37°C, protein expression was induced with 1 mM IPTG while incubating for 16 h at 15°C. Purification steps were all performed either at 4°C or on ice. After cells were centrifuged at 6,000 × g for 15 min, the pellet was resuspended in 80 ml of a solution of 50 mM Tris-HCl (pH 8.0)-200 mM NaCl-1 mM EDTA (buffer A) containing 1 tablet of complete protease inhibitor cocktail (Roche). The cells were then disrupted by sonication; the cell lysate was centrifuged at 31,000 × g for 20 min, and the resulting supernatant was loaded onto a 5-ml glutathione-Sepharose column (Amersham Biosciences) which had been previously equilibrated in buffer A. GST-PBP2a*SGG was eluted with 10 mM reduced glutathione in buffer A. Extensive dialysis against buffer A was performed to eliminate the reduced glutathione from the protein solution. GST-PBP2a*SGG was incubated with 0.16 U of Tev protease/μg of fusion protein in an overnight incubation at room temperature, and the cleaved product was reloaded onto the glutathione-Sepharose column. The resulting flowthrough, containing the isolated PBP2a*SGG, was dialyzed against 25 mM Tris-HCl (pH 8.8)-50 mM NaCl. An anion-exchange chromatography (Resource Q; Amersham Biosciences) was performed at a flow rate of 2 ml/min, using a NaCl gradient (0 to 250 mM) to elute PBP2a*SGG.
The purification of PBP2a* has been previously described (5). Briefly, the postsonication pellet was resuspended in buffer A containing 0.5% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), and the suspension was submitted to rotary shaking. The supernatant containing the GST-PBP2a* fusion protein obtained from a 15-min centrifugation at 20,800 × g was loaded onto a 5-ml glutathione-Sepharose column (Amersham Biosciences) equilibrated in buffer A containing 0.5% CHAPS. The fusion protein was cleaved while bound to the column in the presence of 50 U of thrombin (Sigma) for 1 h at room temperature. The purification of the transpeptidase domain of PBP2a (TP2a) has been previously described (5).
Lipid II binding assay.
Dansylated lipid II at a final concentration of 140 μM was incubated with PBP2a* (2 μM), PBP2a*SGG (2.8 μM), and TP2a (5.5 μM) for 15 min at room temperature before analysis by native polyacrylamide gel electrophoresis (PAGE); buffers corresponded to protein buffers described above. After migration, the fluorescence emitted by the dansyl group of lipid II was detected by illuminating the gel by UV light (Bio-Rad UV transilluminator): both free and bound lipid II forms could be detected. The gel was then stained by Coomassie blue to visualize the protein pattern.
Glycosyltransferase reaction (glycan chain polymerization).
The reaction mix was composed of 40 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES) (pH 6.1), 40 mM MgCl2, 0.16 mM KCl, 0.62 mM NH4Cl, 5 mM ATP, 0.4% CHAPS, and 20% dimethyl sulfoxide (DMSO). PBP2a* was used at a final concentration of 4 μM, and inhibitors (moenomycin and vancomycin) and lipid II were added last at various concentrations (see Results for details). The incubations were performed at 30°C for various periods of time. The reaction products were separated by thin-layer chromatography (TLC) on silica gels deposited on aluminum sheets (Fluka) in chloroform:methanol:water (20:12:3). Fluorescence detected under UV light was quantified with Molecular Analyst (Bio-Rad) software.
RESULTS
Expression and purification of S. pneumoniae PBP2a*SGG.
In a previous article, we proposed that PBP2a* interacts with the cytoplasmic membrane in a region located between Lys 78 and Ser 156 of the GT domain, which is distinct from the transmembrane anchor region (5). In this study, confirmation of this result was sought through the production of the truncated PBP2a*SGG variant (SGG are the N terminus residues). The N-terminal Ser 156 is two residues upstream from the conserved Gly 158 of the second GT motif. The C terminus is Arg 678, corresponding to the C terminus of the isolated TP2a domain (5) (Fig. 1). This variant PBP2a*SGG, lacking the first conserved motif in the GT domain (E131DxxFxxHxG) was tested for its solubility in the absence of detergent. The binding of benzylpenicillin to the truncated PBP2a* form was described previously (5).
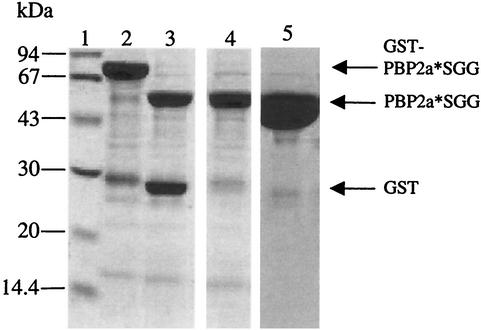
The pDG2aSGG construct allows the expression of PBP2a*SGG fused to the GST tag (Fig. 1), and it is expressed in E. coli cells as a soluble cytoplasmic protein. The GST fusion protein is purified on a glutathione-Sepharose column as an approximately 80-kDa species (Fig. 2, lane 2); the yield was approximately 10 mg of soluble protein per liter of E. coli culture. The fusion protein is cleaved by the Tev protease (Fig. 2, lane 3), and the resulting digestion products are reloaded onto a glutathione-Sepharose column in order to retain the GST contaminant. PBP2a*SGG present in the flowthrough has an apparent molecular mass of approximately 50 kDa, in good agreement with the calculated mass for PBP2a*SGG (57.2 kDa) (Fig. 2, lane 4). An ion-exchange chromatography on Resource Q completed the purification by eliminating minor contaminants (Fig. 2, lane 5). The homogeneity of purified PBP2a*SGG was verified by native PAGE analysis as shown on Fig. 3B, lane 3.
FIG. 2.
Purification of PBP2a*SGG. Proteins were separated by sodium dodecyl sulfate-12.5% PAGE and stained with Coomassie blue. Numbers at the left indicate sizes of standard molecular mass markers. Lanes: 1, molecular mass markers; 2, GST-PBP2a*SGG fusion following purification in a glutathione-Sepharose column; 3, fusion protein cleavage by Tev protease; 4, flowthrough from glutathione-Sepharose (10 μg of protein loaded on the gel); 5, elution from Resource Q column (30 μg of protein loaded on the gel).
FIG. 3.
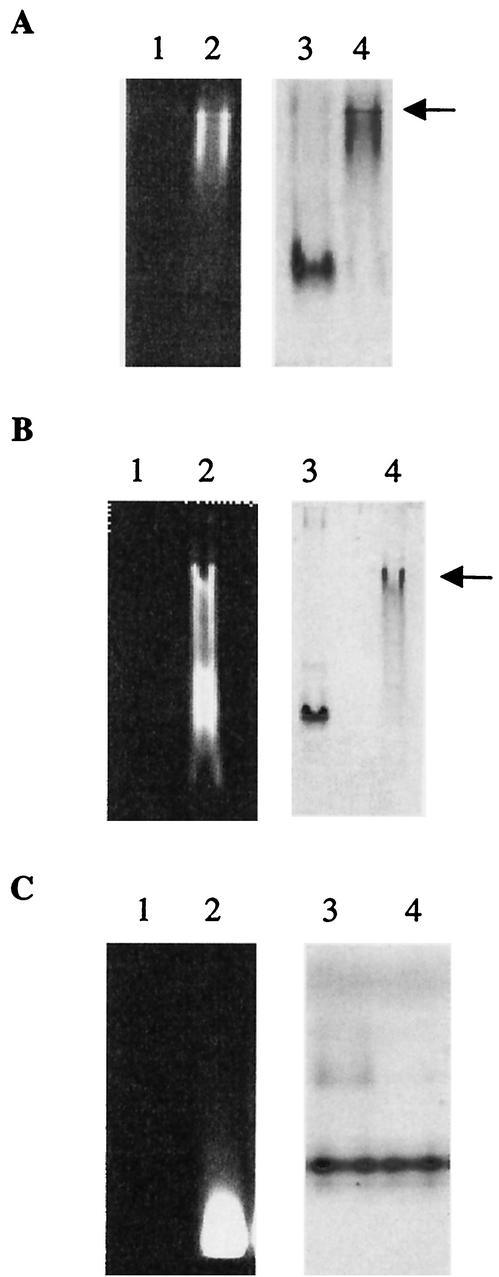
Lipid II binding to PBP2a moieties. The binding was performed at room temperature for 15 min before loading onto native PAGE. Arrows indicate protein/lipid II complex. (A) Lanes: 1 and 3, PBP2a* (2 μM); 2 and 4, PBP2a* (2 μM) and lipid II (140 μM). The same gel was observed under UV light (lanes 1 and 2) before staining with Coomassie blue (lanes 3 and 4). (B) Lanes: 1 and 3, PBP2a*SGG (2.8 μM); 2 and 4, PBP2a*SGG (2.8 μM) and lipid II (140 μM). The same gel was observed under UV light (lanes 1 and 2) before staining with Coomassie blue (lanes 3 and 4). (C) Lanes: 1 and 3, TP2a (5.5 μM); 2 and 4, TP2a (5.5 μM) and lipid II (140 μM). The same gel was observed under UV light (lanes 1 and 2) before staining with Coomassie blue (lanes 3 and 4).
Lipid II binding to the GT domain of S. pneumoniae PBP2a* and PBP2a*SGG.
Lipid II is the natural substrate for the GT activity of class A PBPs (19, 20, 24). The lipid II molecule used in this work has, linked to the amine moiety of the lysine side chain, a dansyl group allowing fluorescence measurements. In order to assay for the recognition of lipid II by the GT domain of PBP2a, protein and substrate were allowed to interact for 15 min at room temperature prior to analysis by native PAGE (Fig. 3) (5). The dansylated lipid II was then revealed by illuminating gels by UV light soon after the completion of the electrophoresis steps (Fig. 3A, B, and C, lanes 1 and 2). Subsequently, gels were stained by Coomassie blue in order to visualize the protein (Fig. 3A, B, and C, lanes 3 and 4). Our objective was to show the binding or absence of binding of lipid II to the various forms of PBP2a. For this purpose, it was important to directly compare the fluorescence and Coomassie blue-stained pattern on the same gels for each individual protein. In the presence of lipid II, PBP2a* and PBP2a*SGG electrophoretic migration on native gels is slowed (Fig. 3A and B, lanes 3 and 4). Under those conditions, both proteins are labeled by fluorescent lipid II (Fig. 3A and B, lane 2), which in the absence of protein migrates at the bottom of the gel (Fig. 3C, lane 2) (3). The protein shift in the presence of lipid II is therefore the result of an interaction between these two components, which is verified for both PBP2a* and PBP2a*SGG. However, despite similar proteins and lipid II concentrations, as well as identical experimental conditions, only PBP2a* can bind to all the lipid II available (Fig. 3A, lane 2). The majority of lipid II is not associated with the shifted PBP2a*SGG (Fig. 3B, lane 2), which indicates a lower affinity of PBP2a*SGG for the lipid II. However, a rather tight binding of lipid II to PBP2a*SGG is indicated by the fact that all the protein is shifted upwards. This discrepancy might result from the presence of multiple binding sites of lipid II on PBP2a*, some of which would have been removed in PBP2a*SGG.
In order to verify the specificity of lipid II binding to the GT domain, the ligand binding test was performed on the TP domain isolated from PBP2a*. On a Coomassie blue-stained native gel, no TP protein shift is observed in the presence of lipid II (Fig. 3C, lane 4), and no modification was observed in the migration of fluorescent lipid II (Fig. 3C, lane 2), showing the absence of binding between the lipid II molecule and the PBP2a* TP domain. The modified migration of PBP2a* and absence of free lipid II (Fig. 3A) do not seem to result from the integration of the protein into the putative lipid II vesicles, since this is not observed for the two other proteins studied in similar conditions (Fig. 3B and C, PBP2a*SGG and TP2a, respectively). In conclusion, the GT domain of PBP2a* specifically binds to lipid II.
Glycosyltransferase activity of PBP2a*.
The specificity of lipid II binding to PBP2a* prompted us to investigate the ability of this bifunctional PBP to polymerize glycan chains from the lipid II substrate. The enzymatic reaction was carried out as described in Materials and Methods. Substrate and polymerized products were detected following TLC under UV light as compounds harboring different Rfs, typically 0.28 to 0.38 and 0, respectively. The assay was carried out with lipid II (28 μM) and with proteins PBP2a* and PBP2a*SGG (4 μM); polymerized product was detected only with PBP2a* (data not shown). This result demonstrates that the sequence between Lys 78 and Ser 156 containing the first conserved motif (E131DxxFxxHxG) is important for the catalytic process.
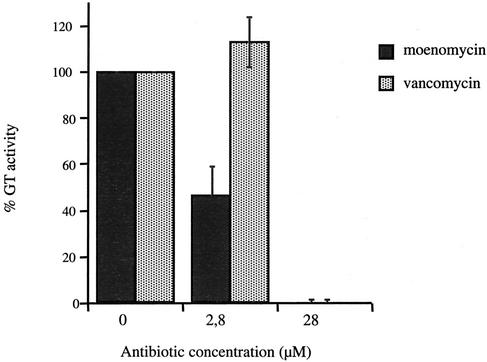
Moenomycin is a competitive inhibitor of the GT activity due to its structural similarity to the substrate lipid II. Vancomycin is a glycopeptide antibiotic which binds to the d-alanyl-d-alanine moiety of the lipid II and the peptidoglycan precursors, inhibiting the two reactions catalyzed by PBPs. Both antibiotics were tested for their ability to inhibit the GT activity of PBP2a*. Moenomycin and vancomycin concentrations were adjusted to reach molar ratios of 0.1:1 and 1:1 in relation to lipid II (28 μM), respectively (Fig. 4). Fifty percent inhibition of the GT activity was reached when 2.8 μM moenomycin was added. No effect was detected with 2.8 μM vancomycin, while complete inhibition was observed with 28 μM vancomycin.
FIG. 4.
Inhibition of PBP2a* GT activity by moenomycin and vancomycin. Reaction mix composition and TLC conditions were as described in Materials and Methods. PBP2a* (4 μM), lipid II (28 μM), and the inhibitors moenomycin and vancomycin (2.8 and 28 μM) were used in the assay.
Kinetic characterization of the glycosyltransferase activity of PBP2a*.
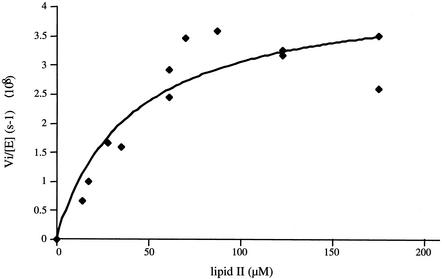
The polymerization of glycan chains by PBP2a* was observed by the TLC method. The disappearance of substrate was monitored through the dansyl fluorescence of lipid II. On the basis of initial rate measurements of fluorescent peptidoglycan synthesis and the Michaelis-Menten equation, the reaction proceeds with a kcat value of 4.3 × 10−8 s−1 (± 0.5 × 10−8), a Km value of 40.6 μM (± 15.5), and, therefore, a kcat/Km efficiency of 1 × 10−3 M−1 s−1 (Fig. 5).
FIG. 5.
Kinetic analysis of PBP2a* GT activity. PBP2a* (4 μM) and lipid II concentrations ranging from 14 to 175 μM were used. The curve fit (as well as the associated errors) was generated by fitting data to the equation Vi/[E] = kcat · [lipid II]/Km + [lipid II] using Kaleidagraph.
DISCUSSION
In previous work with limited proteolysis and reconstitution into lipid vesicles, it was suggested that the membrane association site in S. pneumoniae PBP2a* encompassed only the first conserved GT motif (5). In this work, we have produced PBP2a*SGG, a truncated form of PBP2a* deleted from the sequence between Lys 78 and Ser 156 which contains the first GT conserved motif (E131DxxFxxHxG). This variant protein is expressed and purified from E. coli as a soluble protein, confirming the location of the membrane interaction site. The association with the membrane of bifunctional PBPs from which the transmembrane helix has been deleted is common to certain of the PBPs biochemically studied (2, 5, 15, 25). Even if this putative membrane association site of the class A PBPs' GT domain is consistent with the membrane localization of the substrate lipid II, its precise functional role remains unclear.
Two distinct groups have performed the total synthesis of lipid II, a feat which will help overcome the lack of peptidoglycan synthesis substrate (13, 20, 24). In order to test the binding of lipid II onto PBP2a*, an electrophoretic assay which takes advantage of the fluorescence of dansylated lipid II was developed (3). A shift of the fluorescent band was observed in the presence of full-length PBP2a* but not in the presence of the isolated TP domain, leading to the conclusion that lipid II binds specifically to the GT domain of PBP2a*. Furthermore, PBP2a*SGG conserves the ability to bind to lipid II, although in a way different from that of PBP2a*. It might be that multiple lipid II binding sites are present in PBP2a*, some of which may have been removed in PBP2a*SGG.
To date, the polymerization of glycan chains by a bifunctional PBP had only been described for E. coli enzymes, among which PBP1b is the best characterized molecule from a kinetic point of view (19, 21). The description of the GT catalytic activity of S. pneumoniae PBP2a* reported here will expand upon the functional analysis of this important drug target. The variant PBP2a*SGG does not show GT activity, demonstrating that the sequence between Lys 78 and Ser 156 is important for the catalytic process. This result is in accordance with previous studies performed on E. coli PBP1b: mutation of Glu 233 of motif 1 led to a complete inhibition of the glycan chain elongation, suggesting an active role of the carboxyl group in the catalytic reaction (21). The kinetic parameters of the GT activity of PBP2a* were calculated from the Michaelis-Menten equation. The Km determined was 40.6 μM (± 15.5), which is 20-fold higher than the Km calculated for E. coli PBP1b (21) but still on the micromolar range. The kcat/Km efficiency of PBP2a* is much lower (approximately 7 orders of magnitude) than the E. coli PBP1b value (19, 21). The TLC method used in measuring the peptidoglycan synthesis by PBP2a* cannot be invoked for this discrepancy because similar E. coli PBP1b kcat/Km values were obtained by using either TLC (with radiolabeled lipid II) or a high-performance liquid chromatography-based assay (fluorescent substrate) (19, 21). The low kcat/Km value might be due to the lack of an important component in our in vitro assay. The screening of more reaction conditions will depend on the availability of a larger quantity of lipid II. Furthermore, the PBP2a* form (deleted from the cytoplasmic and transmembrane helix) used in this assay may not be optimal in terms of catalytic efficiency. Indeed, the E. coli PBP1b protein with the high kcat/Km value comprises its transmembrane anchor (21). Alternatively, we cannot exclude that the apparent low kcat/Km value of PBP2a* is due to the unavailability of most of the lipid II substrate, as it may be sequestered into vesicles.
Moenomycin is a well-known inhibitor of GT activity, and its antibiotic property has been well established (23). We and coworkers have previously shown moenomycin binding on the GT domain of PBP1a and PBP1b from S. pneumoniae (3, 4). In this work, we showed that glycan chain polymerization catalyzed by S. pneumoniae PBP2a* is inhibited by moenomycin. When this antibiotic was used at 2.8 μM, i.e., a moenomycin-to-lipid II molar ratio of 0.1:1, the quantity of polymerized material decreased by 50%, showing that the affinity of moenomycin for S. pneumoniae PBP2a* is in the micromolar range.
Vancomycin binds to the d-alanyl-d-alanine terminus of the peptidoglycan precursors (11). Consequently, the peptide cross bridge catalyzed by TP is competitively inhibited, and to a lesser extent, the GT reaction is inhibited through steric hindrance. The lipid II used in the enzymatic assay described in this work bears a dansyl tag on the l-lysine, preventing cross-linking by the TP activity. We observed inhibition of the GT reaction when vancomycin was added, although it was added in a larger quantity than moenomycin (not exceeding 10-fold larger, however). Our in vitro data support direct proof of the inhibition of glycosyltransferase activity by vancomycin.
In this work, the efficient polymerization of peptidoglycan glycan chains catalyzed by S. pneumoniae PBP2a* is described and characterized for the first time. In addition, a membrane-recognizing region, located between Lys 78 and Ser 156 and containing motif 1 of the GT domain, has also been identified. The functional GT domain of PBP2a* should be useful for developing screening procedures aimed at identifying antibiotics against S. pneumoniae.
Acknowledgments
We are very grateful to Larry C. Blaszczak (Infectious Diseases Division, Lilly Research Laboratories) for the generous gift of dansylated lipid II. We thank Nicolas Mouz (Protein'eXpert, Grenoble, France) for encouragement and helpful discussions.
REFERENCES
- 1.Di Guilmi, A. M., and A. Dessen. 2002. New approaches towards the identification of antibiotic and vaccine targets in Streptococcus pneumoniae. EMBO Rep. 3:728-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Di Guilmi, A. M., A. Dessen, O. Dideberg, and T. Vernet. 2002. Bifunctional penicillin-binding proteins: focus on the glycosyltransferase domain and its specific inhibitor moenomycin. Curr. Pharm. Biotechnol. 3:63-75. [DOI] [PubMed] [Google Scholar]
- 3.Di Guilmi, A. M., A. Dessen, O. Dideberg, and T. Vernet. 2003. Functional characterization of penicillin-binding protein 1b from Streptococcus pneumoniae. J. Bacteriol. 185:1650-1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Di Guilmi, A. M., N. Mouz, J. P. Andrieu, J. Hoskins, S. R. Jaskunas, J. Gagnon, O. Dideberg, and T. Vernet. 1998. Identification, purification, and characterization of transpeptidase and glycosyltransferase domains of Streptococcus pneumoniae penicillin-binding protein 1a. J. Bacteriol. 180:5652-5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Di Guilmi, A. M., N. Mouz, L. Martin, J. Hoskins, S. R. Jaskunas, O. Dideberg, and T. Vernet. 1999. Glycosyltransferase domain of penicillin-binding protein 2a from Streptococcus pneumoniae is membrane associated. J. Bacteriol. 181:2773-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goffin, C., and J.-M. Ghuysen. 1998. Multimodular penicillin-binding proteins: an enigmatic family of orthologs and paralogs. Microbiol. Mol. Biol. Rev. 62:1079-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goffin, C., and J. M. Ghuysen. 2002. Biochemistry and comparative genomics of SxxK superfamily acyltransferases offer a clue to the mycobacterial paradox: presence of penicillin-susceptible target proteins versus lack of efficiency of penicillin as therapeutic agent. Microbiol. Mol. Biol. Rev. 66:702-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldman, R. C., and D. Gange. 2000. Inhibition of transglycosylation involved in bacterial peptidoglycan synthesis. Curr. Med. Chem. 7:801-820. [DOI] [PubMed] [Google Scholar]
- 9.Gordon, E., N. Mouz, E. Duee, and O. Dideberg. 2000. The crystal structure of the penicillin-binding protein 2x from Streptococcus pneumoniae and its acyl-enzyme form: implication in drug resistance. J. Mol. Biol. 299:477-485. [DOI] [PubMed] [Google Scholar]
- 10.Hoskins, J., P. Matsushima, D. L. Mullen, J. Tang, G. Zhao, T. I. Meier, T. I. Nicas, and S. R. Jaskunas. 1999. Gene disruption studies of penicillin-binding proteins 1a, 1b, and 2a in Streptococcus pneumoniae. J. Bacteriol. 181:6552-6555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim, S. J., L. Cegelski, D. R. Studelska, R. D. O'Connor, A. K. Mehta, and J. Schaefer. 2002. Rotational-echo double resonance characterization of vancomycin binding sites in Staphylococcus aureus. Biochemistry 41:6967-6977. [DOI] [PubMed] [Google Scholar]
- 12.Kurz, M., W. Guba, and L. Vertesy. 1998. Three-dimensional structure of moenomycin A—a potent inhibitor of penicillin-binding protein 1b. Eur. J. Biochem. 252:500-507. [DOI] [PubMed] [Google Scholar]
- 13.Lazar, K., and S. Walker. 2002. Substrate analogues to study cell-wall biosynthesis and its inhibition. Curr. Opin. Chem. Biol. 6:786-793. [DOI] [PubMed] [Google Scholar]
- 14.Lim, D., and N. C. Strynadka. 2002. Structural basis for the beta-lactam resistance of PBP2a from methicillin-resistant Staphylococcus aureus. Nat. Struct. Biol. 9:870-876. [DOI] [PubMed] [Google Scholar]
- 15.Mahapatra, S., S. Bhakta, J. Ahamed, and J. Basu. 2000. Characterization of derivatives of the high-molecular-mass penicillin-binding protein (PBP) 1 of Mycobacterium leprae. Biochem. J. 350:75-80. [PMC free article] [PubMed] [Google Scholar]
- 16.Paik, J., I. Kern, R. Lurz, and R. Hakenbeck. 1999. Mutational analysis of the Streptococcus pneumoniae bimodular class A penicillin-binding proteins. J. Bacteriol. 181:3852-3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Popham, D. L., and P. Setlow. 1996. Phenotypes of Bacillus subtilis mutants lacking multiple class A high-molecular-weight penicillin-binding proteins. J. Bacteriol. 178:2079-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schiffer, G., and J.-V. Höltje. 2039. 1999. Cloning and characterization of PBP1c, a third member of the multimodular class A penicillin-binding proteins of Escherichia coli. J. Biol. Chem. 274:32031-32033. [DOI] [PubMed] [Google Scholar]
- 19.Schwartz, B., J. A. Markwalder, S. P. Seitz, Y. Wang, and R. L. Stein. 2002. A kinetic characterization of the glycosyltransferase activity of Escherichia coli PBP1b and development of a continuous fluorescence assay. Biochemistry 41:12552-12561. [DOI] [PubMed] [Google Scholar]
- 20.Schwartz, B., J. A. Markwalder, and Y. Wang. 2001. Lipid II: total synthesis of the bacterial cell wall precursor and utilization as a substrate for glycosyltransfer and transpeptidation by penicillin binding protein (PBP) 1b of Escherichia coli. J. Am. Chem. Soc. 123:11638-11643. [DOI] [PubMed] [Google Scholar]
- 21.Terrak, M., T. K. Ghosh, J. van Heijenoort, J. Van Beeumen, M. Lampilas, J. Aszodi, J. A. Ayala, J. M. Ghuysen, and M. Nguyen-Disteche. 1999. The catalytic, glycosyl transferase and acyl transferase modules of the cell wall peptidoglycan-polymerizing penicillin-binding protein 1b of Escherichia coli. Mol. Microbiol. 34:350-364. [DOI] [PubMed] [Google Scholar]
- 22.van Heijenoort, J. 2001. Formation of the glycan chains in the synthesis of bacterial peptidoglycan. Glycobiology 11:25R-36R. [DOI] [PubMed]
- 23.van Heijenoort, Y., M. Leduc, H. Singer, and J. van Heijenoort. 1987. Effects of moenomycin on Escherichia coli. J. Gen. Microbiol. 133:667-674. [DOI] [PubMed] [Google Scholar]
- 24.VanNieuwenhze, M. S., S. C. Mauldin, M. Zia-Ebrahimi, B. E. Winger, W. J. Hornback, S. L. Saha, J. A. Aikins, and L. C. Blaszczak. 2002. The first total synthesis of lipid II: the final monomeric intermediate in bacterial cell wall biosynthesis. J. Am. Chem. Soc. 124:3656-3660. [DOI] [PubMed] [Google Scholar]
- 25.Wang, C.-C., D. E. Schultz, and R. A. Nicholas. 1996. Localization of a putative second membrane association site in penicillin-binding protein 1b of Escherichia coli. Biochem. J. 316:149-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Youssif, S. Y., J. K. Broome-Smith, and B. G. Spratt. 1985. Lysis of Escherichia coli by beta-lactam antibiotics: deletion analysis of the role of penicillin-binding proteins 1a and 1b. J. Gen. Microbiol. 131:2839-2845. [DOI] [PubMed] [Google Scholar]