Abstract
We show that the disruption of one of the mycocerosic acid synthase (mas)-like genes, msl5 (pks8 plus pks17) in Mycobacterium tuberculosis H37Rv generates a mutant incapable of producing monomethyl branched unsaturated C16 to C20 fatty acids that are minor constituents of acyltrehaloses and sulfolipids. The msl5 mutation did not cause any significant change in the acyl lipid composition and also did not affect growth in culture, in mouse alveolar macrophage cell line MH-S, or in the murine lung.
Mycobacterium tuberculosis infection is present in a latent form in one-third of the world population, and 5 to 10% of them will probably develop active tuberculosis some time during their life. This disease accounts for more than a quarter of all preventable adult deaths (32). The high degree of success of this pathogen is, to a large extent, due to its ability to evade the natural host defenses and antimycobacterial therapy. The unusually lipid-rich (50 to 60%) cell wall of the pathogen, which constitutes an impermeable barrier, plays a major role in its ability to successfully infect its host (12, 19, 21, 23). Reflecting the unusual variety and content of lipids in this organism, its genome contains an unusually large number of genes that show homology to genes involved in lipid metabolism (7). Many of these belong to the polyketide synthase (pks) family. These genes encode large multifunctional proteins that contain all of the domains required to catalyze the various steps involved in fatty acid synthesis. One of these, mycocerosic acid synthase, which catalyzes the synthesis of multiple methyl branched fatty acids, has been purified and characterized (22), and the gene that encodes this protein (mas) has been cloned (20). Because isolation and characterization of the many large proteins of similar size and similar functions would pose a technical challenge, a genetic approach has been used to identify the biochemical functions of the pks genes. Based on the lipids missing in the mutants in which specific pks genes have been disrupted, the biochemical functions of some pks genes have been deduced (3, 6, 9, 14, 24, 27-29). However, the biochemical functions of most pks genes remain unknown.
Based on homology to catalytic domains involved in fatty acid synthesis, pks8 would encode ketoacyl synthase (KS), acyl transferase (AT), dehydratase (DH), and enoyl reductase (ER) domains, whereas the adjoining pks17 gene would encode a ketoreductase (KR) and acyl carrier protein (ACP) domain. Thus, the products of pks8 and pks17 together would contain a complete set of domains required to make a saturated fatty acid. Because of its similarity to mas, we designated this combination of pks8 and pks17 as msl5 (27). To elucidate the nature of the branched fatty acids generated by the msl5 gene product, we disrupted this gene and used [1-14C]propionic acid as a radiotracer to identify the fatty acids missing in the msl5 mutant. We report that this approach identified the msl5 product as the one responsible for the synthesis of 2-methyl branched unsaturated C18 fatty acid and homologues that are esterified mainly to acyltrehaloses and sulfated acyltrehaloses as minor constituents. The absence of these fatty acids does not cause any significant change in the composition of the major classes of lipids present in the pathogen and does not significantly decrease the virulence of this pathogen in a murine alveolar macrophage cell line or in mice intranasally infected with the pathogen.
Disruption of the msl5 mutant of M. tuberculosis.
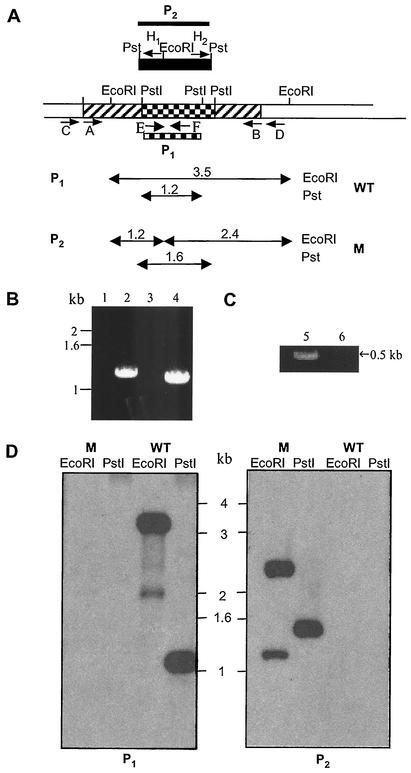
BLAST search of the M. tuberculosis genome for open reading frames (ORFs) with homology to mas and fatty acid synthase (fas) genes identified two adjacent ORFs, annotated as pks8 (Rv1662) and pks17 (Rv1663), both of which together contained one set of active site domains needed to catalyze all of the steps required for the synthesis of a saturated fatty acid. With its homology to mas, we designated pks8 plus pks17 as msl5 (27). In order to determine the nature of the fatty acids produced by the synthase encoded by msl5, a gene-disruption construct was made and used to produce msl5-defective mutants of M. tuberculosis H37Rv (ATCC no. 25618) via homologous recombination using a phage-mediated gene replacement strategy (4, 13, 27). A portion of the msl5 gene (including part of the DH, ER, KR, and ACP domains; bp 4014 of the pks8 coding sequence to bp 631 of the pks17 coding sequence) was amplified from M. tuberculosis genomic DNA by using the sense primer A and antisense primer B (Table 1), introducing BamHI sites at the 5′ and 3′ ends of the sequence. The 3,466-bp PCR product was cloned into BamHI-digested pUC19 vector (in which the PstI site had been eliminated), and a 1,423-bp PstI fragment of msl5 was replaced by a hygromycin resistance gene (hyg). The resulting disrupted msl5 gene and flanking regions were used to produce hygromycin-resistant clones as previously described (27). To confirm the disruption of the msl5 gene, PCR amplification was performed directly on cell lysate obtained by boiling the cells by using standard protocols (25) and Platinum Taq polymerase (Life Technology) along with a set of primers E and F (Table 1) located within the msl5 segment that was replaced with hyg gene. No PCR product was obtained when genomic DNA from the mutant was used, indicating the absence of this segment in the mutant, as expected from msl5 gene replacement. Thus, the hygromycin-resistant mutants failed to amplify a 515-bp PCR product (Fig. 1C). Further PCR analysis confirmed the disruption of the msl5 gene by allelic exchange using two other sets of primers each containing a hyg primer and a primer in the mycobacterial genome directly outside the msl5 sequences used to make the disruption construct: sense primer C and antisense primer H1 for amplification of the 5′-flanking region and sense primer H2 and antisense primer D for amplification of the 3′-flanking region (Table 1). Primer sets C and H1 and D and H2 generated the expected 1,247-bp 5′-flanking and 1,079-bp 3′-flanking products, respectively, whereas the wild type did not yield any products with these primers (Fig. 1B). Southern blot analysis of this mutant confirmed msl5 replacement. Genomic DNA samples from wild-type and mutant strains were digested with EcoRI and PstI and analyzed with probes to both the hygromycin resistance cassette (P2) and the msl5 gene sequence, which was replaced with hyg gene (P1). When P1 was used as a probe, the wild type showed native hybridization bands of the expected sizes (3.5 and 1.2 kb), and the msl5-disrupted mutant did not show hybridization signal, confirming integration by double-crossover recombination (Fig. 1D, P1). Analysis of the same blot with the hyg gene as a probe yielded a hybridization pattern consistent with replacement of the deleted msl5 segment with the hyg gene in the msl5 mutant; a 1.6-kb hybridization band corresponding to the replacement of the internal 1.2-kb PstI fragment with the 1.6-kb hyg cassette in the PstI site, and two EcoRI bands due to the presence of an EcoRI site in the hyg cassette (Fig. 1D, P2). As expected, the wild type did not show any hybridization with probe P2.
TABLE 1.
Sequence of the primers used in the disruption of msl5 in M. tuberculosis
| Primer | Sequence |
|---|---|
| A | 5′-GGATCCCGCTGGAGTGGTGTTGCTC-3′ |
| B | 5′-GGATCCCCAACGTGCACGCGATTGG-3′ |
| C | 5′-CCGGATTCGGACGTGGTGGTACTG-3′ |
| D | 5′-GACGTCGCCAGTAGGCCGCTGATC-3′ |
| E | 5′-GCGCGGGCGCCCTTGAGGAT-3′ |
| F | 5′-GTCCGGCCTCCATCAAGTCG-3′ |
| H1 | 5′-TGGACCTCGACGACCTGCAGGCAT-3′ |
| H2 | 5′-GGAACTGGCGCAGTTCCTCTGGGG-3′ |
FIG. 1.
(A) Schematic representation of the construct used to inactivate the M. tuberculosis H37Rv msl5 gene by allelic exchange. Hatched, checked, and open regions represent msl5 coding sequences, an internal deletion region, and regions of the gene outside those used to make the disruption construct, respectively. A black box indicates the hyg gene used for targeted disruption. Primer pair A/B was used to amplify the msl5 segment used to generate the deletion construct. Primer pairs C/H1, D/H2, and E/F were used for PCR analysis of homologous recombination as described in the text. P1 and P2 are the segments used as probes in Southern blot analysis, P1 represents the msl5 fragment deleted in making the construct, and P2 represents the hyg gene. (B and C) PCR analysis of flanking and internal regions of the msl5 gene showing products expected from homologous recombination. Lanes 1, 3, and 5 contain the wild type; lanes 2, 4, and 6 contain the mutant. Lanes 1 and 2 contain 5′-flanking product with primers C/H1. Lanes 3 and 4 contain 3′-flanking products with primers D/H2. Lanes 5 and 6 contain internal deletion fragments with primers E/F. (D) Southern blot analysis of M. tuberculosis H37Rv wild type (WT) and the msl5 mutant. Genomic DNA was digested with EcoRI and PstI. In the left panel, DNA was hybridized with probe P1, the 1.2-kb fragment that was deleted in making the construct. In the right panel, DNA was hybridized with probe P2, the hyg gene. WT, wild type; M, mutant.
Biochemical characterization of the msl5 gene-disrupted mutant.
To seek the identity of the fatty acids generated by the msl5 product, [1-14C]propionate was used as a precursor to label methyl branched fatty acids in the wild-type and mutant cells, because msl5 is expected to encode a mycocerosic acid synthase-like enzyme that is expected to catalyze the synthesis of methyl branched fatty acids. Na [1-14C]propionate (50 μCi, specific activity, 55 Ci/mol) (American Radiolabeled Chemicals, St. Louis, Mo.) was added to the cultures of M. tuberculosis H37Rv and its msl5 mutant (optical density at 600 nm [OD600] of 1.6 to 1.8), and incubation was continued at 37°C in roller bottles for further 24 h. Cells were used for lipid extraction with an excess of chloroform-methanol (2:1), and the total cellular lipids were assayed for total 14C in a Beckman LA3801 liquid scintillation counter as described previously (27). Both the wild type and the msl5 mutant incorporated similar amounts of radioactivity (20 to 25% of administered [14C]propionate) into total lipids in 24 h. The fractionation of total labeled lipids into nonpolar and polar lipids and the separation of various polar lipids were done exactly in the same manner as previously described (28). The lipids were visualized, and the 14C-labeled lipids were detected by scanning chromatograms in a Berthold Tracemaster 20 automatic thin-layer chromatography (TLC) linear analyzer and by autoradiography as described previously (27).
When the total [14C]propionate-derived lipids were separated by TLC, with 10% ethyl ether in hexane as a solvent, in both the wild type and the msl5 mutant, about 30% of the total radioactivity incorporated into the lipids was found in the nonpolar lipids with an Rf of 0.5 to 0.8, containing dimycocerosyl phthiocerols (DIM) and wax esters. The rest of the label in both cases remained at the origin. When these polar lipids were fractionated by TLC with 10% methanol in chloroform, the labeling pattern of the various classes of lipids in the msl5 mutant was virtually identical to that observed with the wild type, as previously reported (28). In both the wild type and msl5 mutant, the major labeled lipids were sulfolipids (SLs) and two classes of polyacyl trehaloses (PATs), and there was much less label in diacyl trehaloses (DATs).
A detailed analysis of the fatty acids in the major lipid classes derived from [14C]propionate was also done. The labeled lipids were recovered and subjected to alkaline hydrolysis followed by methylation as described previously (27). Methyl esters of the fatty acids and acetylated hydroxy fatty acids were analyzed by radio-gas chromatography (GC) as previously described (27). The methyl esters of fatty acids from the total lipids of both the wild type and msl5 mutant were also separated by argentation-TLC (7.5% AgNO3 in silica gel) by being developed twice with hexane-ethyl ether (9:1 [vol/vol]) as the solvent, and each separated fatty acid methyl ester fraction was analyzed by radio-GC (14). Virtually all of the label in the nonpolar fraction, containing DIM, was found in the mycocerosic acids in both the wild type and the msl5 mutant. A combination of argentation-TLC and radio-GC showed no difference in the profiles of the major fatty acids of individual lipids, because the major labeled fatty acids in the PATs were mycolipanoic, mycolipenic, and mycolipodienoic acids, and there was much less label in methyl branched shorter-chain fatty acids, mycocerosic and phthioceranic acids, as previously reported (14). The argentation-TLC profiles of fatty acid methyl esters derived from SL in the wild type and mutant showed only a small difference. The wild type contained a small amount of label in a fraction, which upon radio-GC was found to contain a mixture of 2-methyl branched unsaturated C16 to C20 fatty acids, whereas this fraction was absent in the msl5 mutant. Besides SLs, these labeled 2-methyl unsaturated fatty acid components were also present in a very small amount in other glycolipids, mainly in the polar PATs and DATs in the wild type (data not shown). There was no significant difference in the 14C distribution in the hydroxy fatty acids (mycolipanolic and hydroxyphthioceranic acids) between the wild type and the msl5 mutant. Radio-GC analyses of acetylated methyl esters of mycolipanolic and hydroxyphthioceranic acids showed that the chain length distributions of label in each fraction were the same in the wild type and the msl5 mutant (data not shown).
To conduct a more detailed examination of the fatty acids derived from [1-14C]propionic acid, we used the combined argentation-TLC and radio-GC analyses of total fatty acid methyl esters. GC-mass spectrometry (MS) analysis was also performed to confirm the identity of the fatty acids. GC-MS analysis of fatty acid methyl esters was done on Thermo-Finnigan Trace MS 2000 (Thermoquest; CE Instrument) using a capillary column (30 m, 0.32-mm inside diameter, 0.25-μm film; XTI-5V/Integra-Guard) with 5% diphenyl-bonded cross-linked phase-95% dimethyl polysiloxane. The column temperature was held for 1 min at 35°C, followed by a program run to 200°C at 16°C/min and finally programmed to run to 360°C at the rate of 4.5°C/min. The methyl esters of the hydroxy acids were analyzed by GC-MS as their trimethylsilyl (TMS) derivative prepared by treatment with N-methyl-N(trimethylsilyl)-trifluroacetamide (MSTFA; Sigma-Aldrich Chemical Co.) for 10 min at room temperature before injection. The unsaturated fatty acid methyl ester fraction, separated by argentation-TLC, was stirred with osmium tetraoxide in ethyl ether-pyridine (8:1 [vol/vol]) for 2 h at room temperature, followed by addition of a saturated solution of sodium sulfite in aqueous methanol (1:1 [vol/vol]) (11). The vicinal diol products were extracted with chloroform three times and subjected to GC-MS analysis as their TMS derivative as described above.
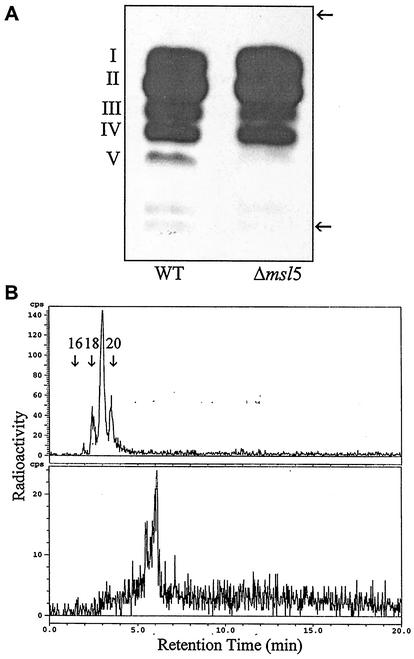
Radio-GC analysis of fatty acid methyl esters from total lipids did not reveal any differences between the wild type and the mutant (data not shown). However, argentation-TLC (7.5% AgNO3 in silica gel) resolved the total fatty acid methyl esters from the wild type into five components, whereas only four components were found in fatty acids from the msl5 mutant (Fig. 2A). Radio-GC and GC-MS analyses of each methyl ester fraction from the wild type showed that the least polar fraction (fraction I, 25% of total labeled fatty acids) contained mycocerosic acids and phthioceranic acids. Fraction II (40% of total labeled fatty acids) contained mycolipanoic and mycolipenic acids. Fraction III (18% of total labeled fatty acids) contained C16 and C18 methyl branched acids. Fraction IV (12% of total labeled fatty acids) contained mainly mycolipodienoic acids, as previously reported (14). The distributions of radioactivity in fractions I to IV were very similar in both the wild type and the msl5 mutant. Fraction V from the wild type, which was absent in the msl5 mutant, contained less than 5% of the label found in the total labeled fatty acids. Radio-GC showed that fraction V contained a mixture of unsaturated methyl branched fatty acids with retention times between those of n-C16 and n-C20 (Fig. 2B top), and after bromination, retention time increased dramatically, showing the presence of unsaturation (Fig. 2B, bottom). The major one had a retention time between those of n-C18 and n-C20 (Fig. 2B, top), indicating that it is 2-methyl C18 acid. The fragmentation pattern in the mass spectrum indicated that it is an unsaturated acid as previously described (16, 26) (Table 2). The fact that the McLafferty ion at m/e 88, characteristic of 2-methyl saturated acids, was not a significant ion indicated that one of the double bonds is at the C-2 position. TLC of the osmylation products of the labeled acid in fraction V showed that all of the 14C was contained in a diol. GC-MS of the TMS derivative of the diol showed that there was a double bond at C-9 of the 2-methyl C18 acid. Thus, this unsaturated acid is identified as 2-methyloctadec-2,9-dienoic acid. The other homologues of the 2-methyl branched acids were relatively minor components (Table 2).
FIG. 2.
(A) Argentation-TLC of total fatty acid methyl esters derived from [1-14 C]propionate in M. tuberculosis H37Rv (wild type [WT]) and its msl5 mutant (Δmsl5). (B) Radio-gas chromatograms of the most polar fraction, V, from the wild type (top) and after bromination of the same fraction (bottom). Arrows show the origin and the solvent front. The 14C distributions in each fraction (except fraction V, missing in the mutant) from the wild type and mutant were identical. n-C16, n-C18, and n-C20 fatty acids are indicated.
TABLE 2.
Composition of monomethyl branched unsaturated fatty acids from methyl ester fraction V of argentation-TLC from wild-type M. tuberculosis H37Rv (missing in the msl5 mutant)
| Fatty acida | Relative amt in methyl ester fraction V (%)b | Diagnostic ions used for identificationc |
|---|---|---|
| 2-Me C16:1 | 16 | 55, 88, 101, 250, 282 |
| 2-Me C17:1 | 7 | 55, 88, 101, 264, 296 |
| 2-Me C18:1 | 3 | 55, 88, 101, 278, 310 |
| 2-Me C18:2 | 58 | 55, 88, 101, 127, 277, 308 |
| 2-Me C19:1 | 4 | 55, 88, 101, 292, 324 |
| 2-Me C19:2 | 3 | 55, 88, 101, 127, 291, 322 |
| 2-Me C20:1 | 2 | 55, 88, 101, 306, 338 |
| 2-Me C20:2 | 7 | 55, 88, 101, 127, 304, 338 |
Identified by GC-MS by their number of carbon atoms and double bond. Me, methyl.
Fraction V was derived from argentation-TLC of total fatty acid methyl ester from the wild type (Fig. 2A).
The ion at m/e 88 was much weaker than that observed with 2-methyl branched acids without a double bond at position C-2.
Na [1-14C]acetate (50 μCi; specific activity, 56.7 Ci/mol) (American Radiolabeled Chemicals, St. Louis, Mo.) was also used as a radiolabeled precursor to examine any changes in the lipid/fatty acid profile. Methyl esters of mycolic acids and n-fatty acids, prepared from cells incubated with [14C]acetate, were also analyzed as described previously (18). Both the wild type and the msl5 mutant incorporated similar amounts of radioactivity (10 to 12% of administered [14C]acetate) into total lipids in 24 h. Saturated and unsaturated n-C16 and n-C18 were the major labeled fatty acids, comprising 65% of the total labeled lipids. A significant amount of 14C (about 6%) was also found in longer homologues, mainly n-C26 and n-C28, with much less found in n-C24 fatty acids. Mycolate methyl esters contained 12.5% of total label. In addition, labeled mycolic acids were also found attached to the cell walls, amounting to about 17% of the total labeled lipids. msl5 mutation did not affect this labeling pattern.
Virulence of the msl5 mutant of M. tuberculosis H37Rv.
Growth in culture was not affected by msl5 mutation. We tested whether the growth of M. tuberculosis in a murine alveolar macrophage cell line, MH-S (ATCC CRL-2019), was affected by the msl5 mutation by using procedures described previously (28). During a 5-day growth period, the wild type and msl5 mutant grew equally well, whereas some other msl mutants we have generated showed a measurable attenuation within this period of growth (28, 29).
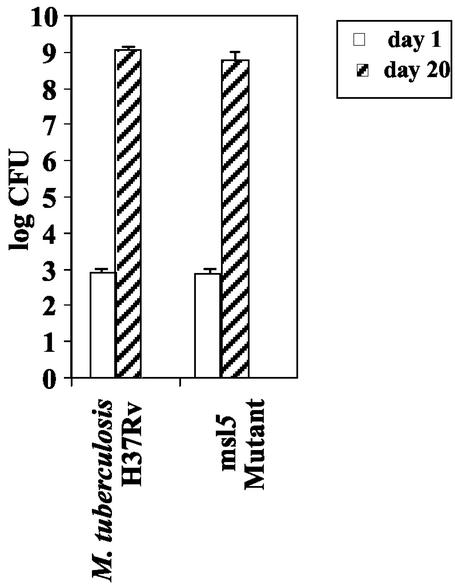
Growth of the wild-type M. tuberculosis strain H37Rv and its msl5 mutant in the murine lung was also measured over a period of 20 days after intranasal administration of the pathogen as described previously (15, 28). The mutant grew as rapidly as the wild type during this period of growth: there was no obvious difference in the growth patterns between the wild type and the msl5 mutant (Fig. 3), whereas some other msl mutants deficient in DIM synthesis showed a measurable attenuation within this period (28, 29). In these experiments, the level of growth of the wild type was found to be very similar to those previously published (15). Even though the number of CFU reached almost 109 per lung, there was no mortality during a 20-day period, although the animals appeared sick.
FIG. 3.
Growth of intranasally administered M. tuberculosis H37Rv and its msl5-disrupted mutant in the lungs of C57BL6/J mice.
Involvement of the msl5 product in the synthesis of monomethyl unsaturated fatty acids.
The lipid-rich nature of the mycobacterial cell wall is reflected in the presence of a large number of pks genes in the genome of this pathogen (7). Some of the pks genes encode proteins that contain all of the catalytic sites required for the synthesis of fatty acids, while others contain only some of the catalytic domains. Two adjacent ORFs, annotated pks8 and pks17, together contain all of the catalytic domains required for the synthesis of a fatty acid. pks8 would encode a protein of 1,602 amino acids with a deduced molecular mass of 167 kDa containing KS, AT, DH, and ER domains, and the adjoining pks17 gene would encode a protein of 502 amino acids with a deduced molecular mass of 53.5 kDa containing KR and ACP domains. Thus, these two ORFs together would constitute a full complement of domains for a fatty acid synthase gene that we designate msl5. msl5 is located between two other pks genes, pks7 (Rv1661) and pks9 (Rv1664). pks7, a 2,126-amino-acid ORF located 19 bp upstream of msl5, contains all the domains required for the synthesis of a branched fatty acid with the same domain organization as mas and shows a 61% identity with pks8. At 5 bp downstream of msl5 is situated pks9, a 1,017-amino-acid ORF containing KS, AT, and ACP domains with 49% identity to pks8. Since msl5 disruption did not prevent DIM synthesis, it is concluded that disruption of msl5 did not affect the expression of the upstream pks7 gene, whose disruption inhibits DIM synthesis (24). Conversely, pks7 disruption did not cause any inhibition of synthesis of the monomethyl branched acids, suggesting that pks7 disruption did not affect msl5 function.
In some cases, two adjoining ORFs found in the genome of M. tuberculosis have been found to be in a single ORF in Mycobacterium bovis BCG, indicating that the organization of genes required for encoding a functional synthase may vary within the M. tuberculosis complex. For example, pks15 and pks1 in M. tuberculosis exist as a single ORF in M. bovis BCG (9, 28). Even within M. tuberculosis, different strains may have different organizations. For example, pks3 and pks4 in the H37Rv strain that was originally sequenced (7) were found to be in a single ORF in H37Rv strain ATCC 25618 (14). In the present case of pks8 and pks17, the same dual-ORF organization is found in three strains of M. tuberculosis (H37Rv ATCC 25618, CDC 1551, and the H37Rv strain originally sequenced) and in M. bovis BCG, whereas these ORFs are presumably lost in Mycobacterium leprae (8). The functional consequence of these variations in genomic organization is not clear. If the split structure encodes two proteins that can together catalyze all of the reactions required for fatty acid synthesis, the split in the ORF would not have any functional consequence. The many reactions involved in fatty acid synthesis are catalyzed by enzymes organized in three different ways in different organisms (17, 31). In most bacteria, such as Escherichia coli, each reaction is catalyzed by a separate enzyme, whereas, in yeast, all of the catalytic activities are organized into two multifunctional peptides—one catalyzing some of the reactions and the other catalyzing the remaining reactions. In vertebrates, all active sites are contained in one large multifunctional protein. M. tuberculosis and closely related bacteria seem to be unique in that they use all three types of organizations for synthesizing different types of the wide variety of fatty acids they generate (17).
Many of the pks gene products that utilize methylmalonyl-coenzyme A (CoA) as the substrate generate multiple methyl branched fatty acids (2, 3, 14, 27, 28). The present case is the first example of elongation by a single methylmalonyl-CoA in M. tuberculosis. The resulting monomethyl branched short-chain acids were found in SLs, PATs, and the more polar DATs. Such unsaturated monomethyl branched acids have been reported in the various glycolipids of Mycobacterium fortuitum (1, 16, 26, 30), but their presence in M. tuberculosis has not been previously noted. The use of [1-14C]propionic acid and careful separation of the methyl esters by argentation chromatography allowed us to detect these minor components.
The major methyl branched acids, such as mycocerosic acids, being essential for the synthesis of DIM, are important for virulence (5, 10). Even though the molecular basis for the requirement of DIM for virulence is not understood, DIM deficiency has been found to cause attenuation. For example, disruption of four different genes, msl4 (pks7) (24), msl6 (pks12) (29), and msl7 (pks1 plus pks15) and pks10 (28), caused DIM deficiency and attenuation. On the other hand, msl5 encodes proteins that catalyze the synthesis of monomethyl branched unsaturated acids that are only minor components of the lipids of M. tuberculosis, and disruption of this gene does not lead to attenuation. Since these acids are not essential for the synthesis of any acyl lipids in M. tuberculosis, the msl5 mutant contains all of the major types of acyl lipids found in the wild type, and therefore it is not surprising that this mutant is not attenuated either in the alveolar macrophage cell line or in the murine model, in which the organism was introduced by the intranasal administration. The experimental condition we used for assessing the virulence might not detect subtle changes in virulence.
Acknowledgments
This work was supported by grants AI46582-03 and AI35272-10 from the National Institutes of Health.
Part of this work was done at the Neurobiotechnology Center at The Ohio State University, Columbus.
REFERENCES
- 1.Ariza, M. A., F. Martin-Luengo, and P. L. Valero-Guillen. 1994. A family of diacyltrehaloses isolated from Mycobacterium fortuitum. Microbiology 140:1989-1994. [DOI] [PubMed] [Google Scholar]
- 2.Azad, A. K., T. D. Sirakova, L. M. Rogers, and P. E. Kolattukudy. 1996. Targeted replacement of the mycocerosic acid synthase gene in Mycobacterium bovis BCG produces a mutant that lacks mycosides. Proc. Natl. Acad. Sci. USA 93:4787-4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Azad, A. K., T. D. Sirakova, N. D. Fernandes, and P. E. Kolattukudy. 1997. Gene knock-out reveals a novel gene cluster for the synthesis of a class of cell wall lipids unique to pathogenic mycobacteria. J. Biol. Chem. 272:16741-16745. [DOI] [PubMed] [Google Scholar]
- 4.Bardarov, S., S. Bardarov Jr., M. S. Pavelka, Jr., V. Sambandamurthy, M. Larsen, J. Tufariello, J. Chan, G. Hatfull, and W. R. Jacobs, Jr. 2002. Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG, and M. smegmatis. Microbiology 148:3007-3017. [DOI] [PubMed] [Google Scholar]
- 5.Camacho, L. R., D. Enserqueix, E. Perez, B. Gicquel, and C. Guilhot. 1999. Identification of a virulence gene cluster of Mycobacterium tuberculosis by signature-tagged transposon mutagenesis. Mol. Microbiol. 34:257-267. [DOI] [PubMed] [Google Scholar]
- 6.Camacho, L. R., P. Constant, C. Raynaud, M. A. Lanéelle, J. A. Triccas, B. Gicquel, M. Daffe, and C. J. Guilhot. 2001. Analysis of the phthiocerol dimycocerosate locus of Mycobacterium tuberculosis. Evidence that this lipid is involved in the cell wall permeability barrier. J. Biol. Chem. 276:19845-19854. [DOI] [PubMed] [Google Scholar]
- 7.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, S. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M.-A. Rajandream, J. Roger, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 8.Cole, S. T., K. Eiglmeier, J. Parkhill, K. D. James, N. R. Thomson, P. R. Wheeler, N. Honore, T. Garnier, C. Churcher, D. Harris, K. Mungall, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. M. Davies, K. Devlin, S. Duthoy, T. Feltwell, A. Fraser, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, C. Lacroix, J. Maclean, S. Moule, L. Murphy, K. Oliver, M. A. Quail, M.-A. Rajandream, K. M. Rutherford, S. Rutter, K. Seeger, S. Simon, M. Simmonds, J. Skelton, R. Squares, S. Squares, K. Stevens, K. Taylor, S. Whitehead, J. R. Woodward, and B. G. Barrell. 2001. Massive gene decay in the leprosy bacillus. Nature 409:1007-1011. [DOI] [PubMed] [Google Scholar]
- 9.Constant, P., E. Perez, W. Malaga, M.-A. Laneelle, O. Saurel, M. Daffe, and C. Guilhot. 2002. Role of the pks15/1 gene in the biosynthesis of phenolglycolipids in the M. tuberculosis complex: evidence that all strains synthesize glycosylated p-hydroxybenzoic methyl esters and that strains devoid of phenolglycolipids harbour a frameshift mutation in the pks 15/1 gene. J. Biol. Chem. 277:38148-38158. [DOI] [PubMed] [Google Scholar]
- 10.Cox, J. S., B. Chen, M. McNeil, and W. R. Jacobs. 1999. Complex lipid determines tissue-specific replication of Mycobacterium tuberculosis in mice. Nature 402:79-83. [DOI] [PubMed] [Google Scholar]
- 11.Croteau, R., and P. E. Kolattukudy. 1974. Direct evidence for the involvement of epoxide intermediates in the biosynthesis of the C18 family of cutin acids. Arch. Biochem. Biophys. 162:471-480. [DOI] [PubMed] [Google Scholar]
- 12.Daffe, M., and P. Draper. 1998. The envelope layers of mycobacteria with reference to their pathogenicity. Adv. Microb. Physiol. 39:131-203. [DOI] [PubMed] [Google Scholar]
- 13.Derbyshire, K. M., and S. Bardarov. 2000. DNA transfer in mycobacteria: conjugation and transduction, p. 93-107. In G. F. Hatfull and W. R. Jacobs, Jr. (ed.), Molecular genetics of mycobacteria. ASM Press, Washington, D.C.
- 14.Dubey, V. S., T. D. Sirakova, and P. E. Kolattukudy. 2002. Disruption of msl 3 abolishes the synthesis of mycolipanoic and mycolipenic acids required for polyacyltrehalose synthesis in Mycobacterium tuberculosis H37Rv and causes cell aggregation. Mol. Microbiol. 45:1451-1459. [DOI] [PubMed] [Google Scholar]
- 15.Edwards, K. M., M. H. Cynamon, R. K. Voladri, C. C. Hager, M. S. DeStefano, K. T. Tham, D. L. Lakey, M. R. Bochan, and D. S. Kernodle. 2001. Iron-cofactored superoxide dismutase inhibits host responses to Mycobacterium tuberculosis. Am. J. Respir. Crit. Care Med. 164:2213-2219. [DOI] [PubMed] [Google Scholar]
- 16.Gautier, N., L. M. Lopez-Marin, M.-A. Laneelle, and M. Daffe. 1992. Structure of mycoside F, a family of trehalose-containing glycolipids of Mycobacterium fortuitum. FEMS Microbiol. Lett. 98:81-88. [DOI] [PubMed] [Google Scholar]
- 17.Kolattukudy, P. E., N. D. Fernandes, A. K. Azad, A. M. Fitzmaurice, and T. D. Sirakova. 1997. Biochemistry and molecular genetics of cell wall lipid biosynthesis in mycobacteria. Mol. Microbiol. 24:263-270. [DOI] [PubMed] [Google Scholar]
- 18.Kremer, L., L. G. Dover, S. Carrere, K. M. Nampoothiri, S. Lesjean, A. K. Brown, P. J. Brennan, D. E. Minnikin, C. Locht, and G. S. Besra. 2002. Mycolic acid biosynthesis and enzymic characterization of the beta-ketoacyl-ACP synthase A-condensing enzyme from Mycobacterium tuberculosis. Biochem. J. 364:423-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu, J., C. E. Barry III, and H. Nikaido. 1999. Cell wall: physical structure and permeability, p. 220-239. In C. Ratledge and J. Dale (ed.), Mycobacteria: molecular biology and virulence. Blackwell Science, Malden, Mass.
- 20.Mathur, M., and P. E. Kolattukudy. 1992. Molecular cloning and sequencing of the gene for mycocerosic acid synthase, a novel fatty acid elongating multi-functional enzyme, from Mycobacterium tuberculosis var. bovis Bacillus Calmette-Guerin. J. Biol. Chem. 267:19388-19395. [PubMed] [Google Scholar]
- 21.Minnikin, D. E. 1982. Lipids: complex lipids, their chemistry, biosynthesis and roles, p. 95-184. In C. Ratledge and J. Stanford (ed.), The biology of the mycobacteria. Academic Press, London, United Kingdom.
- 22.Rainwater, D. L., and P. E. Kolattukudy. 1985. Fatty acid biosynthesis in Mycobacterium tuberculosis var. bovis Bacillus Calmette-Guerin. Purification and characterization of a novel fatty acid synthase, mycocerosic acid synthase, which elongates n-fatty acyl-CoA with methylmalonyl-CoA. J. Biol. Chem. 260:616-623. [PubMed] [Google Scholar]
- 23.Rhoades, E. R., and H. J. Ullrich. 2000. How to establish a lasting relationship with your host: lessons learned from Mycobacterium spp. Immunol. Cell Biol. 78:301-310. [DOI] [PubMed] [Google Scholar]
- 24.Rousseau, C., T. D. Sirakova, V. S. Dubey, Y. Bordat, P. E. Kolattukudy, B. Gicquel, and M. Jackson. 2003. Virulence attenuation of two Mas-like polyketide synthase mutants of Mycobacterium tuberculosis. Microbiology 149:1837-1847. [DOI] [PubMed]
- 25.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 26.Sempere, M. A., P. L. Valero-Guillen, A. E. Godos, and F. Martin-Luengo. 1993. A triacyltrehalose containing 2-methyl-branched unsaturated fatty acyl groups isolated from Mycobacterium fortuitum. J. Gen. Microbiol. 139:585-590. [Google Scholar]
- 27.Sirakova, T. D., A. K. Thirumala, V. S. Dubey, H. Sprecher, and P. E. Kolattukudy. 2001. The Mycobacterium tuberculosis pks2 gene encodes the synthase for the hepta- and octamethyl-branched fatty acids required for sulfolipid synthesis. J. Biol. Chem. 276:16833-16839. [DOI] [PubMed] [Google Scholar]
- 28.Sirakova, T. D., V. S. Dubey, M. H. Cynamon, and P. E. Kolattukudy. 2003. Attenuation of Mycobacterium tuberculosis by disruption of a mas-like gene or a chalcone synthase-like gene that causes deficiency in dimycocerosyl phthiocerol synthesis. J. Bacteriol. 185:2999-3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sirakova, T. D., V. S. Dubey, H.-J. Kim, M. H. Cyanamon, and P. E. Kolattukudy. 2003. The largest open reading frame (pks12) in the Mycobacterium tuberculosis genome is involved in dimycocerosyl phthiocerol synthesis and pathogenesis. Infect. Immun. 71:3794-3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Valero-Guillen, P. L., F. Martin-Luengo, L. Larsson, and J. Jimenez. 1987. Demonstration of 2-methyl branched chain fatty acids in some rapid-growing mycobacteria. FEMS Microbiol. Lett. 44:303-305. [Google Scholar]
- 31.Wakil, S. J. 1989. Fatty acid synthase, a proficient multifunctional enzyme. Biochemistry 28:4523-4530. [DOI] [PubMed] [Google Scholar]
- 32.World Health Organization. 2001. Global tuberculosis control. [Online.] World Health Organization, Geneva, Switzerland. http://www.who.int/gtb/publications/globrep01/.