Abstract
A scaffoldin gene cluster was identified in the mesophilic cellulolytic anaerobe Acetivibrio cellulolyticus. The previously described scaffoldin gene, cipV, encodes an N-terminal family 9 glycoside hydrolase, a family 3b cellulose-binding domain, seven cohesin domains, and a C-terminal dockerin. The gene immediately downstream of cipV was sequenced and designated scaB. The protein encoded by this gene has 942 amino acid residues and a calculated molecular weight of 100,358 and includes an N-terminal signal peptide, four type II cohesions, and a C-terminal dockerin. ScaB cohesins 1 and 2 are very closely linked. Similar, but not identical, 39-residue Thr-rich linker segments separate cohesin 2 from cohesin 3 and cohesin 3 from cohesin 4, and an 84-residue Thr-rich linker connects the fourth cohesin to a C-terminal dockerin. The scaC gene downstream of scaB codes for a 1,237-residue polypeptide that includes a signal peptide, three cohesins, and a C-terminal S-layer homology (SLH) module. A long, ca. 550-residue linker separates the third cohesin and the SLH module of ScaC and is characterized by an 18-residue Pro-Thr-Ala-Ser-rich segment that is repeated 27 times. The calculated molecular weight of the mature ScaC polypeptide (excluding the signal peptide) is 124,162. The presence of the cohesins and the conserved SLH module implies that ScaC acts as an anchoring protein. The ScaC cohesins are on a separate branch of the phylogenetic tree that is close to, but distinct from, the type I cohesins. Affinity blotting with representative recombinant probes revealed the following specific intermodular interactions: (i) an expressed CipV cohesin binds selectively to an enzyme-borne dockerin, (ii) a representative ScaB cohesin binds to the CipV band of the cell-free supernatant fraction, and (iii) a ScaC cohesin binds to the ScaB dockerin. The experimental evidence thus indicates that CipV acts as a primary (enzyme-recognizing) scaffoldin, and the protein was also designated ScaA. In addition, ScaB is thought to assume the role of an adaptor protein, which connects the primary scaffoldin (ScaA) to the cohesin-containing anchoring scaffoldin (ScaC). The cellulosome system of A. cellulolyticus thus appears to exhibit a special type of organization that reflects the function of the ScaB adaptor protein. The intercalation of three multiple cohesin-containing scaffoldins results in marked amplification of the number of enzyme subunits per cellulosome unit. At least 96 enzymes can apparently be incorporated into an individual A. cellulolyticus cellulosome. The role of such amplified enzyme incorporation and the resultant proximity of the enzymes within the cellulosome complex presumably contribute to the enhanced synergistic action and overall efficient digestion of recalcitrant forms of cellulose. Comparison of the emerging organization of the A. cellulolyticus cellulosome with the organizations in other cellulolytic bacteria revealed the diversity of the supramolecular architecture.
The microbial degradation of cellulose is one of the most important processes on Earth, and it affects the human condition in many direct and indirect ways. If it did not occur, there would be an inexhaustible accumulation of plant cell refuse, and herbivorous life forms would largely vanish.
The multienzyme cellulosome complex is a major mechanism by which some cellulolytic bacteria efficiently degrade cellulose and related plant cell wall polysaccharides (2, 4, 5, 8, 9, 18, 20, 32, 58, 59). To date, cellulosomes have been found in several strains of anaerobic bacteria and fungi obtained from very different types of ecosystems. The first cellulosome was discovered in studies on the anaerobic thermophile Clostridium thermocellum (3, 36, 37). The cellulosome system of this organism consists of a variety of different enzymes bound to a noncatalytic scaffoldin subunit, which can, in turn, bind to one of several cell surface anchoring proteins. In this organism, both the attachment of the enzymes to the scaffoldin and the attachment of the scaffoldin to the anchoring proteins are accomplished by a special kind of protein-protein interaction, the cohesin-dockerin interaction. In this context, the enzyme subunits include a dockerin domain, and the scaffoldin contains multiple copies of cohesin modules for collective incorporation into the complex. The scaffoldin subunit itself harbors a single dockerin variant that interacts selectively with corresponding cohesin variants on the anchoring proteins. In C. thermocellum, the primary scaffoldin is trifunctional in that it also contains (in addition to the cohesins and dockerin) a substrate-targeting cellulose-binding domain (CBD). The anchoring proteins are bifunctional; in addition to the cohesins, they have another type of domain, the S-layer homology domain (SLH), which is known to bind strongly to the cell surface (12, 40). Thus, the sequential cohesin-dockerin-mediated set of interactions among the enzymes, scaffoldin, and anchoring protein results in cell surface attachment of the cellulosome and amplification of the number of enzymes in the higher-order complex.
In other cellulosome-producing organisms, the supramolecular organization of the complex and its putative association with the cell surface are less clear. For example, in various mesophilic clostridia, the multicohesin, CBD-containing scaffoldins lack a dockerin (30, 51, 56, 60), and the exact mode of association with the cell surface is not known. Anchoring proteins for these organisms have not been identified. On the other hand, the two known scaffoldins of Ruminococcus flavefaciens lack both a defined CBD and an SLH module, and the molecular mechanism(s) for their interactions with either the cellulose substrate or the cell surface has not been substantiated (17, 52).
As the number of new cellulosomal components increases for different bacterial systems, the diversity of the molecular architecture and the resulting implications for cellulosome assembly become more striking. The cellulosome-producing, anaerobic mesophile Acetivibrio cellulolyticus, for example, is known both for its efficient degradation of crystalline cellulose and for its particularly elaborate cell surface ultrastructure (13, 31, 34, 35, 57). A novel scaffoldin (CipV) of this bacterium has recently been described and sequenced (15). Similar to the CipA scaffoldin of C. thermocellum (23), the A. cellulolyticus CipV scaffoldin contains multiple (seven) cohesin domains, an internally located CBD, and a C-terminal dockerin domain. Unlike all other scaffoldin genes discovered previously, however, cipV is the only gene that has been shown to encode a glycoside hydrolase sequence as an integral part of the deduced polypeptide chain. The analogy between the C-terminal dockerin of the A. cellulolyticus scaffoldin and the C-terminal dockerin of C. thermocellum indicated that there may be cohesin-containing anchoring proteins that are perhaps linked to the cell surface via resident SLH modules. Since a number of C. thermocellum genes that code for anchoring proteins are clustered immediately downstream of the CipA gene on the genome (22), we decided to continue sequencing the A. cellulolyticus genome in a similar manner.
MATERIALS AND METHODS
Preparation of A. cellulolyticus proteins.
Cellulose-binding extracellular proteins and cell-associated protein fractions were prepared from cellobiose-grown cells of A. cellulolyticus ATCC 33288 as described previously (15). Cellulose-binding proteins were obtained by adsorbing cell-free culture supernatant fluids with 0.01 volume of a 10-mg/ml suspension of amorphous cellulose (33).
Isolation of genomic DNA and construction of genomic libraries.
Genomic DNA was isolated by using the protocol of Murray and Thompson (48). A. cellulolyticus genomic libraries were constructed by using a Lambda ZAP II undigested vector kit for an XbaI library and a Uni-ZAP XR vector kit for an EcoRI-XhoI library. Both kits were obtained from Stratagene Cloning Systems (La Jolla, Calif.).
PCR and subcloning.
PCRs were performed with a Master Personal device (Eppendorf, Hamburg, Germany) at various annealing temperatures (50 to 60°C). The resulting PCR fragments were cloned by using pGEM-T Vector System 1 (Promega Corporation, Madison, Wis.). Alternatively, pUC19 was used for cloning the PCR fragments or phage inserts following digestion with appropriate restriction enzymes. Escherichia coli TG1 or XL-1 cells were used as host cells for transformation. DNA samples were purified by using either a QIAquick PCR purification kit (QIAGEN Inc., Valencia, Calif.) or an agarose gel DNA extraction kit (Roche Diagnostics Corporation, Indianapolis, Ind.). Plasmids were purified by using a High Purification plasmid isolation kit (Boehringer, Mannheim, Germany).
Southern blotting.
A. cellulolyticus genomic DNA was digested with various restriction enzymes, including EcoRI, SacI, XbaI, and XhoI, and the DNA fragments were separated on a 1% agarose gel. Relevant DNA fragments were labeled by using a random primed DNA labeling kit (Roche Diagnostics) as instructed by the supplier. Southern blotting was performed by using the protocol described in the DIG Application Manual for Filter Hybridization (Roche Molecular Biochemicals).
Library screening.
Two A. cellulolyticus genomic libraries were screened. The PCR-based library screening method was used for screening of the XbaI library by the procedure described by Israel (29). The primers used for screening were PAC-F1 and PAC-R1, which resulted in a 737-bp PCR product (Table 1). One of the positive plaques was verified by PCR and transferred to a phagemid, which resulted in a 4-kb insert that was subsequently sequenced. For EcoRI-XhoI library screening, a 450-bp PCR fragment obtained with primers ACAnF15 and ACAnR13 (Table 1) was labeled and used as a probe. A positive plaque was identified and transferred to the phagemid, and the resulting 6-kb insert was sequenced.
TABLE 1.
Primers used in this study
| Primer | Nucleotide sequencea | Locationb | Comments |
|---|---|---|---|
| PAC-F1 | GAAGATGTAATGATAGTTGC | Doc, cipV (scaA) | Probe for XbaI library screening |
| PAC-R1 | GTCTATTGCATTTGGAACTGA | Coh-1, scaB | Probe for XbaI library screening |
| ACAnF18 | CAGCTGCAGCTCCTGAACAGAC | Coh-1, scaB | Sequencing |
| pAC-F2 | CAGGAACATTAGCAGTAGTAG | Coh-2, scaB | Sequencing |
| pAC-F3 | GAAGATTCAGAAACAATGCC | Coh-2, scaB | Sequencing |
| ACAnF13 | GAGGATTACGGACCAATAG | Coh-3, scaB | Sequencing |
| pAC-Anc-F4 | CCACTAGAGGGTGAGATACTTGC | Coh-4, scaB | Sequencing |
| ACAn R16 | GAAGTTTAGTAGACCTTCGC | Coh-4, scaB | Sequencing |
| ACAn R15 | GTCTCTTACTAGAACAGCAT | Doc, scaB | Sequencing |
| ACAn F15 | CCCTGTTGAAGAGAAAGAAG | Doc, scaB | Probe for XR library screening |
| ACAnR13 | CTACTACCATCTACTGGG GC | Coh-1, scaC | Probe for XR library screening |
| F-AC-Sca1 | GGCGTTGAATCTGGAAG | Coh-2, scaC | Sequencing |
| F-AC-Sca6 | CTGTAGCAAGTATAGATGCTGGC | Coh-3, scaC | Sequencing |
| F-AC-Sca8 | CATTCTATTATTCTGGAACTGATG | Coh-3, scaC | Sequencing |
| R-AC-SCA10 | GTTACACCTGGTGTTACACTTCCTG | Linker, scaC | Sequencing |
| R-AC-SCA9 | CACATCCGCACCTGATAACTTAGCC | SLH-1, scaC | Sequencing |
| R-AC-SCA8 | CAAAACTGTTCCATATTCAAGTAA | Downstream of scaC | Sequencing |
| R-AC-SCA7 | CCATTGTTATATAAGACTCATCGCTTGC | Downstream of scaC | Sequencing |
| R-AC-SCA6 | CCAACAACAACTGACGTCTTATC | Downstream of scaC | Sequencing |
| F-EX-Coh5A | ATATCCATGGATGGTAAAGTAGAGATCATAGAT | Coh-5, cipV (scaA) | Expression |
| R-EX-Coh5A | AATTCTCGAGCGTTACGTTTCCTACTGTAACAGA | Coh-5, cipV (scaA) | Expression (His tag) |
| Pac-Anc-CohI-N | TAACCATGGCTCCAACATCTAGTATAG | Coh-1, scaB | Expression |
| Pac-Anc-CohI-C | TGTCTCGAGACTTGCTTTAATCATATC | Coh-1, scaB | Expression (His tag) |
| N-AC-Coh3C | CTACCATGGATTTACAGGTTGACATTGGAAGT | Coh-3, scaC | Expression |
| R-New-Coh3C | CAGCTCGAGACTTGCAATTACCTCAATTTTTCC | Coh-3, scaC | Expression (His tag) |
| R-AC-Coh3C | CAGGGATCCACTTGCAATTACCTCAATTTTTCC | Coh-3, scaC | Expression (without His tag) |
| F-9dxyn-docB | TATGGTACCGCCTAAATTTATATATGGTGATGTT | Doc, scaB | Expression, fused to xylanase T6 |
| R-9dxyn-Acdoc | TATGGATCCTTCTTCTTTCTCTTCAACAGGG | Doc, scaB | Expression, fused to xylanase T6 |
| F-9dxyn-docE | ATAGGTACCCCTGCACAATACGTATATGGTGAT | Doc, GH9B | Expression, fused to xylanase T6 |
| R-9dxyn-docE | CAAGGATCCCTTTTGTACCGGAAACTTTGAGAT | Doc, GH9B | Expression, fused to xylanase T6 |
| M13/pUC(-21) | AACAGCTATGACCATGATTACG | Plasmid | Sequencing |
| M13/pUC(-20) | TGTAAAACGACGGCCAGT | Plasmid | Sequencing |
| T7 | CGCGCGTAATACGACTCACTATAG | Plasmid | Sequencing |
| SP6 | CCAAGCTATTTAGGTGACAC | Plasmid | Sequencing |
Restriction sites are underlined in primers used for protein expression.
Doc, dockerin domain; Coh, cohesin domain; GH9B, dockerin from putative family 9 enzyme.
DNA sequencing.
DNA sequencing was performed either directly with PCR products or with cloned fragments by using an ABI PRISM 3100 genetic analyzer (Applied Biosystems, Foster City, Calif.) at the Sequencing Lab of Tel Aviv University, Ramat Aviv, Israel. The resulting sequences were compared to the sequences of known cellulosome-related proteins.
Cloning and overexpression of recombinant proteins.
The appropriate genes were subcloned into expression vectors by PCR (Fig. 1). The PCR products were cloned into either the pET14b, pET28a, or pET9d vector, and their intact sequences were verified by DNA sequencing. The clones were expressed at 37 or 16°C either in E. coli BL21(DE3) or in BL21(DE3)pLysS (Stratagene) grown in the presence of 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Following growth, the cultures were lysed by sonication as described by Ding et al. (17). The expressed proteins were identified by sodium dodecyl sulfate (SDS)-10 or 12% polyacrylamide gel electrophoresis (PAGE) and stained with Coomassie brilliant blue.
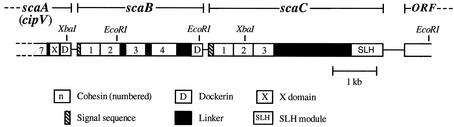
FIG. 1.
Scheme showing the positions on the genome and the domain organization of the scaB and scaC genes of A. cellulolyticus. The two genes are located in tandem immediately downstream of scaA (cipV). scaB and scaC both contain multiple copies of cohesin domains (numbered). scaB harbors a typical dockerin domain at its C terminus. The first two cohesins of scaB and all three scaC cohesins are closely attached, with short or no identifiable linker sequences. In contrast, the linker segments that connect the other modules in scaB are relatively long. The scaC linker sequence that connects cohesin 3 to the SLH module is particularly long and is characterized by an 18-residue repeated sequence. ORF, open reading frame.
Biotinylation.
Expressed CohC3 protein (in pET14b, without a His tag) was purified by heating a preparation at 65°C for 15 min, followed by gel filtration on a Superdex 75 column (Pharmacia, Uppsala, Sweden). The purified protein was labeled with biotin by using an eightfold molar excess of reagent compared to the amount of protein, as described previously (6).
Immunoblotting.
Proteins were subjected to SDS-10% PAGE, and the separated proteins were then transferred onto a nitrocellulose membrane and rinsed with washing buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 25 mM CaCl2). The membranes were then incubated for 2 to 3 h with blocking buffer (3% bovine serum albumin in washing buffer) and rinsed five times with washing buffer. The membranes were then incubated overnight at 4°C with the recombinant His-tagged or biotinylated proteins. The membranes were then treated with either peroxidase-conjugated antibody (anti-His [C-terminal]-horseradish peroxidase mouse antibody) or a streptavidin-horseradish peroxidase detection system used according to the supplier's instructions (Invitrogen Corporation, Carlsbad, Calif.). Bands were visualized by using a chemiluminescent substrate (Supersignal substrate for Western blotting; Pierce Biotechnology, Rockford, Ill.) as recommended by the manufacturer.
Peptide sequencing.
Selected protein bands were excised from SDS-PAGE gels and subjected to proteolysis with Lys-C, and the resultant peptides were resolved by reverse-phase high-performance liquid chromatography, analyzed, and sequenced by Edman degradation (Protein Center, Technion, Haifa, Israel). Alternatively, the bands were treated with trypsin, and the tryptic peptides were identified by matrix-assisted laser desorption ionization mass spectrometry at the Maiman Institute for Proteome Research at Tel Aviv University, Tel Aviv, Israel. The peptide sequence data obtained were compared to the sequences of known genes.
Protein sequence analysis.
Potential signal sequences were determined with the SignalP V2.0 program (49). The parameters for molecular weight, theoretical pI, amino acid composition, and extinction coefficient were computed by using the ProtParam tool (http://www.expasy.org/tools/protparam.html), available at the SWISS-PROT website (1). Multiple-sequence alignment and phylogenetic trees were generated by using the ClustalW program (http://www2.ebi.ac.uk/clustalw/). The SWISS-PROT accession numbers for the C. thermocellum SLH-bearing anchoring proteins OlpA, OlpB, and Orf2p are Q06848, Q06852 and Q06853, respectively. Dockerin and cohesin sequences were obtained from the GenBank website (http://www.ncbi.nlm.nih.gov/) or from the Carbohydrate- Active Enzymes server (CAZy website [http://afmb.cnrs-mrs.fr/∼pedro/CAZY/db.html]), which was designed by Coutinho and Henrissat (10, 14). The ScaB and ScaC cohesins were mapped on a background of cohesin modules, as previously described (15, 16). The ScaB and GH9 dockerins were similarly mapped on a background of dockerins as described previously (52). Secondary structures of the Thr-rich linker sequences were predicted by using the PredictProtein server at Columbia University (http://cubic.bioc.columbia.edu/predictprotein/).
Nucleotide sequence accession numbers.
The nucleotide sequences of the scaB and scaC genes have been deposited in the GenBank database under accession numbers AY221112 and AY221113.
RESULTS
Identification and sequencing of the region downstream of cipV.
Primers PAC-F1 and PAC-R1 (Table 1) were designed from the C-terminal portion of the 10-kb EcoRI fragment of the A. cellulolyticus pUC19 library, which was described previously (15). The resultant 737-bp PCR product was used as a probe for Southern blotting of genomic DNA that was digested with several common restriction enzymes, including XbaI, SacI, XhoI, and EcoRI. XbaI was selected to prepare a lambda ZAPII genomic DNA library, based on the presence of a single 4-kb band and identification of a sole XbaI site in the C-terminal dockerin sequence in the previously described cipV gene (15). The same primers were used for PCR-based high-stringency screening of the XbaI library, performed by the method devised by Israel (29). Positive plaques were confirmed by PCR, and the desired 4-kb insert was identified after transfer to the phagemid. The insert was digested further with EcoRI, and the resultant fragments were subcloned into pUC19 and sequenced. When necessary, internal segments were sequenced either by direct PCR or by prior subcloning into the T vector. The 4-kb insert contained the portion of cipV immediately downstream of the XbaI site, the sequence of a complete gene downstream of cipV, and the beginning of another open reading frame at the C terminus. The sequence of the new open reading frame was extended by designing a PCR-labeled probe (Table 1) based on the 3′ sequence of the 4-kb insert. To do this, a lambda ZAPII (EcoRI-XhoI) library was constructed and screened by using this probe. A 6-kb fragment was identified in the corresponding phagemid from the N and C termini by direct stepwise PCR sequencing. Initially, completion of scaC was hampered because of the repetitive nature of the linker, which prevented design of appropriate primers. In addition, the C-terminal portion of the 6-kb fragment was found to be tainted with two small (222- and 443-bp) noncontiguous segments, both of which were derived from elsewhere on the genome. To further complicate matters, we also determined that the 6-kb fragment had a 0.6-kb deletion within the linker, probably due to its repetitive nature. The problems resulting from the extraneous insertion and deletion events were resolved by sequencing PCR products by using genomic DNA as the template. When this strategy was used, a 5.3-kb portion of the genome was sequenced correctly, and this portion represented the authentic complement of the 6-kb fragment. The sequence was found to contain the terminus of scaB, the entire scaC gene, and the beginning of a new open reading frame.
Description of the genes downstream of cipV (scaA).
The status and modular architecture of scaB and the portion of scaC sequenced are shown in Fig. 1. The scaB gene codes for a 942-residue protein that contains a signal peptide, four cohesin modules, and a C-terminal dockerin domain. The most likely cleavage site of the signal peptide is between residues 27 and 28 (INA-AP). Based on the deduced ScaB sequence, the mature protein (following cleavage of the signal peptide) has a theoretical molecular weight of 97,323 and a calculated pI of 4.84 for the unfolded protein. It is currently not known whether the protein is glycosylated in the native state. Phylogenetic analysis revealed that the four ScaB cohesins can be classified as type II cohesins and are thus most similar to those of the C. thermocellum anchoring proteins and of the Bacteroides cellulosolvens scaffoldin CipBc (Fig. 2A). Little if any linker segment is present between the first two cohesins. In contrast, relatively long Pro-Thr-rich linker sequences separate cohesin 2 from cohesin 3 and cohesin 3 from cohesin 4 (39 amino acid residues each). An even longer Pro-Thr-Ala-rich linker (84 residues) separates cohesin 4 from the dockerin domain. The latter linker is remarkable in that every second residue is a threonine, and the hierarchical frequency of the alternating residues is Ala > Pro > Thr = Gln > Lys (these residues appear 20, 15, 3, and 3 times and only once, respectively, and there is no detectable pattern). Secondary structure analysis (53-55) revealed that unlike the β strands predicted to characterize the long R. flavefaciens cellulosomal linker segments (52), the linker segments of the A. cellulolyticus ScaB protein are predicted to generally lack helices and β strands, but they are expected to assume a loop-like structure. The reliability indices for the predicted loop regions are, however, comparatively low.
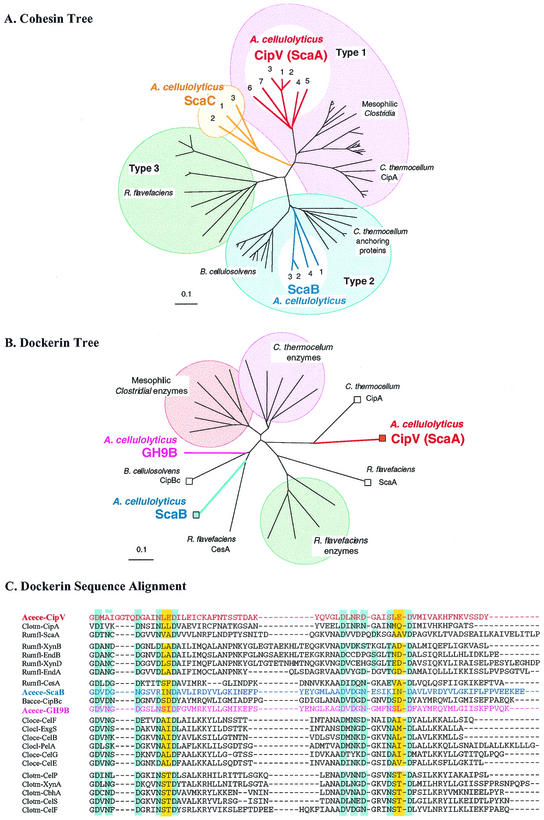
FIG. 2.
Relationship of the A. cellulolyticus cohesins and dockerins to previously described domains. (A) Phylogenetic analysis of the ScaB and ScaC cohesins relative to the known type I, II, and III cohesins. All four ScaB cohesins map together on a separate branch of the type II cohesins. The ScaC cohesins form a new group, which emanates from the central branch and is distinct from the other cohesin types. Scale bar = 0.1% amino acid substitutions. (B) Phylogenetic analysis of the dockerins of ScaB and the GH9B enzyme. Scaffoldin-based dockerins are indicated by squares.(C) Sequence alignment of the ScaB and the GH9B dockerin domains and their relationships to selected type I dockerins from various cellulosomal scaffoldin and enzyme subunits. Presumed calcium-binding residues are indicated by a blue background, and proposed recognition residues are indicated by a yellow background. With the exception of the domains derived from the proteins described in this paper, the sources of sequences used are described in references 15, 16, and 52. Abbreviations: Acece, A. cellulolyticus; Bacce, B. cellulosolvens; Cloce, C. cellulolyticus; Clocl, C. cellulovorans; Clotm, C. thermocellum; Rumfl, R. flavefaciens.
The scaC gene codes for a 1,237-residue protein that includes a signal peptide, three cohesins, an exceptionally long linker, and a C-terminal SLH module. Based on the deduced ScaC sequence, the mature protein (following cleavage of the signal peptide) has a theoretical molecular weight of 124,162 and a calculated pI of 4.99 for the unfolded protein. The ScaC SLH module exhibits very high homology (between 51 and 58% identity and up to 81% overall similarity) with the SLH modules of the C. thermocellum anchoring proteins (22, 38, 41). The presence of the cohesins and the conserved SLH module (Fig. 3) implies that ScaC is a cell surface anchoring protein (12, 19, 40, 42, 47).
FIG. 3.
Multiple-sequence alignment of the SLH module of A. cellulolyticus ScaC (Acece-ScaC) and the three C. thermocellum anchoring proteins (Clotm-OlpA, Clotm-OlpB, and Clotm-Orf2p). The degrees of conservation are indicated as follows: asterisks indicate that all sequences are identical, colons indicate that the residues are conserved, and dots indicate that residues are semiconserved, as defined by the EBI server (http://www2.ebi.ac.uk/clustalw/).
The most likely cleavage site of the scaC signal peptide is between residues 36 and 37 (VQA-AE). Phylogenetic analysis of the ScaC cohesins revealed that they are uniquely separated from the other type I, II, and III cohesins (Fig. 2A). The ScaC cohesins project from the central trunk of the phylogenetic tree, occupying a separate branch that is located between the partition points that separate the type I cohesins from the type II and III cohesins. Because of the growing number of cohesin branches that result from newly sequenced scaffoldins, here we do not classify cohesins, such as those of ScaC, as members of a new type or subtype. Defined linker segments could not be detected between the three ScaC cohesins, indicating that they are closely joined. On the other hand, the ScaC linker segment that separates the third cohesin from the SLH module consists of ∼550 amino acid residues, including 27 repeats of the 18-residue stretch PTPTQSAXPTVTPSATAT, where X is mainly Met (15 copies), with substitutions of Lys (eight copies), Thr (two copies), and Ile (two copies) in no particular order; additional substitutions were also interspersed intermittently at different positions within the 18-residue stretch at lower frequencies. Following the repeated sequence, the linker continues downstream with a ∼70-residue Pro-Thr-rich segment that differs in character from the 18-residue repeat. Like the prediction for the linker segments of A. cellulolyticus ScaB, secondary structure prediction for the long ScaC linker indicated that there is a general lack of α helices and β strands.
Description of the known A. cellulolyticus dockerin sequences.
In the context of the present work, the sequences of three different dockerin domains from A. cellulolyticus were available. These included the previously described CipV (ScaA) dockerin, the ScaB dockerin sequenced in this work, and an enzyme-borne dockerin. In the latter case, a partial sequence of an enzyme was detected in this study. The family 9 enzyme (termed GH9B to distinguish it from the resident GH9A of the CipV polypeptide) was sequenced (Xu, unpublished results) and was found to contain a C-terminal dock-erin domain. The latter dockerin sequence was thus the first representative enzyme-borne cellulosomal dockerin sequence from this organism and consequently complemented the sequences of the two scaffoldins used in this investigation.
Phylogenetic analysis of the three A. cellulolyticus dockerins was carried out with reference to the >100 dockerin domains that have been sequenced to date. The results are shown in Fig. 2B, which shows representative examples of enzyme- and scaffoldin-borne dockerins. The dockerin of CipV (ScaA) is most similar to the C. thermocellum scaffoldin-borne (CipA) dockerin, which is recognized by a type II cohesin. In contrast, the dockerins of A. cellulolyticus ScaB and the GH9B enzyme map on a separate branch of the tree, together with those of the B. cellulosolvens CipBc scaffoldin and the R. flavefaciens CesA enzyme.
The F-hand variation (43, 50) of the EF-hand calcium-binding motif (11, 28) is largely retained in the three A. cellulolyticus dockerins, although there are some deviations. As described previously, the sequence of the A. cellulolyticus CipV (ScaA) dockerin is unusual in that a four-residue insert is present in the first duplicated calcium-binding motif (Fig. 2C). The ScaB dockerin is unusual in terms of another feature: one of the normally conserved residues in the duplicated calcium-binding loop (usually Asn, Asp, or Thr) in each duplicated segment is replaced by a positively charged amino acid (Arg or Lys). It is not clear whether such digressions from the canonical EF-hand calcium-binding motif could interfere with the binding affinity for the ligand. On the other hand, the GH9B enzyme-borne dockerin contains all the essential calcium-binding residues and other requisite features of a conventional dockerin domain (Fig. 2C). It is also interesting that the putative recognition residues of the three A. cellulolyticus dockerins (i.e., LE/LE of ScaA, IN/IN of ScaB, and SI/SL of the GH9B enzyme) are different from each other and from those of all of the other known enzyme- or scaffoldin-borne dockerins derived from the different cellulosome-producing species (44, 46, 50, 52).
Cohesin-dockerin interactions among the A. cellulolyticus proteins.
In order to gain further insight into the interactions among the individual types of protein modules encoded by the genes of the cluster, various gene segments were subcloned and overexpressed. Representative cohesin modules were expressed in pET28a together with a fused C-terminal His tag for subsequent isolation and detection. The dockerin domains were C terminally fused to Geobacillus stearothermophilus xylanase T6 (45) as an appropriate carrier protein. Fusion to this protein ensured enhanced expression, folding, and stability of the attached dockerin domain (A. Mechaly, Y. Barak, T. Handelsman, R. Lamed, Y. Shoham, and E. A. Bayer, unpublished results). The xylanase-dockerin fusion protein also represents an approach for standardization of the dockerin-containing probe. The resultant fusion proteins also included an N-terminal His tag to facilitate subsequent isolation. The expressed proteins used in this study are listed in Table 2. They include the fifth cohesin of CipV (CohA5), the first cohesin of ScaB (CohB1), and the third cohesin of ScaC (CohC3), as well as the two hybrid proteins containing either the dockerin of ScaB (DocB) or the dockerin of the GH9B cellulase fused to the xylanase carrier. For comparative purposes, CohC3 was also expressed without a His tag in pET14b; the purified construct was biotinylated and used as a complementary probe to verify the affinity-based labeling system. The binding specificities of the expressed cohesin probes were investigated by affinity blotting with cell-derived extracts and with the xylanase-dockerin fusion proteins.
TABLE 2.
Expressed proteins prepared in this study
| Protein | Modular content | Plasmid | Comments |
|---|---|---|---|
| CohA5 | Cohesin 5 of CipV (ScaA) | pET28a | C-terminal His tag |
| CohB1 | Cohesin 1 of ScaB | pET28a | C-terminal His tag |
| CohC3 | Cohesin 3 of ScaC | pET28a | C-terminal His tag |
| CohC3′ | Cohesin 3 of ScaC | pET14b | No His tag; biotinylated and used as complementary CohC3 probe |
| Xyn-DocB | Hybrid construct consisting of G. stearothermophilus xylanase T6 harboring the A. cellulolyticus ScaB dockerin at the C terminus | pET9d | N-terminal His tag |
| Xyn-DocGH9 | Hybrid construct consisting of xylanase T6 with the A. cellulolyticus GH9B dockerin at the C terminus | pET9d | N-terminal His tag |
The A. cellulolyticus ScaA (CipV) cohesins interact with cellulosomal enzymes.
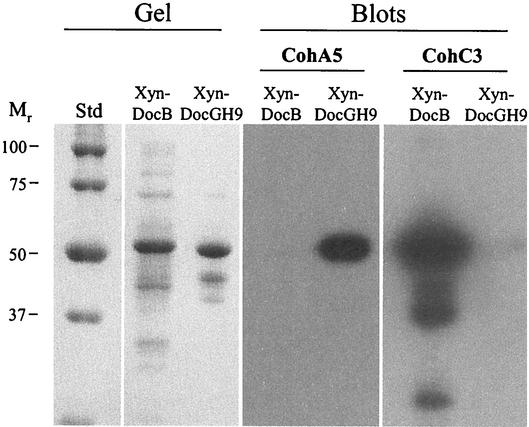
Affinity blotting of supernatant fluids and cell-associated extracts in which ScaA cohesin 5 (CohA5) was used as the probe (Fig. 4) revealed a succession of labeled bands in both cases, consistent with the putative cellulosomal enzyme components (15). Indeed, the same cohesin probe selectively labeled the xylanase hybrid construct (Xyn-DocGH9) that harbored the dockerin domain of a cellulosomal enzyme (Fig. 5). No labeling of the Xyn-DocB construct was apparent, thus confirming the specificity of the interaction.
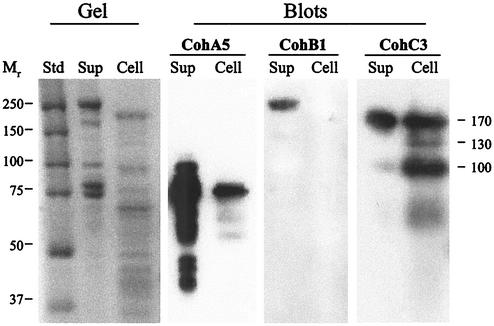
FIG. 4.
Affinity blotting of cell-derived proteins, performed with representative recombinant cohesins from ScaA, ScaB, and ScaC as the probes. A. cellulolyticus cells were grown on cellobiose and separated by centrifugation into supernatant (Sup) and pellet (Cell) fractions. The supernatant fraction was further fractionated by adsorption onto amorphous (phosphoric acid-treated) cellulose prior to subsequent electrophoresis. Samples (cell-associated pellet or cellulose-adsorbed supernatant) were subjected to SDS-PAGE (Gel) and were blotted onto nitrocellulose membranes (Blots). Gels were stained with Coomassie brilliant blue. The blots were probed with different recombinant protein samples, and labeled bands were detected by chemiluminescence by using peroxidase-conjugated, anti-His tag antibody. The probes were CohA5 (the fifth cohesin of ScaA), CohB1 (the first cohesin of ScaB), and CohC3 (the third cohesin of ScaC). Lane Std contained prestained protein molecular weight markers.
FIG. 5.
Affinity blotting of selected dockerin-containing fusion proteins performed with recombinant cohesins from ScaA and ScaC. Dockerins from ScaB and the cellulosomal GH9B enzyme were fused to G. stearothermophilus xylanase T6, and the resultant fusion proteins (Xyn-DocB and Xyn-DocGH9, respectively) were expressed in an appropriate E. coli host cell system. The fusion proteins were subjected to SDS-PAGE (Gel), transferred to nitrocellulose membranes (Blots), and probed with the ScaA and ScaC cohesins (CohA5 and CohC3, respectively), as described in the legend to Fig. 4. Lane Std contained prestained protein molecular weight markers.
The A. cellulolyticus ScaB cohesins interact selectively with ScaA.
The ScaB cohesin 1 (CohB1) probe selectively labeled the ∼240-kDa ScaA band in the supernatant fraction (Fig. 4). This band was identified definitively as ScaA by peptide sequencing of proteolytic digests, as described previously (15). No labeling was observed in the cell-associated fraction. However, it should be noted that the cells were grown on cellobiose, not cellulose, and were harvested in the stationary phase of growth. Attempts to prepare a viable xylanase hybrid protein comprising the ScaA dockerin were not successful, perhaps due to the unusual nature (four-residue insert) of the calcium-binding motif of the first duplicated segment. Consequently, at this stage in our studies, confirmation of the labeling specificity by using an artificial dockerin-containing construct could not be demonstrated.
The A. cellulolyticus ScaC cohesins bind selectively to the ScaB dockerin.
In the cell-associated fraction (Fig. 4), the ScaC cohesin 3 probe (CohC3) labeled three bands having molecular masses of 170, 130, and 100 kDa. The exact identities of the three labeled bands are unclear. The size of the 100-kDa band is consistent with the calculated molecular mass of the ScaB polypeptide. However, the relatively long Thr-rich linker segments could serve as oligosaccharide attachment sites that could result in a higher-molecular-mass glycoprotein, as shown previously for scaffoldins of both C. thermocellum and B. cellulosolvens (24-27). Alternatively, there may be an additional (other than ScaB) dockerin-containing protein(s) that could interact with the ScaC cohesin. In any case, the CohC3 probe appeared to selectively label the ScaB dockerin (Fig. 5), since the corresponding Xyn-DocB construct was heavily labeled with this probe compared to the Xyn-DocGH9 construct, which was essentially not labeled by CohC3.
In the supernatant fraction, the CohC3 probe labeled predominantly a 170-kDa band (Fig. 4), perhaps equivalent to the 170-kDa band of the cell-associated fraction. In addition, a minor band at about 100 kDa was very slightly labeled.
DISCUSSION
In any newly discovered cellulosome system that is characterized by multiple cohesin-containing scaffoldins and dockerin-containing enzymes, a primary interest is to ascertain the specificity of the interactions among the various modules and their parent proteins. This information is especially significant when the genome of a cellulosome-producing organism codes for both scaffoldin and related anchoring proteins, since very few such systems have been verified on the molecular level. By determining the specificities of the resident cohesin and dockerin domains, we can assess the possible modes of interaction and the resulting quaternary structural model for association of the cellulosome with the cell surface.
This approach was accomplished at least partially first with the cellulosome system of C. thermocellum (7) and later with the cellulosome system of R. flavefaciens (17, 52). In the former case, a series of cohesin-containing anchoring proteins (OlpB, Orf2p, and SdbA) were described, which interacted selectively with the C-terminal dockerin of the C. thermocellum CipA scaffoldin (38-40). The anchoring proteins of this bacterium contained a C-terminal SLH module that was used to incorporate the polypeptide into the cell surface. The relevant cohesin-dockerin interactions could thus effect successive binding of the scaffoldin and, in turn, the appropriate enzymes into a higher-order, cell-surface-associated cellulosome system.
The R. flavefaciens system represents another variation on the cellulosome theme, in which the enzyme-based dockerins reflect at least two distinct cohesin-binding specificities. The cellulosome is characterized by two known scaffoldins, ScaA and ScaB. The three ScaA cohesins are used to incorporate some but not all of the dockerin-containing enzymes into the complex, and the C-terminal ScaA dockerin can interact with all seven ScaB cohesins (17, 52). An X domain of unknown function has been suggested to play a role in cell surface attachment, but this suggestion has yet to be experimentally verified. The experimental evidence also suggests that there may be at least one additional type of R. flavefaciens scaffoldin. In any case, elucidation of the modular interactions among the components of each new cellulosome system provides new insight into the diverse supramolecular organization.
The results of the present work demonstrate that in the A. cellulolyticus cellulosome system, both of the genes downstream of cipV contain segments that encode for cohesin domains. The existence of multiple cohesins implies that like cipV, the new genes encode additional scaffoldins. Consequently, cipV was renamed scaA, and its neighboring genes were designated scaB and scaC. It is clear that these three genes constitute an emerging cluster of cellulosome-related genes. The presence of a cluster of multiple scaffoldin-like genes on the chromosome reflects the genomic status previously described for C. thermocellum and R. flavefaciens rather than the genomic status of the scaffoldin-enzyme gene cluster in Clostridium cellulolyticum and cognate bacteria. In its capacity in integration of the dockerin-containing enzymes, ScaA (CipV) can be considered a primary scaffoldin. In contrast, ScaB essentially plays the role of an adaptor protein, which mediates between ScaA (and its attached enzymes) and ScaC. A. cellulolyticus ScaB is the first example of such an adaptor protein. ScaC, on the other hand, clearly plays the role of an anchoring scaffoldin by virtue of its C-terminal SLH module.
Surprisingly, perhaps, the order of the genes in the A. cellulolyticus cluster reflects the recognition properties of the cohesins and dockerins. Thus, the ScaA dockerin binds to the cohesins of ScaB and the ScaB dockerin binds to the cohesins of ScaC (Fig. 6). However, additional, unidentified scaffoldins (e.g., the 170-kDa protein) may also be components of alternative higher-order cellulosome complexes in this bacterium.
FIG. 6.
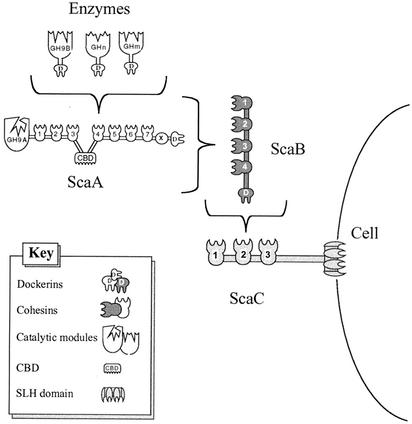
Schematic representation of the proposed cell surface disposition of the A. cellulolyticus cellulosomal components identified. The GH9B enzyme and other putative dockerin-containing enzymes are incorporated into the ScaA scaffoldin by virtue of their interaction with the ScaA cohesins. ScaB plays the role of an adaptor protein that mediates between the dockerin of the primary scaffoldin, ScaA, and the cohesins of the anchoring scaffoldin, ScaC. The entire complex appears to be cell associated via the resident SLH module of ScaC.
Since multiple cohesins are involved in each set of interactions, the net effect is progressive amplification of the number of dockerin-containing enzymes that can be incorporated onto the cell surface. Thus, according to the scheme shown in Fig. 6, seven enzyme subunits can presumably be incorporated into ScaA, four ScaA molecules into ScaB, and three ScaB molecules into ScaC. Altogether, including the resident ScaA enzyme, 96 different enzyme molecules can theoretically be incorporated into each A. cellulolyticus cellulosome. The intricate nature of the putative supramolecular complex is consistent with the exceptionally elaborate cell surface architecture of this bacterium (13, 34, 35). The central factor contributing to the enhanced amplification of the A. cellulolyticus system is the ScaB adaptor protein. In comparison, the available evidence for the C. thermocellum cellulosome system indicates that the amplification factor should be limited to 36 molecules, since nine enzymes can be incorporated into the CipA scaffoldin and up to four CipA molecules can be combined with the OlpB anchoring proteins.
The role of amplified enzyme incorporation into a cellulosome presumably reflects the recently described proximity effect of the cellulosome (21), which is one of the key factors for efficient digestion of recalcitrant forms of cellulose. Concentration of complementary cellulolytic enzymes on the surface of the substrate and in the vicinity of the bacterial cell surface should thus enhance the synergistic action of the enzymes.
Acknowledgments
This research was supported by the Israel Science Foundation (grants 771/01, 446/01, and 250/99), by the U.S.-Israel Binational Agricultural Research and Development Fund (BARD research grant 3106-99C), and by a grant from the United States-Israel Binational Science Foundation, Jerusalem, Israel. Additional support was provided by the Otto Meyerhof Center for Biotechnology, established by the Minerva Foundation (Munich, Germany), and by funds from the Technion-Niedersachsen Cooperation (Hannover, Germany).
REFERENCES
- 1.Bairoch, A., and R. Apweiler. 2000. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 28:45-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bayer, E. A., H. Chanzy, R. Lamed, and Y. Shoham. 1998. Cellulose, cellulases and cellulosomes. Curr. Opin. Struct. Biol. 8:548-557. [DOI] [PubMed] [Google Scholar]
- 3.Bayer, E. A., R. Kenig, and R. Lamed. 1983. Adherence of Clostridium thermocellum to cellulose. J. Bacteriol. 156:818-827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bayer, E. A., E. Morag, and R. Lamed. 1994. The cellulosome—a treasure-trove for biotechnology. Trends Biotechnol. 12:378-386. [DOI] [PubMed] [Google Scholar]
- 5.Bayer, E. A., Y. Shoham, and R. Lamed. September 2000, revision date. Cellulose-decomposing prokaryotes and their enzyme systems. In M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes: an evolving electronic resource for the microbiological community, 3rd ed., release 3.7. [Online.] http://link.springer.de/link/service/books/10125/index.htm. Springer-Verlag, New York, N.Y.
- 6.Bayer, E. A., and M. Wilchek. 1990. Protein biotinylation. Methods Enzymol. 184:138-160. [DOI] [PubMed] [Google Scholar]
- 7.Béguin, P., S. Chauvaux, G. Guglielmi, M. Matuschek, E. Leibovitz, M.-K. Chaveroche, I. Miras, P. Alzari, and P. Gounon. 1999. The Clostridium thermocellum cellulosome: organization and mode of attachment to the cell, p. 437-443. In K. Ohmiya, K. Hayashi, K. Sakka, Y. Kobayashi, S. Karita, and T. Kimura (ed.), Genetics, biochemistry and ecology of cellulose degradation. Uni Publishers Co., Tokyo, Japan.
- 8.Béguin, P., and M. Lemaire. 1996. The cellulosome: an exocellular, multiprotein complex specialized in cellulose degradation. Crit. Rev. Biochem. Mol. Biol. 31:201-236. [DOI] [PubMed] [Google Scholar]
- 9.Belaich, J.-P., A. Belaich, H.-P. Fierobe, L. Gal, C. Gaudin, S. Pagès, C. Reverbel-Leroy, and C. Tardif. 1999. The cellulolytic system of Clostridium cellulolyticum, p. 479-487. In K. Ohmiya, K. Hayashi, K. Sakka, Y. Kobayashi, S. Karita, and T. Kimura (ed.), Genetics, biochemistry and ecology of cellulose degradation. Uni Publishers Co., Tokyo, Japan.
- 10.Bourne, Y., and B. Henrissat. 2001. Glycoside hydrolases and glycosyltransferases: families and functional modules. Curr. Opin. Struct. Biol. 11:593-600. [DOI] [PubMed] [Google Scholar]
- 11.Chauvaux, S., P. Béguin, J.-P. Aubert, K. M. Bhat, L. A. Gow, T. M. Wood, and A. Bairoch. 1990. Calcium-binding affinity and calcium-enhanced activity of Clostridium thermocellum endoglucanase D. Biochem. J. 265:261-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chauvaux, S., M. Matuschek, and P. Béguin. 1999. Distinct affinity of binding sites for S-layer homologous domains in Clostridium thermocellum and Bacillus anthracis cell envelopes. J. Bacteriol. 181:2455-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Colvin, J. R., L. C. Sowden, G. B. Patel, and A. W. Khan. 1982. The ultrastructure of Acetivibrio cellulolyticus, a recently isolated cellulolytic anaerobe. Curr. Microbiol. 7:13-17. [Google Scholar]
- 14.Coutinho, P. M., and B. Henrissat. 1999. Carbohydrate-active enzymes: an integrated database approach, p. 3-12. In H. J. Gilbert, G. J. Davies, B. Henrissat, and B. Svensson (ed.), Recent advances in carbohydrate bioengineering. The Royal Society of Chemistry, Cambridge, United Kingdom.
- 15.Ding, S.-Y., E. A. Bayer, D. Steiner, Y. Shoham, and R. Lamed. 1999. A novel cellulosomal scaffoldin from Acetivibrio cellulolyticus that contains a family 9 glycosyl hydrolase. J. Bacteriol. 181:6720-6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ding, S.-Y., E. A. Bayer, D. Steiner, Y. Shoham, and R. Lamed. 2000. A scaffoldin of the Bacteroides cellulosolvens cellulosome that contains 11 type II cohesins. J. Bacteriol. 182:4915-4925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ding, S.-Y., M. T. Rincon, R. Lamed, J. C. Martin, S. I. McCrae, V. Aurilia, Y. Shoham, E. A. Bayer, and H. J. Flint. 2001. Cellulosomal scaffoldin-like proteins from Ruminococcus flavefaciens. J. Bacteriol. 183:1945-1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doi, R. H., and Y. Tamura. 2001. The Clostridium cellulovorans cellulosome: an enzyme complex with plant cell wall degrading activity. Chem. Rec. 1:24-32. [DOI] [PubMed] [Google Scholar]
- 19.Engelhardt, H., and J. Peters. 1998. Structural research on surface layers: a focus on stability, surface layer homology domains, and surface layer-cell wall interactions. J. Struct. Biol. 124:276-302. [DOI] [PubMed] [Google Scholar]
- 20.Felix, C. R., and L. G. Ljungdahl. 1993. The cellulosome—the exocellular organelle of Clostridium. Annu. Rev. Microbiol. 47:791-819. [DOI] [PubMed] [Google Scholar]
- 21.Fierobe, H.-P., A. Mechaly, C. Tardif, A. Belaich, R. Lamed, Y. Shoham, J.-P. Belaich, and E. A. Bayer. 2002. Designer nanosomes: selective engineering of dockerin-containing enzymes into chimeric scaffoldins to form defined nanoreactors, p. 113-123. In T. T. Teeri, B. Svensson, H. J. Gilbert, and T. Feizi (ed.), Carbohydrate bioengineering: interdisciplinary approaches. The Royal Society of Chemistry, Cambridge, United Kingdom.
- 22.Fujino, T., P. Béguin, and J.-P. Aubert. 1993. Organization of a Clostridium thermocellum gene cluster encoding the cellulosomal scaffolding protein CipA and a protein possibly involved in attachment of the cellulosome to the cell surface. J. Bacteriol. 175:1891-1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerngross, U. T., M. P. M. Romaniec, T. Kobayashi, N. S. Huskisson, and A. L. Demain. 1993. Sequencing of a Clostridium thermocellum gene (cipA) encoding the cellulosomal SL-protein reveals an unusual degree of internal homology. Mol. Microbiol. 8:325-334. [DOI] [PubMed] [Google Scholar]
- 24.Gerwig, G., P. de Waard, J. P. Kamerling, J. F. G. Vliegenthart, E. Morgenstern, R. Lamed, and E. A. Bayer. 1989. Novel O-linked carbohydrate chains in the cellulase complex (cellulosome) of Clostridium thermocellum. J. Biol. Chem. 264:1027-1035. [PubMed] [Google Scholar]
- 25.Gerwig, G., J. P. Kamerling, J. F. G. Vliegenthart, E. Morag (Morgenstern), R. Lamed, and E. A. Bayer. 1992. Novel oligosaccharide constituents of the cellulase complex of Bacteroides cellulosolvens. Eur. J. Biochem. 205:799-808. [DOI] [PubMed]
- 26.Gerwig, G., J. P. Kamerling, J. F. G. Vliegenthart, E. Morag, R. Lamed, and E. A. Bayer. 1991. Primary structure of O-linked carbohydrate chains in the cellulosome of different Clostridium thermocellum strains. Eur. J. Biochem. 196:115-122. [DOI] [PubMed] [Google Scholar]
- 27.Gerwig, G., J. P. Kamerling, J. F. G. Vliegenthart, E. Morag, R. Lamed, and E. A. Bayer. 1993. The nature of the carbohydrate-peptide linkage region in glycoproteins from the cellulosomes of Clostridium thermocellum and Bacteroides cellulosolvens. J. Biol. Chem. 268:26956-26960. [PubMed] [Google Scholar]
- 28.Herzberg, O., and M. N. James. 1988. Refined crystal structure of troponin C from turkey skeletal muscle at 2.0 Å resolution. J. Mol. Biol. 203:761-779. [DOI] [PubMed] [Google Scholar]
- 29.Israel, D. I. 1993. A PCR-based method for high stringency screening of DNA libraries. Nucleic Acids Res. 21:2627-2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kakiuchi, M., A. Isui, K. Suzuki, T. Fujino, E. Fujino, T. Kimura, S. Karita, K. Sakka, and K. Ohmiya. 1998. Cloning and DNA sequencing of the genes encoding Clostridium josui scaffolding protein CipA and cellulase CelD and identification of their gene products as major components of the cellulosome. J. Bacteriol. 180:4303-4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khan, A. W. 1980. Cellulolytic enzyme system of Acetivibrio cellulolyticus, a newly isolated anaerobe. J. Gen. Microbiol. 121:499-502. [DOI] [PubMed] [Google Scholar]
- 32.Lamed, R., and E. A. Bayer. 1988. The cellulosome of Clostridium thermocellum. Adv. Appl. Microbiol. 33:1-46. [Google Scholar]
- 33.Lamed, R., R. Kenig, E. Setter, and E. A. Bayer. 1985. Major characteristics of the cellulolytic system of Clostridium thermocellum coincide with those of the purified cellulosome. Enzyme Microb. Technol. 7:37-41. [Google Scholar]
- 34.Lamed, R., J. Naimark, E. Morgenstern, and E. A. Bayer. 1987. Scanning electron microscopic delineation of bacterial surface topology using cationized ferritin. J. Microbiol. Methods 7:233-240. [Google Scholar]
- 35.Lamed, R., J. Naimark, E. Morgenstern, and E. A. Bayer. 1987. Specialized cell surface structures in cellulolytic bacteria. J. Bacteriol. 169:3792-3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lamed, R., E. Setter, and E. A. Bayer. 1983. Characterization of a cellulose-binding, cellulase-containing complex in Clostridium thermocellum. J. Bacteriol. 156:828-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lamed, R., E. Setter, R. Kenig, and E. A. Bayer. 1983. The cellulosome—a discrete cell surface organelle of Clostridium thermocellum which exhibits separate antigenic, cellulose-binding and various cellulolytic activities. Biotechnol. Bioeng. Symp. 13:163-181. [Google Scholar]
- 38.Leibovitz, E., and P. Béguin. 1996. A new type of cohesin domain that specifically binds the dockerin domain of the Clostridium thermocellum cellulosome-integrating protein CipA. J. Bacteriol. 178:3077-3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leibovitz, E., H. Ohayon, P. Gounon, and P. Béguin. 1997. Characterization and subcellular localization of the Clostridium thermocellum scaffoldin dockerin binding protein SdbA. J. Bacteriol. 179:2519-2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lemaire, M., I. Miras, P. Gounon, and P. Béguin. 1998. Identification of a region responsible for binding to the cell wall within the S-layer protein of Clostridium thermocellum. Microbiology 144:211-217. [DOI] [PubMed] [Google Scholar]
- 41.Lemaire, M., H. Ohayon, P. Gounon, T. Fujino, and P. Béguin. 1995. OlpB, a new outer layer protein of Clostridium thermocellum, and binding of its S-layer-like domains to components of the cell envelope. J. Bacteriol. 177:2451-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lupas, A., H. Engelhardt, J. Peters, U. Santarius, S. Volker, and W. Baumeister. 1994. Domain structure of the Acetogenium kivui surface layer revealed by electron crystallography and sequence analysis. J. Bacteriol. 176:1224-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lytle, B., B. F. Volkman, W. M. Westler, and J. H. D. Wu. 2000. Secondary structure and calcium-induced folding of the Clostridium thermocellum dockerin domain determined by NMR spectroscopy. Arch. Biochem. Biophys. 379:237-244. [DOI] [PubMed] [Google Scholar]
- 44.Mechaly, A., H.-P. Fierobe, A. Belaich, J.-P. Belaich, R. Lamed, Y. Shoham, and E. A. Bayer. 2001. Cohesin-dockerin interaction in cellulosome assembly: a single hydroxyl group of a dockerin domain distinguishes between non-recognition and high-affinity recognition. J. Biol. Chem. 276:9883-9888. [DOI] [PubMed] [Google Scholar]
- 45.Mechaly, A., A. Teplitsky, V. Belakhov, T. Baasov, G. Shoham, and Y. Shoham. 2000. Overproduction and characterization of seleno-methionine xylanase T-6. J. Biotechnol. 78:83-86. [DOI] [PubMed] [Google Scholar]
- 46.Mechaly, A., S. Yaron, R. Lamed, H.-P. Fierobe, A. Belaich, J.-P. Belaich, Y. Shoham, and E. A. Bayer. 2000. Cohesin-dockerin recognition in cellulosome assembly: experiment versus hypothesis. Proteins 39:170-177. [DOI] [PubMed] [Google Scholar]
- 47.Mesnage, S., E. Tosi-Couture, M. Mock, and A. Fouet. 1999. The S-layer homology domain as a means for anchoring heterologous proteins on the cell surface of Bacillus anthracis. J. Appl. Microbiol. 87:256-260. [DOI] [PubMed] [Google Scholar]
- 48.Murray, M. G., and W. F. Thompson. 1980. Rapid isolation of high-molecular-weight plant DNA. Nucleic Acids Res. 8:4321-4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nielsen, H., S. Brunak, and G. von Heijne. 1999. Machine learning approaches for the prediction of signal peptides and other protein sorting signals. Protein Eng. 12:3-9. [DOI] [PubMed] [Google Scholar]
- 50.Pagès, S., A. Belaich, J.-P. Belaich, E. Morag, R. Lamed, Y. Shoham, and E. A. Bayer. 1997. Species-specificity of the cohesin-dockerin interaction between Clostridium thermocellum and Clostridium cellulolyticum: prediction of specificity determinants of the dockerin domain. Proteins 29:517-527. [PubMed] [Google Scholar]
- 51.Pagès, S., A. Belaich, H.-P. Fierobe, C. Tardif, C. Gaudin, and J.-P. Belaich. 1999. Sequence analysis of scaffolding protein CipC and ORFXp, a new cohesin-containing protein in Clostridium cellulolyticum: comparison of various cohesin domains and subcellular localization of ORFXp. J. Bacteriol. 181:1801-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rincon, M. T., S.-Y. Ding, S. I. McCrae, J. C. Martin, V. Aurilia, R. Lamed, Y. Shoham, E. A. Bayer, and H. J. Flint. 2003. Novel organization and divergent dockerin specificities in the cellulosome system of Ruminococcus flavefaciens. J. Bacteriol. 185:703-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rost, B. 1996. PHD: predicting one-dimensional protein structure by profile based neural networks. Methods Enzymol. 266:525-539. [DOI] [PubMed] [Google Scholar]
- 54.Rost, B., and C. Sander. 1994. Combining evolutionary information and neural networks to predict protein secondary structure. Proteins 19:55-72. [DOI] [PubMed] [Google Scholar]
- 55.Rost, B., and C. Sander. 1993. Prediction of protein secondary structure at better than 70% accuracy. J. Mol. Biol. 232:584-599. [DOI] [PubMed] [Google Scholar]
- 56.Sabathe, F., A. Belaich, and P. Soucaille. 2002. Characterization of the cellulolytic complex (cellulosome) of Clostridium acetobutylicum. FEMS Microbiol. Lett. 217:15-22. [DOI] [PubMed] [Google Scholar]
- 57.Saddler, J. N., and A. W. Khan. 1980. Cellulase production by Acetivibrio cellulolyticus. Can. J. Microbiol. 26:760-765. [Google Scholar]
- 58.Schwarz, W. H. 2001. The cellulosome and cellulose degradation by anaerobic bacteria. Appl. Microbiol. Biotechnol. 56:634-649. [DOI] [PubMed] [Google Scholar]
- 59.Shoham, Y., R. Lamed, and E. A. Bayer. 1999. The cellulosome concept as an efficient microbial strategy for the degradation of insoluble polysaccharides. Trends Microbiol. 7:275-281. [DOI] [PubMed] [Google Scholar]
- 60.Shoseyov, O., M. Takagi, M. A. Goldstein, and R. H. Doi. 1992. Primary sequence analysis of Clostridium cellulovorans cellulose binding protein A. Proc. Natl. Acad. Sci. USA 89:3483-3487. [DOI] [PMC free article] [PubMed] [Google Scholar]