Abstract
Secondary RNA polymerase sigma factors in many bacteria are responsible for regulating a vast range of processes including virulence. A protein (σX) in the gram-positive human pathogen Streptococcus pyogenes (the group A Streptococcus or GAS) was recently shown to function in vitro as a secondary sigma factor. We report here the isolation of a mutant in which both sigX genes are inactivated, show that σX functions in GAS cells, and show that the amount of σX is controlled at two levels. Primer extension analysis indicates that sigX transcription is low in GAS cells grown in Todd-Hewitt yeast broth, and immunoblot assays with a σX-specific polyclonal antibody demonstrate that the protein does not accumulate in these cells. To increase the level of sigX transcription in GAS, we constructed a strain that constitutively expresses the sigX gene from a heterologous promoter. Expression of sigX from this promoter led to transcription of the σX-dependent cinA promoter in GAS cells. We found that expression of the sigX gene in a clpP mutant strain resulted in greater accumulation of σX protein, which resulted in higher levels of transcription from the σX-dependent promoters cinA, smf, and cglA. In addition, a clpP mutant containing sigX only at its wild-type loci on the chromosome generated more transcription from the σX-dependent cinA promoter than did the wild-type parental strain. Therefore, σX activity in GAS is limited by low-level transcription of the sigX structural genes and by clpP, which appears to negatively regulate σX accumulation.
Streptococcus pyogenes (the group A Streptococcus [GAS]) is a gram-positive pathogen that is responsible for a wide range of diseases in people. GAS can cause localized infections of the throat or skin, such as pharyngitis and impetigo, or deep tissue infections, such as fasciitis and myositis. GAS can also cause systemic infections, including toxic shock syndrome and septicemia (for a review see reference 7). For its success as a pathogen, GAS must be able to sense and respond to the different and changing environments that it encounters during infection, and its response to these probably ultimately controls disease outcome. Thus, understanding of regulation of GAS gene expression is critical for understanding and predicting GAS disease outcome.
Currently, two global transcriptional regulators have been identified in GAS. The multiple gene activator Mga regulates transcription of several virulence genes, including the gene encoding the M protein, which is considered the major virulence factor of this pathogen (6, 18, 29, 31). Transcription of mga is responsive to changes in environmental conditions (4, 25). In addition, CovR represses transcription of about 15% of the GAS genome, either directly or indirectly (14). Among the operons that CovR regulates directly is the has operon, which contains the biosynthetic genes necessary for the production of the hyaluronic acid capsule, another important virulence factor of this organism (3, 12, 15, 22). Because CovR appears to be the response regulator of a two-component signal transduction system, it seems likely that it facilitates alterations in gene expression in the GAS as it encounters different environments.
In addition to specific transcriptional regulators whose activity may vary in response to environmental conditions, secondary RNA polymerase sigma factors play a critical role in a wide variety of bacteria in regulating gene transcription in an environmentally sensitive way. Such secondary sigma factors control a vast range of processes in different bacteria and respond to changes in environmental conditions including pH, osmotic shock, the presence of specific ions, and temperature. In Escherichia coli, Pseudomonas aeruginosa, and Salmonella enterica serovar Typhimurium, secondary sigma factors are essential for the transcription of virulence factors (10, 11, 38).
The sequence of the GAS genome indicates that it encodes the primary or housekeeping sigma factor, sigA, and two copies of a gene with homology to comX of Streptococcus pneumoniae (21). We have named the latter genes sigX1 and sigX2 because we have recently shown that their product acts in vitro as a sigma factor (27). Purified recombinant σX directs RNA polymerase from GAS to specifically use promoters (e.g., cinA and femB) that contain a sequence similar to that recognized by ComX of S. pneumoniae (called the “cin-box”) in vitro (27). However, whether GAS σX can function in vivo has not been tested, and its role in GAS has not been investigated. Because two identical copies of the structural gene for σX are found in all GAS strains for which data are available, it seems likely that σX plays an important role in GAS biology.
To begin to understand the role of σX in GAS, we deleted both copies of the σX structural gene and found that sigX is dispensable for growth under standard laboratory conditions. Moreover, we found that the level of sigX transcription is very low when the GAS is grown under these conditions. Therefore, we expressed the sigX gene in GAS from a heterologous promoter to study its transcriptional effects. We show that expression of sigX in GAS leads to transcription from three different promoters containing Cin box sequences. Furthermore, we have identified ClpP as a negative regulator of the σX protein.
MATERIALS AND METHODS
Bacterial strains and plasmids.
All GAS strains are derivatives of the M6 serotype strain JRS4 (35) and were grown in Todd-Hewitt medium supplemented with 0.2% yeast extract (THY).
A fragment of DNA directly upstream of the sigX gene (this sequence is identical for sigX1 and sigX2) was PCR amplified from JRS4 chromosomal DNA with primers 5′CACACAGAATTCACGCTGCTCTTGTCTCTATCGACA3′ and 5′CACACAGGATCCGTTGGAAATCAATCGCAGAG3′ and cloned into the EcoRI and BamHI restriction sites of pLitmus28 (New England Biolabs) to created plasmid pJO106. A fragment of DNA directly downstream of sigX1 was PCR amplified from JRS4 chromosomal DNA with primers 5′CACACAAGATCTCACAGCAGTTGTAAGCAAGACC3′ and 5′CACACAACTAGTGGATCCGGTCATACCTTATGTAAGTCGC3′ and cloned into the BglII and SpeI restriction sites of pJO106 to create pJO112. The Ω kanamycin resistance cassette of pUC4ΩKan (29) was removed by BamHI digestion and cloned into the BglII restriction site of pJO112 to create pJO117. The BamHI restriction fragment from pJO117 was then cloned into pJRS233 (30), creating plasmid pJO118.
A DNA fragment directly downstream of sigX2 was PCR amplified from JRS4 chromosomal DNA with primers 5′CACACAAGATCTCACAGCAGTTGTAAGCAAGACC3′ and 5′CACACAACTAGTGGATCCCTCATTGATACGCCCAATCA3′ and cloned into the BglII and SpeI restriction sites of pJO106 to create pJO119. A DNA fragment of the cat86 gene was PCR amplified from plasmid pLZ12 (9) with primers 5′CACACAGGATCCAGTTCAACAAACGAAAATTG3′ and 5′CAGCGGGATCCCATCTAGGCCTCCTCATATTATAAAAGCCAGTC3′ and cloned into the BglII restriction site of pJO119 to create plasmid pJO133. The gusA gene was PCR amplified from plasmid pMLK99 (17) with primers 5′ACACACATGCATCGACGGTATCGATAAGCTTG3′ and 5′ACACACATGCATAAGCTTCCCCACCGAGGCTGTAGC3′ and cloned into the NsiI restriction site of pJO133 to create pJO138. The BamHI restriction fragment from pJO139 was then cloned into the BamHI restriction site of pJRS233 to create pJO140.
A DNA fragment directly downstream of sigX2 was PCR amplified from AM3 (37) chromosomal DNA with primers 5′CACACAAGATCTCACAGCAGTTGTAAGCAAGACC3′ and 5′CACACAACTAGTGGATCCGTCCACTGAAATCAGTTGATG3′ and cloned into the BglII and SpeI restriction sites of pJO106 to create pJO120. A DNA fragment of the cat86 gene was PCR amplified from plasmid pLZ12 (9) with primers 5′CACACAGGATCCAGTTCAACAAACGAAAATTG3′ and 5′CAGCGGGATCCCATCTAGGCCTCCTCATATTATAAAAGCCAGTC3′ and cloned into the BglII restriction site of pJO120 to create plasmid pJO137. The BamHI restriction fragment from pJO137 was then cloned into the BamHI restriction site of pJRS233 to create pJO141.
pJO118 and pJO140 were then used to delete sigX1 and sigX2, respectively, in the M6 strain JRS4. pJO118 and pJO141 were used to delete sigX1 and sigX2, respectively, in M3 strain AM3. Plasmids pJO118, pJO140, and pJO141 were introduced into GAS cells by electroporation, and cells were plated on THY agar with 200 μg of kanamycin/ml or 2.5 μg of chloramphenicol/ml and grown at 30°C, which is a permissive temperature for the replication of these three plasmids. Single colonies were picked, inoculated into THY broth, and grown overnight at 37°C, which is nonpermissive for replication of these plasmids. Cells were then plated for single colonies on THY agar plates with 200 μg of kanamycin/ml or 2.5 μg of chloramphenicol/ml and grown at 37°C. Resulting colonies contained the plasmid integrated into the chromosome by homologous recombination. Individual recombinants were screened for the loss of the plasmid by their sensitivity to 0.5 μg of erythromycin/ml while maintaining kanamycin or chloramphenicol resistance on THY agar plates at 37°C. The resulting sigX double deletion strain in M6 was named JOS21, and the sigX double deletion strain in M3 was named JOS24.
The plasmid pJO162 was constructed to constitutively express the sigX gene. The veg promoter (26) was PCR amplified from Bacillus subtilis MB24 (28) chromosomal DNA with primers 5′GTCCAATTAACAGTTGAAAAC3′ and 5′CACCTCACTACATTTATTG3′. The sigX gene was PCR amplified with primers 5′AATGTAGTGAGGTGAAAGGAGACTCAAAATGTCG3′ and 5′GATTTACCCCGAATTCCTTATAGG3′. The veg promoter and the sigX gene were then fused together by PCR with primers 5′CACAAAGCTTCTGCATAGGAGAGCTATGCG3′ and 5′CACACAGAATTCTTACAACTGCTGTGCAAATTCCTT3′ and cloned into the EcoRI and HindIII restriction sites of plasmid pLZ12-Spec (1). pJO162 was introduced into GAS cells by electroporation, and transformants were grown on THY agar plates containing 100 μg of spectinomycin/ml.
An internal fragment of the clpP gene was PCR amplified using Pfu DNA polymerase (Stratagene) from JRS4 chromosomal DNA with primers 5′CACAGGATCCCCTGTTGTTATTGAACAAAC3′ and 5′CACAGGATCCTGCTGCGATAGCCATATCCG3′ and blunt end cloned into the EcoRV restriction site of pSK-erm, creating pJO164. pSK-erm contains an erythromycin resistance cassette from Tn1545 (39) cloned to replace the bla gene of pBluescript SK (Stratagene). pJO164 was introduced into GAS cells by electroporation, and transformants resulting in the insertional inactivation of the clpP gene by pJO164 were selected on plates containing 0.5 μg of erythromycin/ml. A transformant was picked and named JOS34. Southern blot analysis was used to confirm the mutation in clpP.
RNA purification.
Bacteria were grown at 37°C in THY broth to an optical density at 600 nm (OD600) of 0.7. Cells were treated with 2 mg of lysozyme/ml and 1% sodium dodecyl sulfate and then boiled to lyse the bacteria. RNA was harvested as described in the work of Macdonald et al. by CsCl2 purification and resuspended in a final volume of 400 μl of H2O to a final concentration of ∼3.0 μg/μl (24).
Primer extension reactions.
Primers cinA-R (5′CCGACAGAAATTGAGCATTGG3′), smf-R (5′TTTGGTAGTCAAGAATATTGAGAATG3′), sigX-R2 (5′GTCATCTCTATCCCACAATTG3′), and cglA-PE2 (5′GATCATATTGATCTGCTCTTGGC3′) were end labeled with [γ-32P]ATP as described in the T4 polynucleotide kinase user manual (Promega). Then, 1.5 pmol of labeled primer was added to 20 μg of total RNA along with annealing buffer to final concentrations of 200 mM KCl and 20 mM Tris (pH 8.3) in a final volume of 10 μl. The RNA and primer were heated to 80°C for 5 min and then slowly cooled to 42°C. Elongation buffer was added to a final concentration of 100 mM Tris (pH 8.3)-10 mM MgCl2-10 mM dithiothreitol in a final volume of 20 μl. Two hundred units of avian myeloblastosis virus reverse transcriptase (Promega) was added, and the reaction mixture was incubated at 42°C for 30 min. The primer extension product was ethanol precipitated, resuspended in 5 μl of formamide sequencing loading buffer, and subjected to electrophoresis in a 6% polyacrylamide gel containing 7 M urea alongside a sequencing reaction ladder generated with primer cinA-R or sigX-R2 and template pJO96 or pJO162.
Immunoblotting.
Cultures were grown in THY broth at 37°C to an OD600 of 0.7, harvested by centrifugation, and resuspended in 1 ml of phosphate-buffered saline, pH 7.0. Samples were then added to lysing matrix (Applied Biosystems) and lysed in a Fast Prep cell disruptor (Savant). To obtain the soluble protein fraction, samples were centrifuged at 13,000 × g, and the supernatant was saved. Twenty micrograms of total protein was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and immunoblotting was done as described in the work of Ozin et al. (28). The blot was reacted with a 1:5,000 dilution of rabbit anti-σX antibody raised against recombinant σX purified from E. coli (27) and detected with anti-rabbit-horseradish peroxidase by enhanced chemiluminescence.
RESULTS
Disruption of sigX1 and sigX2.
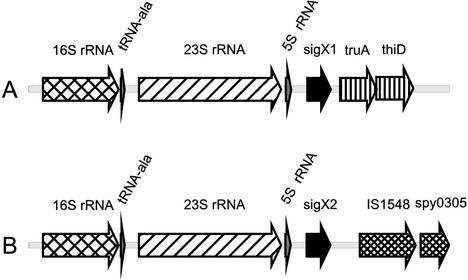
There are two identical copies of the sigX gene at unlinked regions of the chromosome of the GAS strains for which the genome sequence is available (M types 1, 3, and 18) (2, 13, 36). In addition, Southern blot analysis showed that the M6 strain JRS4, used in our studies, also contains two copies of sigX (M. J. Federle and J. R. Scott, unpublished data). In all of these strains, both copies of the sigX genes reside directly downstream of 6 kb of identical sequence encoding an rRNA operon (Fig. 1). The conserved sequence at the sigX loci terminates approximately 30 bp downstream from the sigX coding sequence.
FIG. 1.
Genetic organization of the sigX1 and sigX2 loci. Shown are 9-kb segments of GAS DNA surrounding sigX1 (A) and sigX2 (B) from M1 strain SF370. The two sigX genes are represented by black arrows. Six kilobases of sequence upstream of sigX are conserved for each copy of the two genes and contain two of the six rRNA operons. The sequence downstream from each copy of sigX is unique at each locus. IS1548 is present in all three sequenced strains but not in JRS4.
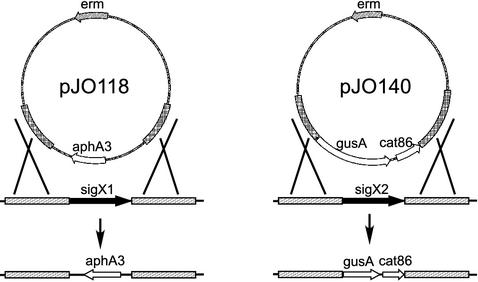
Homologous recombination was used to sequentially delete each copy of the sigX gene in an M6 and an M3 strain (Fig. 2). The sigX1 gene was replaced with aphA3, conferring kanamycin resistance, and sigX2 was replaced with a fragment containing cat86, which confers chloramphenicol resistance (Fig. 2). The resulting strains were confirmed by Southern blot analysis to have both copies of the sigX gene deleted (data not shown). The growth rates of the parent and the double sigX mutant at 37°C in THY broth for both the M6 strain and the M3 strain showed no obvious difference (data not shown).
FIG. 2.
Deletion of the sigX1 and sigX2 genes. Plasmid pJO118 was used to delete sigX1, and plasmid pJO140 was used to delete sigX2. Shaded regions indicate sequences upstream and downstream from sigX1 and sigX2 that were cloned into pJO118 and pJO140 to target homologous recombination with the GAS chromosome.
To examine σX accumulation in these cultures, we used a σX-specific polyclonal antibody raised against recombinant σX purified from E. coli (27) in Western immunoblotting assays. The sensitivity of this antibody was tested by performing blotting assays on serial dilutions of purified recombinant σX premixed with GAS protein extracts. Although 0.5 ng of recombinant σX was detected on the immunoblots (data not shown), we did not detect σX in cell extracts from the GAS cultures (data not shown). Therefore, there were fewer than 75 molecules of σX per cell in the JRS4 strain.
Expression of sigX from a heterologous promoter leads to expression of σX-dependent genes in GAS.
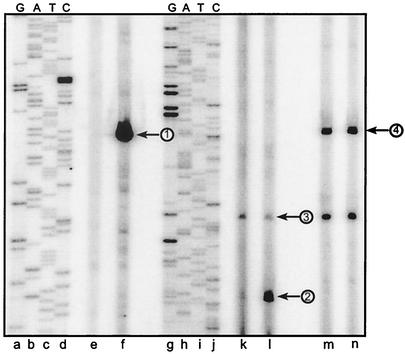
Since there was little expression of sigX during growth of GAS in THY broth, we expressed the sigX gene from a heterologous promoter in GAS. The sigX gene was cloned downstream from the B. subtilis veg promoter to create pJO162 (see Materials and Methods), a multicopy plasmid that replicates in GAS. The veg promoter was expected to be active in GAS because it contains a σA consensus −35 sequence (TTGACA) and a near-consensus −10 sequence (TACAAT) with 17 bp between the two elements (26). To assess sigX transcription from this promoter, primer extension analysis was used. RNA was isolated late in exponential phase (OD600 of 0.7) and 2 h after the beginning of the stationary phase. A sigX-specific transcript was detected from strain JRS4/pJO162 but not from JRS4 without the sigX expression plasmid (Fig. 3, lanes e and f). No effect of growth phase on the amount of sigX transcript was detected (data not shown). Confirmation that the product produced originated from the veg promoter at a G residue 5 nucleotides downstream from the −10 sequence was obtained by comparison of the sigX product size with the sequence of pJO162 generated with the identical primer used in the primer extension reaction (Fig. 3, lane f).
FIG. 3.
Primer extension analysis of transcription products in GAS expressing sigX from the veg promoter. Shown is an autoradiograph of radiolabeled primer extension products subjected to electrophoresis on a 6% polyacrylamide gel containing 7 M urea. Twenty micrograms of GAS RNA from strains JRS4 (wild type) (lanes e, k, and m) and JRS4/pJO162 (sigX expression plasmid) (lanes f, l, and n) was subjected to primer extension analysis with primers sigX-R2 (lanes e and f), cinA-R (lanes k and l), and Pemm-PE (lanes m and n). DNA template pJO162 (lanes a to d) or pJO96 (cinA promoter cloned in pUC19 [27]) (lanes g to j) was used for sequencing with radiolabeled primers sigX-R2 (lanes a to d) or cinA-R (lanes g to j). Arrows show the positions of the sigX transcript originating from pJO162 (1), the σX-dependent cinA product (2), the σA-dependent cinA product (3), and the σX-independent emm product (4).
Purified σX directs the utilization of a promoter located upstream from the GAS cinA gene by GAS core RNA polymerase in in vitro transcription reactions (27). However, it is not known whether σX functions within GAS cells. Therefore, we tested whether expression of sigX on the expression plasmid in GAS strain JRS4/pJO162 would result in production of a transcript from the σX-dependent cinA promoter. Primer extension reactions demonstrated that strain JRS4 grown in THY broth to late exponential phase produced little σX-dependent transcript from the cinA promoter (Fig. 3, lane k). However, when sigX was expressed in GAS from the veg promoter, a significant amount of σX-dependent cinA transcript was detected (Fig. 3, lane l). We mapped the 5′ end of this transcript to an A residue that was shown previously to be the start point of σX-dependent transcription from this promoter in vitro (Fig. 3, lane l). As noted previously, this transcription start point is 8 nucleotides downstream from the Cin box-like sequence.
We also observed a second cinA transcript that appeared to start at an A residue positioned 6 nucleotides downstream from a −10 promoter sequence (TAAAAT) similar to those recognized by σA-RNA polymerase (Fig. 3, lanes k and l). As expected for a σA-dependent transcript, the appearance of this transcript was not dependent upon the presence of the σX expression plasmid. Primer extension reactions from the σX-independent emm gene showed equivalent transcripts, indicating that equivalent amounts of RNA from the two strains had been used in each reaction (Fig. 3, lanes m and n).
σX accumulates in a clpP mutant.
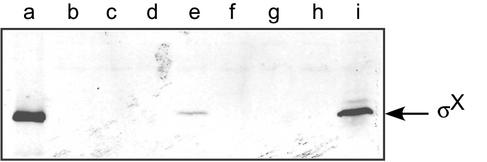
The amount of σX protein in the GAS sigX expression strain was examined by immunoblotting with the σX-specific antiserum. Although we found that JRS4/pJO162 contained a significant amount of a sigX transcript (Fig. 3, lane l), no σX protein was detected in this strain with our antibody (Fig. 4, lanes c and g). As described above, this assay would detect as few as 75 molecules of σX per cell. This suggested the possibility that σX is unstable in the GAS cell.
FIG. 4.
Immunoblot analysis of σX in a clpP mutant. Soluble fractions (lanes b to e) and whole-cell fractions (lanes f to i) of strains JRS4 (lanes b and f), JRS4/pJO162 (sigX expression plasmid) (lanes c and g), JOS34 (clpP mutant) (lanes d and h), and JOS34/pJO162 (lanes e and i) were subjected to electrophoresis on a sodium dodecyl sulfate-15% polyacrylamide gel and immunoblotted with anti-σX antiserum. Five nanograms of purified recombinant σX protein (lane a) ran at the predicted molecular mass of 19.6 kDa. The position of the σX protein is indicated.
One of the major proteases affecting cytoplasmic protein stability in many bacteria is ClpP, which is conserved in most gram-negative and gram-positive bacteria (32). Inactivation of clpP in S. pneumoniae results in an extended period of competence, consistent with the idea that it affects the amount of ComX in this organism (33). To test whether the ClpP protease of GAS plays a role in controlling σX protein level, we generated a clpP mutant in JRS4 (strain JOS34) and in JRS4/pJO162 (strain JOS34/pJO162). These strains were then tested for the presence of the σX protein by Western blotting with σX antiserum. No σX protein was detected in the clpP mutant (strain JOS34) as previously seen for JRS4 and JRS4/pJO162 (Fig. 4, lanes b, c, d, f, g, and h). However, a strain that contained both the sigX expression plasmid and the clpP mutation produced enough σX protein to be detected easily (Fig. 4, lanes e and i). Some σX protein was detected in the soluble fraction of the cell lysate; however, most accumulated in the insoluble fraction (Fig. 4, lanes e and i). This suggests that, under conditions of overexpression of sigX, ClpP negatively regulates σX by directly or indirectly controlling the level of the protein in the cell.
Accumulation of σX leads to increased transcription of σX-dependent genes.
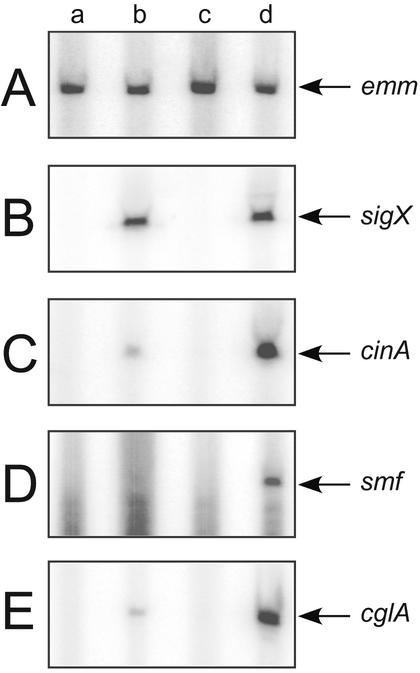
To assess the effect of σX accumulation in a clpP mutant, we used primer extension analysis. We assayed transcription from three different cin-box promoters, cinA, smf, and cglA, as well as promoters Pveg-sigX and emm, which do not contain cin-boxes, in GAS strains JRS4, JRS4/pJO162, JOS34, and JOS34/pJO162. The emm gene, used as a σX-independent control, displayed equivalent amounts of transcript from all four strains, indicating that equal amounts of mRNA were used as template in each reaction (Fig. 5A, lanes a to d). Primer extension analysis also showed that the clpP mutation had no effect on sigX transcription from the expression plasmid (Fig. 5B, lanes b and d). As expected, the two strains constitutively expressing sigX, JRS4/pJO162 and JOS34/pJO162, produced more cinA, smf, and cglA transcript than the other two strains (Fig. 5B to D, lanes a to d). In addition, strain JOS34/pJO162 produced significantly more σX-dependent transcript from cinA, smf, and cglA than did JRS4/ppJO162 (Fig. 5C to E, lanes b and d). These data indicate that increasing the effective concentration of σX protein in the cell by mutation of clpP is sufficient to increase transcription from these three Com box promoters.
FIG. 5.
Transcription products in the clpP mutant. Equal amounts of RNA from strains JRS4 (lane a), JRS4/pJO162 (sigX expression plasmid) (lane b), JOS34 (clpP mutant) (lane c), and JOS34/pJO162 (lane d) were subjected to primer extension analysis with primers Pemm-PE (A), sigX-R2 (B), cinA-R (C), smf-R (D), and cglA-PE2 (E) and subjected to electrophoresis on a 6% polyacrylamide gel containing 7 M urea. The position of each transcript is indicated.
ClpP controls the activity of σX expressed from its native promoter.
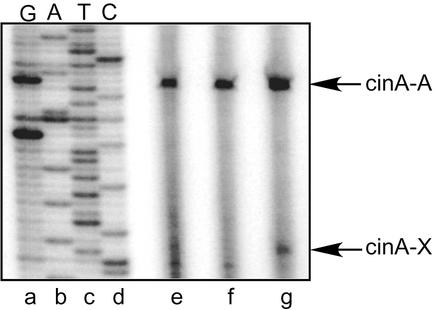
The previous experiments showed that clpP affects the amount of σX protein and σX transcriptional activity when sigX is overexpressed from a strong promoter on a multicopy plasmid. To assess the effect of ClpP on σX activity when sigX is expressed from its wild-type locus on the chromosome, we used primer extension reactions to detect cinA transcripts. σX-dependent cinA transcript was not detected in the wild-type strain or in the sigX mutant strain (Fig. 6, lanes e and f). However, a weak σX-dependent cinA transcript was detected in the clpP mutant strain (Fig. 6, lane g). These experiments indicate that ClpP inhibits σX activity when sigX is expressed from its normal chromosomal positions under its own promoter.
FIG. 6.
Primer extension analysis of cinA transcription in a clpP mutant. RNA from strains JRS4 (lane e), JOS21 (sigX deletion mutant) (lane f), and JOS34 (clpP mutant) (lane g) was subjected to primer extension analysis with primer cinA-R. DNA template pJO96 (cinA promoter cloned in pUC19 [27]) (lanes a to d) was used for sequencing with radiolabeled primer cinA-R (lanes a to d). Indicated in the figure are the positions of the σX-dependent cinA product (cinA-X) and the σA-dependent cinA product (cinA-A).
DISCUSSION
An important result of this work is that σX directs transcription of at least three genes when expressed in GAS. However, the amount of σX in GAS is limited at two levels. The sigX genes are not highly transcribed in JRS4 cultured in THY medium. It is unknown whether the sigX genes in this strain are transcribed more actively under other conditions. Analysis of the DNA upstream from the sigX open reading frame yields no candidate sequences resembling a classical σA-dependent promoter that could direct sigX expression. Therefore, if the sigX genes are transcribed in this strain under some, as yet unknown conditions, they may be transcribed from a novel promoter located directly upstream from sigX that is activated by an unknown signal, or from a promoter far upstream of sigX within the rRNA operon, possibly from the rRNA promoters themselves.
The sigX genes may be more actively transcribed in other GAS strains. S. pneumoniae and Streptococcus mutans contain σX homologs, ComX, that are essential for competence (21, 23). Although most GAS strains are not known to become competent for DNA uptake, an M14 GAS strain was found to be receptive for genetic exchange (16). It is unknown whether this process is transformation of competent cells by DNA; however, this genetic exchange required the sil locus (streptococcal invasion locus), which exhibits striking similarities to the ComDE two-component system that is required for competence development in S. pneumoniae (16). If this sil locus-dependent genetic exchange in GAS is similar to competence development in S. pneumoniae, it probably requires σX, which would serve a role homologous to that of ComX in S. pneumoniae. The sil locus was also shown to be essential for virulence in a mouse model of GAS infection (16). Therefore, if σX expression is activated by sil, σX may play a role in the virulence of this strain. While sil may be important for sigX expression in the M14 strain, multiple strains of GAS, including the M6 strain used in this study, do not contain the sil locus. Therefore, there may be strain-specific alternative mechanisms for regulating sigX.
ClpP limits the activity of σX, since disruption of clpP caused greater accumulation and activity of σX. Negative regulation of global transcriptional regulators by proteolysis is common among bacteria. ClpP is known to control the protein levels of transcriptional regulators in both E. coli and B. subtilis. The stationary-phase sigma factor, σS, in E. coli is specifically degraded by the ClpP protease during logarithmic growth when σS is not needed by the cell (34). This process relies on a protein, RssB, to bind to σS and deliver it to the proteolytic complex, thereby targeting it for degradation (41, 42). The use of a trans-acting targeting protein such as RssB adds specificity to the proteolytic process while not affecting the degradation of other proteins by ClpP (42). ClpP from B. subtilis also degrades a global regulator of transcription, ComK (40). ComK is a transcriptional activator that controls competence induction in this organism. Similar to σS in E. coli, ComK is targeted for degradation by a trans-acting protein, MecA (40). ClpP mutants in S. pneumoniae were shown to have an extended state of competence (33). One explanation for such an effect is that ClpP degrades the σX homolog, ComX, in wild-type S. pneumoniae and thus limits the duration of competence. Our immunoblot assays using a σX-specific polyclonal antibody showed increased accumulation of σX protein when the clpP gene was inactivated. Most likely ClpP in GAS also functions by directly degrading σX, although this has not yet been demonstrated. Degradation of σX by ClpP in GAS is a mechanism that would allow for σX activity to be negatively regulated in an irreversible fashion. Moreover σX activity may be induced by cellular conditions that reduce ClpP-dependent degradation of σX.
The amount of ClpP in both E. coli and B. subtilis is controlled at transcription. Transcription of clpP in E. coli is induced during heat shock by the secondary sigma factor σ32 (19). Gram-positive bacteria do not contain a σ32 homolog, and instead a transcriptional repressor, CtsR, negatively regulates clpP transcription in B. subtilis (8). ClpP degrades CtsR under stress conditions, which results in loss of repression of clpP (20). Consensus binding sites have been identified for CtsR in B. subtilis, and these binding sites have been shown to be conserved upstream of the clpP gene for many gram-positive organisms (8). A homolog of the ctsR gene is present in GAS, and the CtsR binding sites are present at the clpP promoter, suggesting the possibility of a similar mode of regulation (5, 8). The ctsR gene is negatively regulated in GAS by CovR, a global regulator of many genes including some needed for virulence (14). When CovR is inactive, CtsR would be produced in GAS cells, and this would lead to down-regulation of clpP. This in turn would increase σX activity coordinately with increased expression of virulence factors. However, as yet no direct experimental result demonstrates a role for σX in virulence of any GAS strain.
In conclusion, we found that σX functions in an M6 strain of GAS. However, σX function is limited by low-level transcription of its structural genes and by clpP, which appears to negatively regulate σX accumulation. Although the sigX genes are conserved among several GAS strains, the mechanisms regulating their transcription may be strain specific. It is important to determine whether σX plays different roles in virulence, survival, and possibly competence in different GAS strains.
Acknowledgments
This work was supported by PHS grant AI049013 from the National Institutes of Health. Jason Opdyke was partially supported by training grant AI07470 from the National Institutes of Health.
REFERENCES
- 1.Andersson, G., K. McIver, L. O. Heden, and J. R. Scott. 1996. Complementation of divergent mga genes in group A Streptococcus. Gene 175:77-81. [DOI] [PubMed] [Google Scholar]
- 2.Beres, S. B., G. L. Sylva, K. D. Barbian, B. Lei, J. S. Hoff, N. D. Mammarella, M. Y. Liu, J. C. Smoot, S. F. Porcella, L. D. Parkins, D. S. Campbell, T. M. Smith, J. K. McCormick, D. Y. Leung, P. M. Schlievert, and J. M. Musser. 2002. Genome sequence of a serotype M3 strain of group A Streptococcus: phage-encoded toxins, the high-virulence phenotype, and clone emergence. Proc. Natl. Acad. Sci. USA 99:10078-10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernish, B., and I. van de Rijn. 1999. Characterization of a two-component system in Streptococcus pyogenes which is involved in regulation of hyaluronic acid production. J. Biol. Chem. 274:4786-4793. [DOI] [PubMed] [Google Scholar]
- 4.Caparon, M. G., R. T. Geist, J. Perez-Casal, and J. R. Scott. 1992. Environmental regulation of virulence in group A streptococci: transcription of the gene encoding M protein is stimulated by carbon dioxide. J. Bacteriol. 174:5693-5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chastanet, A., and T. Msadek. 2003. ClpP of Streptococcus salivarius is a novel member of the dually regulated class of stress response genes in gram-positive bacteria. J. Bacteriol. 185:683-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, C., N. Bormann, and P. P. Cleary. 1993. VirR and Mry are homologous trans-acting regulators of M protein and C5a peptidase expression in group A streptococci. Mol. Gen. Genet. 241:685-693. [DOI] [PubMed] [Google Scholar]
- 7.Cunningham, M. W. 2000. Pathogenesis of group A streptococcal infections. Clin. Microbiol. Rev. 13:470-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Derre, I., G. Rapoport, and T. Msadek. 1999. CtsR, a novel regulator of stress and heat shock response, controls clp and molecular chaperone gene expression in gram-positive bacteria. Mol. Microbiol. 31:117-131. [DOI] [PubMed] [Google Scholar]
- 9.de Vos, W. M. 1986. Genetic improvement of starter streptococci by cloning and expression of the gene encoding for a non-bitter proteinase, p. 465-472. In E. Magnien (ed.), Biomolecular engineering programme—final report. Biomolecular engineering in the European Community: achievements in the research programme (1982-1986)—final report. Martinus Nijhoff, Lancaster, England.
- 10.Dineen, S. S., K. Takeuchi, J. E. Soudah, and K. J. Boor. 1998. Persistence of Escherichia coli O157:H7 in dairy fermentation systems. J. Food Prot. 61:1602-1608. [DOI] [PubMed] [Google Scholar]
- 11.Fang, F. C., S. J. Libby, N. A. Buchmeier, P. C. Loewen, J. Switala, J. Harwood, and D. G. Guiney. 1992. The alternative sigma factor katF (rpoS) regulates Salmonella virulence. Proc. Natl. Acad. Sci. USA 89:11978-11982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Federle, M. J., K. S. McIver, and J. R. Scott. 1999. A response regulator that represses transcription of several virulence operons in the group A Streptococcus. J. Bacteriol. 181:3649-3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferretti, J. J., W. M. McShan, D. Ajdic, D. J. Savic, G. Savic, K. Lyon, C. Primeaux, S. Sezate, A. N. Suvorov, S. Kenton, H. S. Lai, S. P. Lin, Y. Qian, H. G. Jia, F. Z. Najar, Q. Ren, H. Zhu, L. Song, J. White, X. Yuan, S. W. Clifton, B. A. Roe, and R. McLaughlin. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 98:4658-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graham, M. R., L. M. Smoot, C. A. Migliaccio, K. Virtaneva, D. E. Sturdevant, S. F. Porcella, M. J. Federle, G. J. Adams, J. R. Scott, and J. M. Musser. 2002. Virulence control in group A Streptococcus by a two-component gene regulatory system: global expression profiling and in vivo infection modeling. Proc. Natl. Acad. Sci. USA 99:13855-13860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heath, A., V. J. DiRita, N. L. Barg, and N. C. Engleberg. 1999. A two-component regulatory system, CsrR-CsrS, represses expression of three Streptococcus pyogenes virulence factors, hyaluronic acid capsule, streptolysin S, and pyrogenic exotoxin B. Infect. Immun. 67:5298-5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hidalgo-Grass, C., M. Ravins, M. Dan-Goor, J. Jaffe, A. E. Moses, and E. Hanski. 2002. A locus of group A Streptococcus involved in invasive disease and DNA transfer. Mol. Microbiol. 46:87-99. [DOI] [PubMed] [Google Scholar]
- 17.Karow, M. L., and P. J. Piggot. 1995. Construction of gusA transcriptional fusion vectors for Bacillus subtilis and their utilization for studies of spore formation. Gene 163:69-74. [DOI] [PubMed] [Google Scholar]
- 18.Kihlberg, B. M., J. Cooney, M. G. Caparon, A. Olsen, and L. Bjorck. 1995. Biological properties of a Streptococcus pyogenes mutant generated by Tn916 insertion in mga. Microb. Pathog. 19:299-315. [DOI] [PubMed] [Google Scholar]
- 19.Kroh, H. E., and L. D. Simon. 1990. The ClpP component of Clp protease is the sigma 32-dependent heat shock protein F21.5. J. Bacteriol. 172:6026-6034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kruger, E., D. Zuhlke, E. Witt, H. Ludwig, and M. Hecker. 2001. Clp-mediated proteolysis in Gram-positive bacteria is autoregulated by the stability of a repressor. EMBO J. 20:852-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, M. S., and D. A. Morrison. 1999. Identification of a new regulator in Streptococcus pneumoniae linking quorum sensing to competence for genetic transformation. J. Bacteriol. 181:5004-5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levin, J. C., and M. R. Wessels. 1998. Identification of csrR/csrS, a genetic locus that regulates hyaluronic acid capsule synthesis in group A Streptococcus. Mol. Microbiol. 30:209-219. [DOI] [PubMed] [Google Scholar]
- 23.Li, Y. H., N. Tang, M. B. Aspiras, P. C. Lau, J. H. Lee, R. P. Ellen, and D. G. Cvitkovitch. 2002. A quorum-sensing signaling system essential for genetic competence in Streptococcus mutans is involved in biofilm formation. J. Bacteriol. 184:2699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Macdonald, P. M., E. Kutter, and G. Mosig. 1984. Regulation of a bacteriophage T4 late gene, soc, which maps in an early region. Genetics 106:17-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McIver, K. S., A. S. Heath, and J. R. Scott. 1995. Regulation of virulence by environmental signals in group A streptococci: influence of osmolarity, temperature, gas exchange, and iron limitation on emm transcription. Infect. Immun. 63:4540-4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moran, C. P., Jr., N. Lang, S. F. LeGrice, G. Lee, M. Stephens, A. L. Sonenshein, J. Pero, and R. Losick. 1982. Nucleotide sequences that signal the initiation of transcription and translation in Bacillus subtilis. Mol. Gen. Genet. 186:339-346. [DOI] [PubMed] [Google Scholar]
- 27.Opdyke, J. A., J. R. Scott, and C. P. Moran, Jr. 2001. A secondary RNA polymerase sigma factor from Streptococcus pyogenes. Mol. Microbiol. 42:495-502. [DOI] [PubMed] [Google Scholar]
- 28.Ozin, A. J., A. O. Henriques, H. Yi, and C. P. Moran, Jr. 2000. Morphogenetic proteins SpoVID and SafA form a complex during assembly of the Bacillus subtilis spore coat. J. Bacteriol. 182:1828-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perez-Casal, J., M. G. Caparon, and J. R. Scott. 1991. Mry, a trans-acting positive regulator of the M protein gene of Streptococcus pyogenes with similarity to the receptor proteins of two-component regulatory systems. J. Bacteriol. 173:2617-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perez-Casal, J., J. A. Price, E. Maguin, and J. R. Scott. 1993. An M protein with a single C repeat prevents phagocytosis of Streptococcus pyogenes: use of a temperature-sensitive shuttle vector to deliver homologous sequences to the chromosome of S. pyogenes. Mol. Microbiol. 8:809-819. [DOI] [PubMed] [Google Scholar]
- 31.Podbielski, A., A. Flosdorff, and J. Weber-Heynemann. 1995. The group A streptococcal virR49 gene controls expression of four structural vir regulon genes. Infect. Immun. 63:9-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Porankiewicz, J., J. Wang, and A. K. Clarke. 1999. New insights into the ATP-dependent Clp protease: Escherichia coli and beyond. Mol. Microbiol. 32:449-458. [DOI] [PubMed] [Google Scholar]
- 33.Robertson, G. T., W. L. Ng, J. Foley, R. Gilmour, and M. E. Winkler. 2002. Global transcriptional analysis of clpP mutations of type 2 Streptococcus pneumoniae and their effects on physiology and virulence. J. Bacteriol. 184:3508-3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schweder, T., K. H. Lee, O. Lomovskaya, and A. Matin. 1996. Regulation of Escherichia coli starvation sigma factor (σs) by ClpXP protease. J. Bacteriol. 178:470-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scott, J. R., P. C. Guenthner, L. M. Malone, and V. A. Fischetti. 1986. Conversion of an M− group A Streptococcus to M+ by transfer of a plasmid containing an M6 gene. J. Exp. Med. 164:1641-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smoot, J. C., K. D. Barbian, J. J. Van Gompel, L. M. Smoot, M. S. Chaussee, G. L. Sylva, D. E. Sturdevant, S. M. Ricklefs, S. F. Porcella, L. D. Parkins, S. B. Beres, D. S. Campbell, T. M. Smith, Q. Zhang, V. Kapur, J. A. Daly, L. G. Veasy, and J. M. Musser. 2002. Genome sequence and comparative microarray analysis of serotype M18 group A Streptococcus strains associated with acute rheumatic fever outbreaks. Proc. Natl. Acad. Sci. USA 99:4668-4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stamp, T. C., and E. B. Hendry. 1937. The immunizing activity of certain chemical fractions isolated from haemolytic streptococci. Lancet i:257-259. [Google Scholar]
- 38.Suh, S. J., L. Silo-Suh, D. E. Woods, D. J. Hassett, S. E. West, and D. E. Ohman. 1999. Effect of rpoS mutation on the stress response and expression of virulence factors in Pseudomonas aeruginosa. J. Bacteriol. 181:3890-3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trieu-Cuot, P., C. Poyart-Salmeron, C. Carlier, and P. Courvalin. 1990. Nucleotide sequence of the erythromycin resistance gene of the conjugative transposon Tn1545. Nucleic Acids Res. 18:3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Turgay, K., J. Hahn, J. Burghoorn, and D. Dubnau. 1998. Competence in Bacillus subtilis is controlled by regulated proteolysis of a transcription factor. EMBO J. 17:6730-6738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou, Y., and S. Gottesman. 1998. Regulation of proteolysis of the stationary-phase sigma factor RpoS. J. Bacteriol. 180:1154-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou, Y., S. Gottesman, J. R. Hoskins, M. R. Maurizi, and S. Wickner. 2001. The RssB response regulator directly targets sigma(S) for degradation by ClpXP. Genes Dev. 15:627-637. [DOI] [PMC free article] [PubMed] [Google Scholar]