Abstract
Nucleotide sequence analysis of an ∼80-kb genomic region revealed an ∼65-kb locus that bears hallmarks of a pathogenicity island. This locus includes homologues of a type IV secretion system, mobile genetic elements, and known virulence factors. Comparative studies with other Legionella pneumophila strains and serogroups indicated that this ∼65-kb locus is unique to L. pneumophila serogroup 1 Philadelphia-1 strains.
The genus Legionella is a diverse group of gram-negative gamma proteobacteria that inhabit most aquatic environments as intracellular parasites of protozoa (for reviews, see references 23 and 40). Of more than 48 named species and their many serogroups, the dimorphic (12, 19) bacterium Legionella pneumophila serogroup 1 is most frequently associated with Legionnaires' disease, a sometimes fatal pneumonia that occurs following inhalation of bacterium-laden aerosols by susceptible individuals (6, 15).
Previous molecular studies identified several genomic loci associated with the pathogenesis of L. pneumophila serogroup 1. The dot/icm (for “defect in organelle trafficking/intracellular multiplication”) loci are required for intracellular growth in human macrophages and are homologous to the tra-trb genes of the Shigella flexneri IncI plasmid collb-P9 and to some of the genes coding for the type IV protein secretory system of the plant pathogen Agrobacterium tumefaciens (4, 8, 36, 37). Type IV protein secretory systems are composed of membrane-associated and transmembrane proteins, which are involved in the delivery of effector proteins or protein-DNA complexes to the host cell cytoplasm (8). Several bacterial pathogens use type IV systems to deliver virulence factors to modulate host defenses (3). Recently, another type IV secretion system, termed lvh, was described in L. pneumophila (38, 8); however, unlike the dot/icm system, the lvh system is dispensable for intracellular growth in macrophages and in the protozoan Acanthamoeba castellanii (38).
In addition to the dot-icm system, genes encoding adhesins and invasins have also been reported. The enh locus contains the rtxA gene, a member of the RTX (repeats in structural toxin) family, and is not only involved in adherence but appears to play a role in cytotoxicity and pore formation of the host cell (9). The htpAB locus encodes Hsp10 and the chaperone protein Hsp60, a member of the GroEL family of heat shock proteins. Most cellular Hsp60 is located in the periplasm and on the surface of L. pneumophila, where it mediates both adherence and invasion of nonphagocytic cells (17, 18). The roles of Hsp60 and the Dot/Icm system may be related because mutations in various dot/icm genes lead to a reduction in surface-associated Hsp60 (25).
Virulence differences have been noted clinically among the various serogroups and subtypes of L. pneumophila; however, the genetic basis for these variations has not been identified (15, 35). It has been noted that L. pneumophila serogroup 1 strains reacting with monoclonal antibody 2, including strains Philadelphia-1 and 130b (AA100), are isolated from more than 90% of cases of community-acquired Legionnaires' disease (11, 21). Alternatively, molecular phase variation in the virulence of L. pneumophila serogroup 1 strain Olda (nonreactive to monoclonal antibody 2) was attributed to an unstable ∼30-kb genetic element (27).
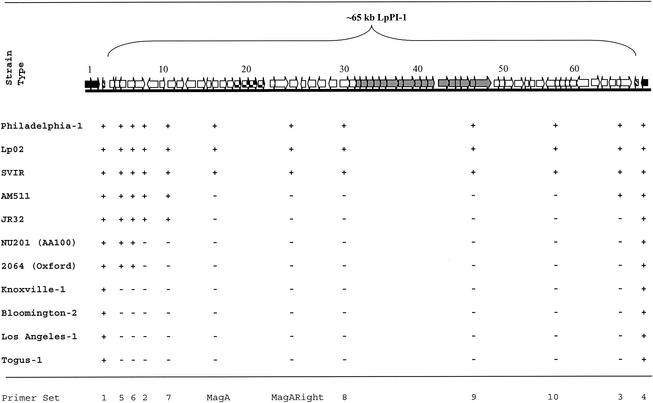
Here, we report the identification and genetic characterization of a 65-kb chromosomal region that is unique to the Philadelphia-1 strains (type strain and strains Svir and Lp02) of serogroup 1 L. pneumophila. The locus is absent in non-Philadelphia-1 serogroup 1 strains NU201 (AA100), Oxford, and Knoxville-1 and in some strains previously reported as Philadelphia-1, including AM511 and its derivative JR32. The locus is flanked by tRNA genes, contains regions rich in GC content, and is predicted to encode approximately 70 proteins, including MagA, a previously described developmental marker (19; unpublished results from our laboratory). Other genes are homologues of known virulence factors (carbon storage regulator [csrA] and peptide methionine sulfide reductases [msrAB]), and two gene clusters bear strong similarity in gene arrangement to Escherichia coli plasmid F genes encoding a type IV secretion system and to Ralstonia solanacearum megaplasmid genes encoding transmembrane proteins, respectively. The evolutionary origin of the putative pathogenicity island, here designated LpPI-1, is unknown but likely occurred recently on the basis of the conserved gene arrangement of the aforementioned two gene clusters observed within the locus.
Sequence compilation, PCR, Southern blotting, and comparative nucleotide sequence analysis of the ∼65-kb LpPI-1 locus in L. pneumophila strains.
The L. pneumophila strains used in this study are listed in Table 1, and the oligonucleotide primer pairs used in assembly of the ∼65-kb locus are listed in Table 2. The sequence of the ∼80-kb chromosomal region containing the ∼65-kb locus, designated LpPI-1, was compiled as follows. An ∼5-kb locus that included magA (19; unpublished results from our laboratory) was sequenced via in-house manual sequencing and automated sequencing (Institute for Marine Biosciences Joint Laboratory, National Research Council, Halifax, Nova Scotia, Canada), after which the peripheral ends of the magA locus sequence were matched to contigs taken from the Columbia Legionella Genome Project website (http://genome3.cpmc.columbia.edu/∼legion/). The ∼70 genes within the LpPI-1 locus were identified on the basis of protein homology searches with ORF Finder (National Center for Biotechnology Information website) and BLASTP (Fig. 1), and the identities of the majority of ∼70 genes were confirmed by the Columbia Legionella Genome Project website. Validation of gene content, as well as the flanking DNA sequence, of the LpPI-1 locus (common in other non-Philadelphia-1 strains) was determined by combinations of PCR walking (34) and Southern blot hybridizations with magA and the upstream sequence of transposase AB as probes (Table 2) (data not shown). The LpPI-1 locus and gene content among various L. pneumophila strains are depicted in Fig. 2. Standard PCRs (34) with each primer set were performed with 50-μl reaction volumes in a GeneAmp system 2400 (Perkin-Elmer) or a PTC programmable thermocycler (MJ Research, Inc.) for 25 cycles, and the products were subjected to agarose gel electrophoresis for analysis. The presence or absence of appropriate DNA bands was considered to be a positive or negative result, respectively, and the results are tabulated in Fig. 2. The results were confirmed by Southern blot analysis with the ECL direct nucleic acid labeling and detection system kit (Amersham Pharmacia Biotech), and probes were PCR amplified with the oligonucleotides listed in Table 2.
TABLE 1.
Bacterial strains used in this study and their reactivity to monoclonal antibody 2
| L. pneumophila strain, serogroup | Genotype and ATCCa no. (if applicable) | Monoclonal antibody 2 reactivity | Reference |
|---|---|---|---|
| Philadelphia-1, 1 | Type strain isolated from 1976 Philadelphia outbreak, 33152 | + | 6 |
| Lp02, 1 | rpsL thy hsdR derivative of Philadelphia-1 strain | Unknown | 4 |
| Svir, 1 | Spontaneous Strr isolate of Philadelphia-1 strain | Unknown | 24 |
| AM511, 1 | Strr r− m+ derivative of Philadelphia-1 strain | Unknown | 28 |
| JR32, 1 | Isogenic isolate of AM511 | Unknown | 32 |
| NU201, 1 | AA100 (spontaneous Strr isolate of clinical isolate 130b [Pontiac subtype 1]) | + | 29 |
| 2064, 1 | Oxford 4032E, 43110 | − | 13 |
| Knoxville-1, 1 | Clinical isolate, 33153 | + | 6 |
| Bloomington-2, 3 | Clinical isolate, 33155 | − | 6 |
| Los Angeles-1, 4 | Clinical isolate, 33156 | − | 6 |
| Togus-1, 2 | Clinical isolate, 33154 | − | 6 |
ATCC, American Type Culture Collection.
TABLE 2.
List of oligonucleotides used in this study
| Primer namea | Oligonucleotide sequence (5′ to 3′) | Amplified L. pneumophila Philadelphia-1 gene or region (reference) | Size of amplified region (bp) |
|---|---|---|---|
| F 1 | GCTTACTGAGTTACAACCTGA | Putative activation protein | 1,198 |
| R 1 | GCAATTTGGCAGCGTCAA | ||
| F 5 | GCGCTCAAGACGCTGATTCA | Intergenic region of tRNA valine and acetyltransferase | 562 |
| R 5 | GCACAGATCCACATTAATA | ||
| F 6 | ATCTTGAATTCCTCCACTGTA | Intergenic region of acetyltransferase and putative protein | 872 |
| R 6 | GATTAGCGGATCCTAACAGAA | ||
| F 2 | GCAACCCGGGATGAGTTTGA | Putative phage protein | 1,457 |
| R 2 | GCTGAGCAATTCTTGGTACGA | ||
| F 7 | GCATCGGCACTCTGTATA | 900 bp upstream of transposase AB | 869 |
| R 7 | GCTGATGATATGAGGATA | ||
| F MagA 5134 | CTCTATCGCTAACGCACAAGG | magA | 469 |
| R MagA 5603 | CGTTGAAGTAGTTAGTGAAAG | ||
| F MagA Right | GCCTTCGCCGAGGTGGACAGA | Intergenic region of mercuric reductase and hypothetical protein | 541 |
| R MagA Right | GCTATGGTTAGTTAGACGAGCA | ||
| F 8 | GCTTTATGTGGGATGCCACA | lvrA | 784 |
| R 8 | GCCTGATTTACGCTAAGGA | ||
| F 9 | GCTTATCATCACTTGCCCTTTA | traD | 633 |
| R 9 | GCAGAGATACACCACCAATCCGA | ||
| F 10 | GCAAGATTCCAACCTTCCGCA | Tn5 transposase WO | 894 |
| R 10 | GCGTTGGATCGAGTCTCTCGA | ||
| F 3 | GCGTATTGCGACTCAGGTTA | Putative prophage repressor | 705 |
| R 3 | GCAGTGCAGTTGGCTGAAGTCA | ||
| F 4 | GCCCTAAAGAGGCTGAAATCATA | Mevalonate kinase | 1,013 |
| R 4 | GCTTATGACCTATTAGCTGA | ||
| F VirB4 | GCTTTTATTTCGGGGAGTGC | virB4 (38) | 677 |
| R VirB4 | GTTTCCTCCGGAAAATGGCGG | ||
| F VirD4 | CATAGGCTTTACCGATGATG | virD4 (38) | 414 |
| R VirD4 | AGACTCGGAATAACCTTGGG |
F, forward; R, reverse.
FIG. 1.
Identification of genes within the L. pneumophila serogroup 1 strain Philadelphia-1 ∼65-kb LpPI-1 locus on the basis of protein homology searches. Striped arrow boxes indicate the borders of the LpPI-1 locus, checkered arrow boxes refer to the conserved gene arrangement also observed on R. solanacearum megaplasmid, and gray solid arrow boxes refer to the conserved gene arrangement observed also in E. coli plasmid F. Arrow boxes are not drawn to scale. See text for details.
FIG. 2.
Comparative nucleotide sequence analysis of L. pneumophila strains and serotypes. The PCR amplification result of each primer set (see Table 2) is represented by a plus or a minus sign for the presence or absence of amplified DNA, respectively. The gene numbers in the ∼65-kb LpPI-1 locus and the patterned and shaded boxes correspond to the schematic in Fig. 1.
We determined that L. pneumophila serogroup 1 strain Philadelphia-1 and its derivative strains Lp02 and SVIR contained the entire LpPI-1 locus (Fig. 2). However, ∼55 kb of the LpPI-1 locus is absent in AM511 and its derivative JR32. It should be noted that the region flanking the right end of LpPI-1, containing the L. pneumophila putative prophage repressor (gene 64), is present in the AM511 strain but absent in JR32, suggesting that the region may be unstable (Fig. 2). The Salmonella enterica serovar Typhimurium phage protein homologue (gene 7) defines the border of the ∼60-kb region absent in NU201 (AA100) and 2064 (Oxford 4032E) (Fig. 2). The LpPI-1 locus was completely absent in the L. pneumophila serogroup 1 Knoxville-1, serogroup 2 Togus-1, serogroup 3 Bloomington-2, and serogroup 4 Los Angeles-1 strains, where the valyl-tRNA synthetase, putative protein, and acetyltransferase homologues (genes 3, 4, and 5, respectively) are also absent (Fig. 2). The nucleotide sequence variability in this region is suggestive of a hypervariable or “plasticity zone” function, as noted in other bacteria, such as Helicobacter pylori (2).
Pathogenicity island features.
The LpPI-1 locus contains a variety of genes coding for hypothetical proteins, transport proteins acetyltransferases, and reductases (Fig. 1). The region also contains a large number of transposase, integrase, and phage-related genes located at the periphery of the LpPI-1 locus, hallmark elements defining pathogenicity islands (Fig. 1) (20). In addition, the LpPI-1 locus is also bordered by tRNA genes (genes 3 and 67), which are often insertion sites for laterally acquired DNA (Fig. 1) (20). The overall G+C content of the LpPI-1 locus is ∼38% (data not shown), which is not distinct from the overall 38% G+C content of the L. pneumophila genome (5). However, the region containing only the plasmid F gene homologues (Fig. 1) comprise a G+C content of ∼44%, which is substantially higher than that of the rest of the genome.
The magA-msrA1 locus.
The L. pneumophila magA (gene 16) gene encodes a 20- to 24-kDa protein that is expressed intracellularly during the transition from exponential phase to poststationary phase (19; unpublished results from our laboratory), suggesting a role in maturation of the cyst-like mature intracellular form (Fig. 1). Located upstream of magA is msrA1 (for peptide methionine sulfoxide reductase) (unpublished data; gene 17), which is transcribed in the opposite direction. MsrA1 has 80% homology with methionine sulfoxide reductases in other bacteria (41), some of which have been annotated as PilB transcription regulators (39) (Fig. 1). Two additional msrA genes, msrA2 and msrA3 (genes 26 and 27, respectively), which are orthologs of msrA and msrB of E. coli (41), are located within LpPI-1. As proteins can lose biological activity when methionines are oxidized by reduced oxygen intermediates (30), the antioxidant repair enzyme MsrA catalyzes the reduction of methionine sulfoxide residues in proteins to methionine and are established virulence determinants in E. coli (30), Neisseria gonorrhoeae (39), and Mycoplasma genitalium (10). In addition to the msrA genes in LpPI-1, it was observed that three other orthologs of msrA are located elsewhere in the L. pneumophila serogroup1 strain Philadelphia-1 genome sequence (Columbia Legionella Genome Project website).
Conserved gene cluster.
A gene cluster located immediately upstream of the magA-msrA1 locus contains four genes (genes 19, 20, 21, and 22) predicted to code for protein homologues (∼50% similarity) of signal peptide, hypothetical, and transmembrane proteins (Fig. 1) of R. solanacearum GMI1000 (33). The beta proteobacterium plant pathogen R. solanacearum has a 5.8-Mb genome that is organized into two replicons: a 3.7-Mb chromosome and a 2.1-Mb megaplasmid. The megaplasmid carries all of the virulence hypersensitive response (hrp) genes, as well as several primary metabolic enzymes, suggesting that the megaplasmid has a significant function in overall fitness and adaptation to environmental conditions (33). The aforementioned four proteins are encoded on the megaplasmid in the same gene arrangement as observed in the L. pneumophila LpPI-1 locus; however, the functional role of these proteins in virulence, if any, is unknown.
A second type IV protein secretory system.
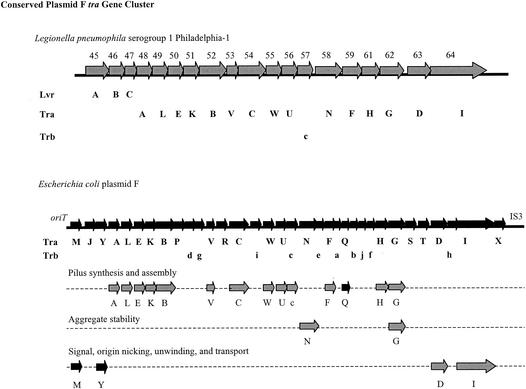
Another conserved gene cluster, located centrally in the LpPI-1 locus, encodes homologues (∼55% similarity) of the Tra proteins associated with plasmid F of E. coli (Fig. 1) (16). An alignment map of the L. pneumophila tra genes (no. 33 to 48) with those of plasmid F is presented in Fig. 3. The LpPI-1 tra gene homologues are arranged identically to those of plasmid F, with the exception of traM, traY, and traX, which are missing in L. pneumophila. In addition, all of the trb genes of the plasmid F transfer region, with the exception of trbC, are absent from LpPI-1. The trbC gene and the majority of the tra genes retained by the LpPI-1 locus have predicted functional roles in F pilus synthesis and assembly (16).
FIG. 3.
Conserved gene arrangement alignment of the tra gene cluster from the L. pneumophila serogroup 1 strain Philadelphia-1 LpPI-1 locus and the transfer region gene cluster of E. coli plasmid F. Genes from the LpPI-1 tra gene cluster that are homologous to the genes in the E. coli plasmid F transfer region and in an identical gene arrangement are gray, and genes that are present in the plasmid F transfer region but absent from the LpPI-1 tra gene cluster are black. The map of the transfer gene region of E. coli plasmid F is adapted from Frost et al. (16). See the text for details.
The prpA-lvrA-lvrB-lvrC gene cluster.
In the LpPI-1 locus, a cluster of four genes (no. 29 to 32) located upstream of traM are homologues of the putative prophage repressor protein (PrpA) and of the LvrA, LvrB, and LvrC (CsrA) proteins described previously in L. pneumophila (35, 38). The lvh-associated prpA-lvrA-lvrB-lvrC gene cluster previously described by Segal et al. (38) and Samrakandi et al. (35) is located upstream of the lvh region in L. pneumophila serogroup 1 strain Philadelphia-1 (Columbia Legionella Project Genome website), its derivative strain JR32 (39), and L. pneumophila serogroup 1 strain AA100 (36). A sequence alignment with ClustalW was performed with the amino acid sequences of the LvrA, LvrB, and LvrC (CsrA) proteins encoded by the lvh-associated prpA-lvrA-lvrB-lvrC gene cluster and the LpPI-1 prpA-lvrA-lvrB-lvrC gene cluster (Fig. 4), and the quantitative values are presented in Table 3. It should be noted that the JR32 prpA, lvrA, lvrB, and lvrC genes are identical, on both the protein and nucleotide levels, to those annotated in the L. pneumophila serogroup 1 strain Philadelphia-1 genome sequence at the Columbia Genome Center website. Comparison of the lvh-associated prpA-lvrA-lvrB-lvrC genes in the two strains (AA100 and JR32) revealed >87% amino acid identity, with the exception of PrpA (Table 3). However, comparison of the genes from the LpPI-1 prpA-lvrA-lvrB-lvrC gene cluster and the lvh-associated prpA-lvrA-lvrB-lvrC gene cluster showed significant differences at the amino acid identity level (Table 3). Therefore, it appears that the lvh-associated prpA-lvrA-lvrB-lvrC gene cluster described in association with the lvh locus is genetically distinct from the prpA-lvrA-lvrB-lvrC gene cluster described in association with the ∼65-kb LpPI-1 locus. Thus, although the lvh and LpPI-1 loci are both present in L. pneumophila serogroup 1 strain Philadelphia-1, it appears that recent gene duplication is not the origin of the LpPI-1 prpA-lvrA-lvrB-lvrC gene cluster.
FIG. 4.
ClustalW analysis of the amino acid sequences of the LvrA, LvrB, and LvrC (CsrA-LgrA) proteins encoded by the genomic prpA-lvrA-lvrB-lvrC gene cluster of L. pneumophila serogroup 1 strains AA100 and JR32 and the LpPI-1 prpA-lvrA-lvrB-lvrC gene cluster. Asterisks indicate identical residues, colons indicate similar residues with conserved codon base pair substitutions, and dots indicate residues with reduced similarity, as the codon base pair substitutions are semiconserved). The JR32 prpA-lvrA-lvrB-lvrC sequences are identical to those in the lvh locus of L. pneumophila serogroup 1 strain Philadelphia-1 currently being sequenced by the Columbia Legionella Genome Project. Note the genetic variability of the genes in the LpPI-1 locus in comparison to the genomic genes of strains AA100 and JR32.
TABLE 3.
ClustalW analysis of the proteins encoded by the genomic pprA-lvrA-lvrB-lvrC gene cluster located upstream of the lvh region in L. pneumophila serogroup 1 strain Philadelphia-1 derivative JR32 and L. pneumophila serogroup 1 strain AA100 in comparison to those encoded by the the LpPI-1 pprA-lvrA-lvrB-lvrC gene clustera
| Strains and protein | % Protein identity/similarityb
|
|||||||
|---|---|---|---|---|---|---|---|---|
| JR32
|
LpPI-1
|
|||||||
| PprA | LvrA | LvrB | LvrC | PprA | LvrA | LvrB | LvrC | |
| AA100 | ||||||||
| PprA | 35/56 | 40/57 | ||||||
| LvrA | 93/96 | 54/66 | ||||||
| LvrB | 88/89 | 40/52 | ||||||
| LvrC | 98/98 | 41/58 | ||||||
| LpPI-1 | ||||||||
| PprA | 54/77 | |||||||
| LvrA | 54/66 | |||||||
| LvrB | 40/52 | |||||||
| LvrC | 40/61 | |||||||
Note that the nucleotide sequences of the lvh-associated pprA-lvrA-lvrB-lvrC gene cluster are identical for L. pneumophila serogroup 1 strain Philadelphia-1 and its derivative strain J32.
See the text for details.
It should be noted that L. pneumophila serogroup Philadelphia-1-derivative strain Lp02 does not possess the lvh system, as determined by PCR analysis with primer sets for virB4 and virD4 (data not shown). Coincidentally, Samrakandi et al. (35) observed the absence of the lvh locus in Lp01 (a restriction-minus streptomycin-resistant variant derivative of L. pneumophila serogroup 1 strain Philadelphia-1) (4). Lp02 is a thymidine auxotroph derivative of Lp01 (4). They further suggested that loss of the lvh locus from Lp01 most likely occurred during the selection process for the restriction-minus phenotype, as the parental strain possessed the lvh locus.
The LpPI-1 LvrC protein is ∼85% homologous to the E. coli CsrA protein, whereas the lvh-encoded LvrC protein is only 63% homologous. E. coli CsrA is an RNA-binding protein that negatively regulates stationary-phase and virulence genes (31). A third csrA homologue gene (lgrA) is associated with the lipopolysaccharide biosynthesis gene cluster in L. pneumophila serogroup 1 strain Philadelphia-1 (14). LgrA is 88% homologous to E. coli CsrA; however, it has only 38 and 63% amino acid identity to Lvh LvrC and LpPI-1 LvrC, respectively (Fig. 4). While the functions of Lvh LvrC and LpPI-1 LvrC are not known, a function of LgrA has been elucidated. lgrA can complement E. coli csrA null mutants, and overexpression of lgrA in L. pneumophila strain Philadelphia-1 leads to reduction of flagellation, pigmentation, and altered cell morphology, suggesting that LgrA may facilitate rapid adaptation of the bacterium to the external environment (14).
Approximately 1,100 cases of Legionnaires' disease are reported each year in the United States, the majority of which are attributed to infections with L. pneumophila serogroup 1 strain Philadelphia-1 (15). Several subgroups of serogroup 1 are more often associated with human disease; however, a genetic correlation has not been established. We have identified an ∼65-kb putative pathogenicity island (LpPI-1) that bears hallmark features typical of pathogenicity islands as defined by Heuner and colleagues (22), and it appears that this LpPI-1 locus is unique to L. pneumophila serogroup 1 strain Philadelphia-1. This putative pathogenicity island encodes genes that are homologues of virulence genes in other bacterial pathogens, as well as homologues of the genes encoding the type IV system of the transfer region of plasmid F, and may contribute to the increased virulence associated with L. pneumophila serogroup 1 strain Philadelphia-1. Alternatively, genes associated with the LpPI-1 locus may contribute to increased fitness in natural environments or the stability of cyst-like MIF, in which MagA is abundant.
Genetic differences pertaining to virulence have been previously observed in L. pneumophila serogroup 1 strain Olda (27), in L. pneumophila serogroup 1 strain Philadelphia-1-derivative strains Lp01 and JR32, and in L. pneumophila serogroup 1 strain AA100 (35). In strain Olda, a 30-kb unstable genetic element identified in the chromosome has been shown to play a role in molecular phase variation (27). Excision of the genetic element (believed to be a nonfunctional phage remnant) mediated by a RecA-independent pathway attenuates virulence, and virulence is restored upon chromosomal insertion of the genetic element (27). It appears that this genetic element is unique to strain Olda, as it is not present in the L. pneumophila serogroup 1 strain Philadelphia-1 genome sequence (Columbia Legionella Genome Project website). Currently, 3,410,037 bp of the ∼4-Mb genome have been sequenced and organized into 73 contigs (Columbia Legionella Genome Project website). Another unique genetic region, designated the tra-1 locus, was identified in strain AA100 but was not present in strains JR32 and Lp01. The tra-1 locus encodes the traH, -I, -J, K, -L, and -M genes and thus was deemed to be involved in the type IV secretion system in strain AA100 (35). The lvh and rtxA regions were frequently found to be associated with Legionnaires' disease, whereas the tra-1 locus was not. In addition, the tra-1 locus was not found in the L. pneumophila serogroup 1 strain Philadelphia-1 genome sequence (Columbia Legionella Genome Project website) by a BLASTN search (data not shown). Our studies confirm the findings of Samrakandi et al. (35) indicating that of the three L. pneumophila strains examined, it appears that the tra-1 locus is unique to strain AA100.
LpPI-1 appears to be a unique marker for L. pneumophila serogroup 1 strain Philadelphia-1. Our studies also reveal that strains AM511 and JR32, reported to be derivative strains of L. pneumophila serogroup 1 strain Philadelphia-1, may have a different origin, although the possibility of loss of the LpPI-1 locus by culturing cannot be ruled out. The extensive genetic variability observed in the prpA-lvrA-lvrB-lvrC gene cluster and other previous observations of genetic variability in the dotA and rpoB genes (7, 26) further support our hypothesis that identical serogroup typing of strains does not imply genetic identity. However, it remains to be seen whether this putative LpPI-1 locus of L. pneumophila serogroup 1 strain Philadelphia-1 contributes to the virulence of this strain. Recently, Alli et al. (1) were unsuccessful in determining a correlation among five virulence traits (cytopathogenicity, intracellular replication, apoptosis, pore formation, and presence of the dot/icm loci) of seven L. pneumophila serogroup 1 strains in U937 human-derived macrophages. It is difficult to assess the level of virulence of those strains with and without the ∼65-kb LpPI-1 locus, as different definitions of virulence exist with regard to the experimental model used (i.e., aquatic protozoa, macrophages, nonphagocytic cells, and guinea pigs). In addition, virulence is largely dependent on the passage history of the particular strain in question. With this in mind, we are attempting to ascertain the importance of the LpPI-1 locus to the overall virulence of L. pneumophila serogroup 1 strain Philadelphia-1 by furthering our studies of the LpPI-1 locus through deletion analysis. Nevertheless, the presence or absence of this putative ∼65-kb LpPI-1 locus should be taken into account when analyzing the data obtained from reported derivative strains of L. pneumophila serogroup 1 strain Philadelphia-1.
Acknowledgments
This study was support by CIHR grant MT14443 to P.S.H. and R.A.G. and NSERC and Killam scholarships to M.F.H.
REFERENCES
- 1.Alli, O. A. T., S. Zink, K. von Lackum, and Y. Abu-Kwaik. 2003. Comparative assessment of virulence traits in Legionella spp. Microbiology 149:631-641. [DOI] [PubMed]
- 2.Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 3.Baron, C., D. O'Callaghan, and E. Lanka. 2002. Bacterial secrets of secretions: EuroConference on the biology of type IV secretion processes. Mol. Microbiol. 43:1359-1365. [DOI] [PubMed] [Google Scholar]
- 4.Berger, K. H., and R. R. Isberg. 1993. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol. Microbiol. 7:7-19. [DOI] [PubMed] [Google Scholar]
- 5.Brenner, D. J., A. G. Steigerwalt, R. E. Weaver, J. E. McDade, J. C. Feeley, and M. Mandel. 1978. Classification of the Legionnaires' disease bacterium: an interim report. Curr. Microbiol. 1:71-75. [Google Scholar]
- 6.Brenner, D. J., A. G. Steigerwalt, and J. E. McDade. 1979. Classification of the Legionnaires' disease bacterium: Legionella pneumophila, genus novum, species nova, of the family Legionellaceae, familia nova. Ann. Intern. Med. 90:656-658. [DOI] [PubMed] [Google Scholar]
- 7.Bumbaugh, A. C., E. A. McGraw, K. L. LePage, R. K. Selander, and T. S. Whittam. 2002. Sequence polymorphism of dotA and mip alleles mediating invasion and intracellular replication of Legionella pneumophila. Curr. Microbiol. 44:314-322. [DOI] [PubMed] [Google Scholar]
- 8.Christie, P. J., and J. P. Vogel. 2000. Bacterial type IV secretion: conjugation systems adapted to deliver effector molecules to host cells. Trends Microbiol. 8:354-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cirillo, S. L. G., L. E. Bermudez, S. El-Etr, G. E. Duhamel, and J. D. Cirillo. 2001. Legionella pneumophila entry gene rtxA is involved in virulence. Infect. Immun. 69:508-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dhandayuthapani, S., M. W. Blaylock, C. M. Bebear, W. G. Rasmussen, and J. B. Baseman. 2001. Peptide methionine sulfoxide reductase (MsrA) is a virulence determinant in Mycoplasma genitalium. J. Bacteriol. 183:5645-5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dournon, E., W. F. Bibb, P. Rajagopalan, N. Desplaces, and R. M. McKinney. 1988. Monoclonal antibody reactivity as a virulence marker for Legionella pneumophila serogroup 1 strains. J. Infect. Dis. 157:496-501. [DOI] [PubMed] [Google Scholar]
- 12.Faulkner, G., and R. A. Garduno. 2002. Ultrastructural analysis of differentiation in Legionella pneumophila. J. Bacteriol. 184:7025-7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fernandez, R. C., S. M. Logan, S. H. Lee, and P. S. Hoffman. 1998. Elevated levels of Legionella pneumophila stress protein Hsp60 early in infection of human monocytes and L929 cells correlate with virulence. Infect. Immun. 64:1968-1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fettes, P. S., V. Forsbach-Birk, D. Lynch, and R. Marre. 2001. Overexpression of a Legionella pneumophila homologue of the E. coli regulator csrA affects cell size, flagellation, and pigmentation. Int. J. Med. Microbiol. 291:353-360. [DOI] [PubMed] [Google Scholar]
- 15.Fields, B. S., R. F. Benson, and R. E. Besser. 2002. Legionella and Legionnaires' disease: 25 years of investigation. Clin. Microbiol. Rev. 15:506-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frost, L. S., K. Ippen-Ihler, and R. A. Skurray. 1994. Analysis of the sequence and gene products of the transfer region of the F sex factor. Microbiol. Rev. 58:162-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garduno, R. A., E. Garduno, and P. S. Hoffman. 1998a. Surface-associated Hsp60 chaperonin of Legionella pneumophila mediates invasion in a HeLa cell model. Infect. Immun. 66:4602-4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garduno, R. A., F. D. Quinn, and P. S. Hoffman. 1998b. HeLa cells as a model to study the invasiveness and biology of Legionella pneumophila. Can. J. Microbiol. 44:430-440. [PubMed] [Google Scholar]
- 19.Garduno, R. A., E. Garduno, M. Hiltz, and P. S. Hoffman. 2002. Intracellular growth of Legionella pneumophila gives rise to a differentiated form dissimilar to stationary-phase forms. Infect. Immun. 70:6273-6283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hacker, J., and J. B. Kaper. 2000. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 54:641-679. [DOI] [PubMed] [Google Scholar]
- 21.Helbig, J. H., S. Bernander, M. Castellani, Pastoris, J. Etienne, V. Gaia, S. Lauwers, D. Lindsay, P. C. Luck, T. Marques, S. Mentula, M. F. Peeters, C. Pelaz, M. Struelens, S. A. Uldum, G. Wewalka, and T. G. Harrison. 2002. Pan-European study on culture-proven Legionnaires' disease: distribution of Legionella pneumophila serogroups and monoclonal subgroups. Eur. J. Clin. Microbiol. Infect. Dis. 21:710-716. [DOI] [PubMed] [Google Scholar]
- 22.Heuner, K., M. Steinert, R. Marre, and J. Hacker. 2002. Genomic structure and evolution of Legionella species. Curr. Top. Microbiol. Immunol. 264:61-78. [PubMed] [Google Scholar]
- 23.Hoffman, P. S. 1997. Invasion of eukaryotic cells by Legionella pneumophila: a common strategy for all hosts? Can. J. Infect. Dis. 8:139-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoffman, P. S., C. A. Butler, F. D. Quinn. 1989. Cloning and temperature-dependent expression in Escherichia coli of a Legionella pneumophila gene coding for a genus-common 60-kilodalton antigen. Infect. Immun. 57:1731-1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoffman, P. S., and R. A. Garduno. 1999. Surface-associated heat shock proteins of Legionella pneumophila and Helicobacter pylori: roles in pathogenesis and immunity. Infect. Dis. Obstet. Gynecol. 7:58-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ko, K. S., H. K. Lee, M. Y. Park, M. S. Park, K. H. Lee, S. Y. Woo, Y. J. Yun, and Y. H. Kook. 2002. Population genetic structure of Legionella pneumophila inferred from RNA polymerase gene (rpoB) and DotA gene (dotA) sequences. J. Bacteriol. 184:2123-2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lüneberg, E., B. Mayer, N. Daryab, O. Kooistra, U. Zähringer, M. Rohde, J. Swanson, and M. Frosch. 2001. Chromosomal insertion and excision of a 30 kb unstable genetic element is responsible for phase variation of lipopolysaccharide and other virulence determinants in Legionella pneumophila. Mol. Microbiol. 39:1259-1271. [DOI] [PubMed] [Google Scholar]
- 28.Marra, A., S. J. Blander, M. A. Horwitz, and H. A. Shuman. 1992. Identification of a Legionella pneumophila locus required for intracellular multiplication in human macrophages. Proc. Natl. Acad. Sci. USA 89:9607-9611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moffat, J. F., P. H. Edelstein, D. P. Regula, Jr., J. D. Cirillo, and L. S. Tompkins. 1994. Effects of an isogenic Zn-metalloprotease-deficient mutant of Legionella pneumophila in a guinea-pig pneumonia model. Mol. Microbiol. 12:693-705. [DOI] [PubMed] [Google Scholar]
- 30.Moskovitz, J., M. A. Rahman, J. Strassman, S. O. Yancey, S. R. Kushner, N. Brot, and H. Weissbach. 1995. Escherichia coli peptide methionine sulfoxide reductase gene: regulation of expression and role in protecting against oxidative damage. J. Bacteriol. 177:502-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Romeo, T. 1998. Global regulation by the small RNA-binding protein CsrA and the non-coding RNA molecule CsrB. Mol. Microbiol. 29:1321-1330. [DOI] [PubMed] [Google Scholar]
- 32.Sadosky, A. B., L. A. Wiater, and H. A. Shuman. 1993. Identification of Legionella pneumophila genes required for growth within and killing of human macrophages. Infect. Immun. 61:5361-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salanoubat, M., S. Genin, F. Artiguenave, J. Gouzy, S. Mangenot, M. Arlat, A. Billault, P. Brottier, J. C. Camus, L. Cattolico, M. Chandler, N. Choisne, C. Claudel-Renard, S. Cunnac, N. Demange, C. Gaspin, M. Lavie, A. Moisan, C. Robert, W. Saurin, T. Schiex, P. Siguier, P. Thebault, M. Whalen, P. Wincker, M. Levy, J. Weissenbach, and C. A. Boucher. 2002. Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 415:497-501. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 35.Samrakandi, M. M., S. L. Cirillo, D. A. Ridenour, L. E. Bermudez, and J. D. Cirillo. 2002. Genetic and phenotypic differences between Legionella pneumophila strains. J. Clin. Microbiol. 40:1352-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Segal, G., M. Purcell, and H. A. Shuman. 1998. Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc. Natl. Acad. Sci. USA 95:1669-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Segal, G., and H. A. Shuman. 1999. Legionella pneumophila utilize the same genes to multiply within Acanthamoeba castellanii and human macrophages. Infect. Immun. 67:2117-2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Segal, G., J. J. Russo, and H. A. Shuman. 1999. Relationships between a new type IV secretion system and the icm/dot virulence system of Legionella pneumophila. Mol. Microbiol. 34:799-809. [DOI] [PubMed] [Google Scholar]
- 39.Skaar, E. P., D. M. Tobiason, J. Quick, R. C. Judd, H. Weissbach, F. Etienne, N. Brot, and H. S. Seifert. 2002. The outer membrane localization of the Neisseria gonorrhoeae MsrA/B is involved in survival against reactive oxygen species. Proc. Natl. Acad. Sci. USA 99:10108-10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Swanson, M. S., and B. K. Hammer. 2000. Legionella pneumophila pathogenesis: a fateful journey from amoebae to macrophages. Annu. Rev. Microbiol. 54:567-613. [DOI] [PubMed] [Google Scholar]
- 41.Taha, M. K., M. So, H. S. Seifert, E. Billyard, and C. Marchal. 1988. Pilin expression in Neisseria gonorrhoeae is under both positive and negative transcriptional control. EMBO J. 7:4367-4378. [DOI] [PMC free article] [PubMed] [Google Scholar]