Abstract
A psychrophilic bacterium, Cytophaga sp. strain KUC-1, that abundantly produces a NAD+-dependent l-threonine dehydrogenase was isolated from Antarctic seawater, and the enzyme was purified. The molecular weight of the enzyme was estimated to be 139,000, and that of the subunit was determined to be 35,000. The enzyme is a homotetramer. Atomic absorption analysis showed that the enzyme contains no metals. In these respects, the Cytophaga enzyme is distinct from other l-threonine dehydrogenases that have thus far been studied. l-Threonine and dl-threo-3-hydroxynorvaline were the substrates, and NAD+ and some of its analogs served as coenzymes. The enzyme showed maximum activity at pH 9.5 and at 45°C. The kinetic parameters of the enzyme are highly influenced by temperatures. The Km for l-threonine was lowest at 20°C. Dead-end inhibition studies with pyruvate and adenosine-5′-diphosphoribose showed that the enzyme reaction proceeds via the ordered Bi Bi mechanism in which NAD+ binds to an enzyme prior to l-threonine and 2-amino-3-oxobutyrate is released from the enzyme prior to NADH. The enzyme gene was cloned into Escherichia coli, and its nucleotides were sequenced. The enzyme gene contains an open reading frame of 939 bp encoding a protein of 312 amino acid residues. The amino acid sequence of the enzyme showed a significant similarity to that of UDP-glucose 4-epimerase from Staphylococcus aureus and belongs to the short-chain dehydrogenase-reductase superfamily. In contrast, l-threonine dehydrogenase from E. coli belongs to the medium-chain alcohol dehydrogenase family, and its amino acid sequence is not at all similar to that of the Cytophaga enzyme. l-Threonine dehydrogenase is significantly similar to an epimerase, which was shown for the first time. The amino acid residues playing an important role in the catalysis of the E. coli and human UDP-glucose 4-epimerases are highly conserved in the Cytophaga enzyme, except for the residues participating in the substrate binding.
Various psychrophilic and psychrotrophic microorganisms widely occur in natural and artificial environments, such as in cold rooms and refrigerated transport systems. They take part in the natural turnover of a variety of organic and inorganic compounds under cold conditions (13). In addition to l-threonine dehydratase and l-threonine aldolase, which are pyridoxal enzymes, l-threonine dehydrogenase (l-ThrDH; EC 1.1.1.103) plays an important role in l-threonine catabolism.
l-ThrDH catalyzes the NAD-dependent dehydrogenation of l-threonine to l-2-amino-3-oxobutyrate, which spontaneously decomposes to aminoacetone and CO2 (15) or is cleaved thiolytically by 2-amino-3-oxobutyrate coenzyme A lyase to glycine and acetyl coenzyme A (14). d-ThrDH exclusively catalyzes an analogous reaction with d-threonine (16). The dehydrogenation catalyzed by l-ThrDH occurs at the β-position of l-threonine, although other amino acid dehydrogenases (3, 8, 18-20) catalyze the α-deamination reactions. l-ThrDH is regarded as a kind of alcohol dehydrogenase in this respect. l-ThrDHs were purified to homogeneity from Escherichia coli K-12 (5), Clostridium sticklandii (26), chicken liver mitochondria (1), goat liver mitochondria (21), and porcine liver mitochondria (11). Only the E. coli enzyme has been intensively characterized, and the primary structure was determined (2). This enzyme belongs to the medium-chain alcohol dehydrogenase family, is composed of four identical subunits with a molecular weight of 35,000, and contains 1 g-atom of zinc per subunit (10). However, until now, no information on l-ThrDH from a psychrophile was available.
Recently, for the first time, we found the occurrence of NAD-dependent l-ThrDH in cells of a psychrophile, Cytophaga sp. strain KUC-1, isolated from Antarctic seawater. The enzyme is structurally unique among several l-ThrDHs so far studied. In particular, the amino acid sequence is similar to that of UDP-glucose 4-epimerase, which belongs to the short-chain dehydrogenase-reductase superfamily, but the enzyme does not resemble l-ThrDH from E. coli and d-ThrDH from Pseudomonas cruciviae.
We here describe the purification, characterization, and determination of the primary structure of the Cytophaga l-ThrDH, with emphasis on comparison with other l-ThrDHs and d-ThrDH.
MATERIALS AND METHODS
Materials.
Amino acids, their isomers and substituted derivatives, NAD+, and NADP+ were purchased from Wako Chemical Co. DEAE-Toyopearl 650M and Phenyl-Toyopearl 650M were products of Tosoh, and blue Sepharose CL6B and 5′-AMP Sepharose 4B were purchased from Pharmacia. The plasmid purification kit and gel extraction kit were purchased from Bio-Rad Laboratories Inc., and long and accurate PCR reagents were products of the Takara Shuzo Co.
Organism and culture conditions.
Throughout the experiments we used a psychrophilic bacterium, Cytophaga sp. strain KUC-1, which was isolated from Antarctic seawater. The organism is an atypical psychrophile because it grows optimally at 15°C and can grow even at temperatures over 25°C, though slowly (17, 20). The cells were grown aerobically at 15°C in a medium containing 2% polypeptone and 1% yeast extract (pH 7.0). The medium (7.0 liters) was inoculated with a seed culture (200 ml) of the cells grown at 15°C for 48 h (turbidity at 660 nm, about 10), and the cells were grown in a jar fermenter (10 liters; Marubishi Bioengineering, Tokyo, Japan) at 15°C for 48 h. The cells were harvested by centrifugation at 4°C, washed twice with a chilled 10 mM potassium phosphate buffer (pH 7.0) containing 0.75% NaCl, and then suspended in a 10 mM potassium phosphate buffer (pH 7.0; 0.5 g [wet weight] of cells/ml). E. coli (NovaBlue) was obtained from Novagen and grown aerobically at 37°C in a Luria-Bertani medium supplemented with ampicillin (100 μg/ml).
Purification of l-ThrDH.
All operations were carried out at temperatures in the range of 0 to 4°C. The cells were disrupted six times by ultrasonication for 5 min at intervals of 5 min. After centrifugation at 27,600 × g for 30 min, the supernatant solution was dialyzed against a 10 mM potassium phosphate buffer (pH 7.0) containing 0.01% 2-mercaptoethanol (the standard buffer).
The enzyme solution was applied to a DEAE-Toyopearl column (2.5 by 27 cm) previously equilibrated with the standard buffer, and the column was washed with 650 ml of the standard buffer. The enzyme was eluted with a 0 to 0.15 M potassium chloride gradient, and the active fractions were combined and concentrated by ultrafiltration (Advantec ultrafilter; PO200 membrane).
Ammonium sulfate powder was added to the enzyme solution to yield a concentration of 1.0 M. The mixture was cooled in an ice bath for 30 min and centrifuged at 27,600 × g for 30 min. The supernatant solution was applied to a Phenyl-Toyopearl column (2.5 by 14 cm) equilibrated with the standard buffer containing 1.0 M ammonium sulfate. After the column was washed with 450 ml of the standard buffer containing 1.0 M ammonium sulfate, the adsorbed protein was eluted with a 1.0 to 0 M ammonium sulfate gradient in the standard buffer. The active fractions were combined and concentrated.
After dialysis against the standard buffer, the enzyme solution was applied to a blue Sepharose column (2.5 by 6 cm) equilibrated with the standard buffer. After the column was washed with 150 ml of the standard buffer, the enzyme was eluted with a 0 to 0.4 M potassium chloride gradient. The active fractions were combined and concentrated.
The enzyme solution was dialyzed against the standard buffer and applied to a 5′-AMP Sepharose column (2.5 by 3.5 cm) equilibrated with the standard buffer. After the column was washed with 100 ml of the standard buffer containing 0.15 M KCl and 10 mM l-threonine, the enzyme was eluted with 100 ml of 0.2 mM NAD+ in the standard buffer containing 0.15 M KCl and 10 mM l-threonine.
The enzyme solution was concentrated and mixed with the same volume of the standard buffer containing 60% (wt/vol) sucrose and stored at −20°C. NAD+, l-threonine, KCl, and sucrose were removed from the solution by HW-50 gel filtration chromatography before use.
Enzyme assay of l-ThrDH.
The enzyme was assayed by spectrophotometrically monitoring NADH formed at 340 nm with a reaction mixture (3 ml) consisting of 25 mM l-threonine, 1 mM NAD+ in a 100 mM glycine-NaOH buffer (pH 9.0), and the enzyme at 30°C (the standard assay). The reactions were initiated by the addition of enzyme. The initial steady-state rates were determined from the initial linear portions of reaction progress curves. One unit of enzyme was defined as the amount of enzyme that catalyzes the formation of 1 μmol of NADH per min. The inhibition of Cytophaga l-ThrDH by UDP-galactose was tested under assay conditions that were standard except for the addition of 10 mM UDP-galactose.
The UDP-glucose 4-epimerase activity of the Cytophaga l-ThrDH was determined with a reaction mixture (150 μl) consisting of 10 mM UDP-galactose, 0.5 mM NAD+, and 0.03 U of E. coli UDP-glucose dehydrogenase (Calbiochem)/ml in a 100 mM glycine-NaOH buffer (pH 8.8) including 0.2 U of Cytophaga l-ThrDH/ml at 25°C.
Enzyme assay of UDP-glucose 4-epimerase.
The l-ThrDH activity of Streptococcus thermophilus UDP-glucose 4-epimerase (Calbiochem) was determined with a reaction mixture (150 μl) consisting of 25 mM l-threonine, 0.5 mM NAD+, and 0.03 U of Streptococcus thermophilus UDP-glucose 4-epimerase/ml in a 100 mM glycine buffer (pH 9.0) at 25°C. The inhibition of UDP-glucose 4-epimerase by l-threonine was tested with a reaction mixture (150 μl) consisting of 10 mM UDP-galactose, 0.5 mM NAD+, 0.03 U of UDP-glucose dehydrogenase/ml and 25 mM l-threonine in a 100 mM glycine buffer (pH 8.8), and 0.03 U of Streptococcus thermophilus UDP-glucose 4-epimerase/ml at 25°C.
Effects of temperature and pH.
The effects of temperature on enzyme activity were examined under assay conditions that were standard except for the temperatures (which ranged from 5 to 60°C). The effects of temperature on enzyme stability were examined by measurement of the residual activities under standard assay conditions after incubation of the enzyme (1 mg/ml) at various temperatures and at pH 7.0 for 60 min. The effects of pH on enzyme activity were examined at pHs in the range of 3.0 to 11.5 with the following 100 mM buffers: citrate (pH 3.0 to 6.0), potassium phosphate (pH 6.0 to 7.5), Tris-HCl (pH 7.5 to 9.0), and glycine-NaOH (pH 9.0 to 11.5).
Kinetics.
Kinetic constants were determined at various temperatures by measurement of initial velocities at various concentrations of one substrate at several fixed levels of the other substrate. Apparent maximum velocities (V′maxs) and apparent Michaelis constants (K′ms) were determined by Lineweaver-Burk double reciprocal plots. The maximum velocities (Vmaxs) and the Michaelis constants (Kms) for l-threonine were determined by plotting 1/V′max against the reciprocals of NAD+ and l-threonine concentrations, respectively. The concentrations of the substrate and coenzyme were as follows: l-threonine, 2, 3, 5, 8, and 20 mM; NAD+, 0.05, 0.1, 0.2, 0.4, and 1 mM.
Metal analysis.
The enzyme was dialyzed at 4°C for 8 h against three changes of 150 volumes of a Tris-HCl buffer (pH 7.0) from which divalent cations were removed by Chelex 100 resin (Bio-Rad) and then analyzed for six elements (Ni, Mn, Fe, Cu, Mg, and Zn) by using a polarized Zeeman atomic absorption spectrophotometer (model Z-8200; Shimadzu, Kyoto, Japan). The dialysate obtained by the last dialysis was used as a blank.
N-terminal and internal amino acid sequence determination.
The N-terminal and internal amino acid sequences were determined by Edman degradation with an automated sequencer (model 477A; Applied Biosystems). For N-terminal sequence analysis, approximately 15 μg of protein was transferred to a polyvinylidene difluoride membrane and then analyzed. Approximately 15 μg of protein was digested by trypsin in the presence of 2 M urea, 100 mM CaCl2, and 100 mM Tris-HCl (pH 8.0) at 37°C for 18 h. Peptides separated by reverse-phase high-performance liquid chromatography were transferred to a polyvinylidene difluoride membrane and analyzed.
Cloning and sequence analysis of the gene encoding l-ThrDH.
On the basis of the partial N-terminal sequence, N-MNPKI-C, and the internal sequence, N-DYAVDI-C, two oligonucleotides, 5′-GCCATGAAYCCDAARAT-3′ and 5′-ATRTCIACTGCRTARTC-3′, were synthesized and used as the forward and reverse PCR primers, respectively. A PCR was performed with 100 pmol of each of the above primers and 70 ng of chromosomal DNA isolated from the Cytophaga cells. The thermal profiles involved 30 cycles of denaturation at 94°C for 30 s, annealing at 37°C for 1 min, and extension at 72°C for 1 min. The resulting 550-bp DNA fragment was ligated using a pT7 Blue T-Vector (Novagene) and sequenced. The Big Dye Terminator Cycle Sequencing Ready Reaction kit and a model 377A DNA sequencing system gel apparatus (Applied Biosystems) were used according to the manufacturer's instructions. Genome-walking PCR was used to obtain sequences upstream and downstream from the 550-bp insert because sequence analysis of the 550-bp insert did not reveal in-frame start and termination codons. Genome-walking PCR was performed using the TaKaRa long and accurate PCR in vitro cloning kit according to the manufacturer's instructions. Two primers, 5′-CAGGGTTTTTCTCGGCTGTAG-3′ and 5′-CATCCAGTATTGCGGTTTTTGG-3′, were designed to obtain the sequences upstream and downstream, respectively, of the 550-bp insert. The Cytophaga chromosomal DNA extracted was digested with EcoRI and ligated to the EcoRI cassette. The obtained DNA fragments were used as templates for PCR. The thermal profiles involved 25 cycles of denaturation at 94°C for 30 s, annealing at 57°C for 30 s, and extension at 72°C for 1 min. The resulting fragments, 750 bp for the upstream fragment and 500 bp for the downstream fragment, were sequenced as described above. The start codon of the gene encoding the l-ThrDH (the ltd gene) was included in the 750-bp fragment, but the termination codon was inert in the 500-bp one. A primer, 5′-GCGACGATTAACATTATGAAAGCG-3′, was synthesized to obtain the sequence of the region farther downstream. The Cytophaga chromosomal DNA extracted was digested with XbaI and ligated to the XbaI cassette. The DNA fragments obtained were used as templates for PCR. The thermal profiles involved 30 cycles of denaturation at 94°C for 30 s, annealing at 58°C for 30 s, and extension at 72°C for 1 min. The resulting 1-kb fragment was sequenced as described above.
Other methods.
Protein concentrations were measured by the method of Bradford (6) with bovine serum albumin as a standard. With column chromatography, protein concentrations were determined by the measurement of A280. The molecular weight of the enzyme was estimated by gel filtration with a Superose 12 HR 10/30 column (1.0 by 30 cm; Pharmacia) at 4°C and at a flow rate of 0.4 ml/min. The potassium phosphate buffer (50 mM; pH 7.0) containing 150 mM NaCl was used as an eluant. The following proteins were used as molecular weight standards: RNase (13,700), ovalbumin (43,000), aldolase (158,000), and ferritin (440,000). Polyacrylamide gel electrophoresis (PAGE) and sodium dodecyl sulfate (SDS)-PAGE were performed by the methods of Tulchin et al. (25) and Laemmli (12), respectively. The LMN marker kit (Pharmacia) was used as a molecular weight marker in SDS-PAGE.
Nucleotide sequence accession number.
The nucleotide sequence of the ltd gene has been submitted to the DDBJ, EMBL, and GenBank data banks with accession number AB088140.
RESULTS
Enzyme purification.
The enzyme was purified to homogeneity with a yield of 9.1% by DEAE-Toyopearl, Phenyl-Toyopearl, blue Sepharose, and 5′-AMP Sepharose column chromatography: a single protein band was detected by both PAGE and SDS-PAGE. The specific activity of the enzyme was 151 U/mg. The enzyme makes up about 0.57% of the soluble cellular protein. It was stored at −20°C in a 10 mM potassium phosphate buffer (pH 7.0) containing 30% sucrose, 0.1 mM NAD+, 5 mM l-threonine, and 75 mM KCl without loss of activity for several months.
Molecular weight of the enzyme and subunit and metal content.
The molecular weight of the enzyme was determined to be about 139,000 by Superose 12 gel chromatography. SDS-PAGE showed a subunit molecular weight of 35,000. This shows that the enzyme has a homotetrameric structure. Atomic absorption analysis showed that no metals were found in the enzyme.
Substrate specificity and coenzyme.
The enzyme was specific for the l-isomer of threonine, and three other isomers, l-allo-threonine, d-threonine, and d-allo-threonine were inert. The enzyme acts on dl-threo-3-hydroxynorvaline as well, though slowly (Table 1); however, it is presumed that the l-isomer could be a substrate. Other l-amino acids and glycine were inactive as substrates. NAD+ (100%) and its various analogs (oxidized form), such as 3-acetylpyridine adenine dinucleotide (60.6%), thionicotinamide adenine dinucleotide (5.1%), 3-pyridinealdehyde adenine dinucleotide (7.2%), and nicotinamide guanine dinucleotide (5.1%), served as coenzymes, but nicotinic acid adenine dinucleotide, α-NAD, and nicotinamide hypoxanthine dinucleotide were inert. Although d-ThrDH from P. cruciviae required either NADP (100%) or NAD (4.5%) (16), the Cytophaga l-ThrDH did not react with NADP. The coenzyme specificity of the Cytophaga enzyme is similar to that of the E. coli enzyme (4) but is lower than those of the Clostridium (26) and chicken (1) enzymes.
TABLE 1.
Comparison of substrate specificities
| Substratea | Relative activityb (%) of l-ThrDH from:
|
|||
|---|---|---|---|---|
| Cytophaga sp. strain KUC-1 | E. coli K-12 | C. sticklandti | Chicken liver | |
| l-Threonine | 100 | 100 | 100 | 100 |
| l-allo-Threonine | 0 | 0 | ND | 0 |
| d-Threonine | 0 | 63.7 | ND | 0 |
| d-allo-Threonine | 0 | ND | ND | ND |
| dl-allo-Threonine | ND | 95 | ND | 0 |
| l-Serine | 0 | 4 | 0 | 0 |
| dl-threo-3-Hydroxynorvaline | 31 | ND | 20 | ND |
| dl-threo-3-Phenylserine | 0 | 3 | 0 | ND |
The following compounds were inert: l-serine, l-phenylalanine, l-leucine, l-isoleucine, l-proline, l-tryptophan, l-valine, l-histidine, l-methionine, l-aspartic acid, l-asparagine, l-glutamine, l-tyrosine, glycine, l-threo-3-thionylserine, dl-norvaline, malonic acid, pyruvic acid, R-(−)-2-butanol, S-(+)-2-butanol, R-(−)-3-hydroxybutylic acid, and S-(+)-3-hydroxybutylic acid.
The activities were determined under conditions that were standard except for the use of 1 mM substrate. Data were obtained from the following sources: for cytophage sp. strain Kuc-1, this study; for E. coli K-12, references 4 and 5, for C. sticklandii, reference 26; and for chicken liver, reference 11. ND, not determined.
Effects of temperature and pH.
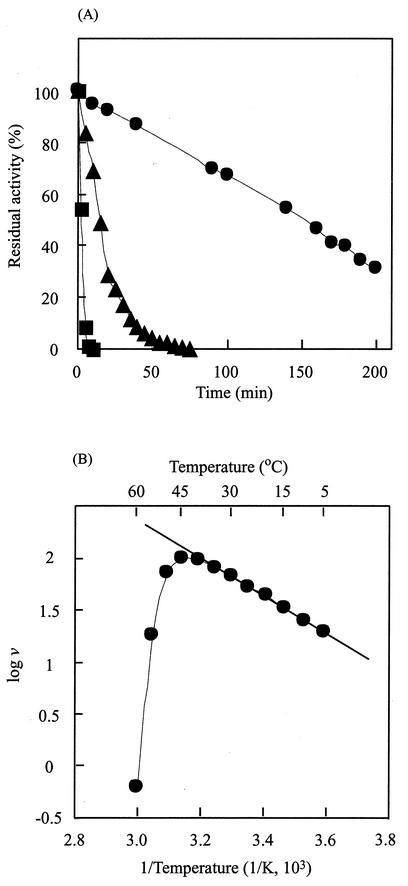
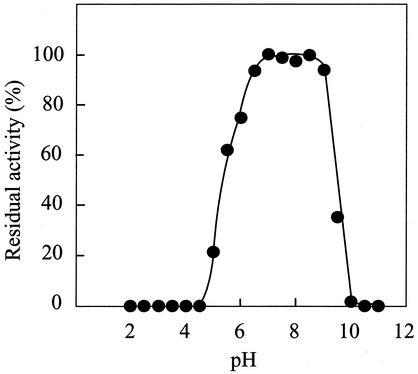
When the enzyme was incubated at temperatures in the range of 10 to 30°C for 60 min, more than 90% of the initial activity was retained. The half-lives were estimated to be 160 and 14 min at 40 and 45°C, respectively, and the enzyme was rapidly inactivated at 50°C (Fig. 1A). The Cytophaga enzyme was more thermolabile than the P. cruciviae d-ThrDH, which is stable at up to 40°C (16). The enzyme showed maximum activity at about 45°C. The activation energy was calculated by the Arrhenius plot to be 34.4 kJ/mol (Fig. 1B). The enzyme showed activity at pHs in the range of 5.5 to 11. The optimum pH was 9.5, and relative activities of about 74 and 31% of the initial activity were found at pHs 8.5 and 10.5, respectively. When the enzyme was incubated at pHs in the range of 6.5 to 9.0 for 60 min, more than 90% of the initial activity remained (Fig. 2). The half-lives were found to be 224 and 44.2 min at pHs 9.0 and 9.5, respectively.
FIG. 1.
Effects of temperature on enzyme activity and stability. (A) Effects of temperature on stability. The enzyme was incubated in a 10 mM potassium phosphate buffer, pH 7.0, at various temperatures. At intervals, 3-μl aliquots of the incubated 1-mg/ml enzyme solution were withdrawn and used for the enzyme assay under the standard assay conditions. •, ▴, and ▪ represent the residual activities at 40, 45, and 50°C, respectively. (B) Arrhenius plot. The activation energy for the oxidation of l-threonine was calculated from the Arrhenius plot. v, velocity.
FIG. 2.
The effects of pH on enzyme stability were examined by measurement of the residual activities under standard assay conditions after incubation of the enzyme at various pHs at 30°C for 30 min. The following 100 mM buffers were used: citrate (pH 3.0 to 5.5), potassium phosphate (pH 6.0 to 7.5), Tris-HCl (pH 8.0 to 9.0), and glycine-NaOH (pH 9.5 to 11.5).
Kinetic properties.
The double reciprocal plots of the initial velocities against the concentrations of a substrate and a coenzyme in the presence of various fixed concentrations of a coenzyme and a substrate, respectively, gave sets of straight lines, and the lines converged. The results show that the reaction proceeds through the formation of a ternary complex of the enzyme with a substrate and a coenzyme but not via a Ping-Pong Bi Bi mechanism. The kinetic parameters for l-threonine at various temperatures were calculated from the secondary plots of intercepts versus the reciprocal concentrations of the other substrate. The kinetic parameters of the enzyme were highly influenced by the reaction temperatures, and the lowest Km for l-threonine was found at 20°C (Table 2). The Vmax and Vmax/Km values were maximized at 40°C.
TABLE 2.
Effects of temperature on kinetic parameters of l-threoninea
| Temp (°C) | Km (mM) | Vmax (U/mg) | Vmax/Km (U/mg/mM) |
|---|---|---|---|
| 10 | 3.81 ± 0.39 | 91.4 ± 3.0 | 24.0 |
| 20 | 3.56 ± 0.34 | 156 ± 2.2 | 43.7 |
| 30 | 5.96 ± 0.22 | 284 ± 3.6 | 47.7 |
| 40 | 11.2 ± 1.1 | 722 ± 65 | 64.5 |
| 50 | 19.5 ± 3.2 | 611 ± 82 | 31.3 |
Data are means ± standard deviations of results from four determinations.
Product inhibition studies were carried out with NADH alone, because 2-amino-3-oxobutyrate is too unstable to be converted to aminoacetone and CO2 (15). NADH gave a competitive inhibition with respect to NAD+ and a noncompetitive inhibition with respect to l-threonine. To determine the substrate binding order in a sequential mechanism, dead-end inhibition experiments were performed as described by Aoyama and Motokawa (1). Pyruvate and adenosine-5′-diphosphoribose were used as likely dead-end competitive inhibitors for l-threonine and NAD+, respectively. Pyruvate was found to be a competitive inhibitor with respect to l-threonine (Ki = 23.2 mM). When the concentrations of NAD+ were varied, the reciprocal plots of the initial velocities gave a series of parallel lines indicative of uncompetitive inhibition (Ki = 50.7 mM). Adenosine-5′-diphosphoribose gave competitive inhibition with respect to NAD+ (Ki = 0.37 mM) and noncompetitive inhibition with respect to l-threonine (Ki = 1.42 mM). These inhibition patterns indicate that the enzyme reaction proceeds through an ordered Bi Bi mechanism in which NAD+ binds to the enzyme prior to l-threonine. After NAD+ and l-threonine are converted to NADH and 2-amino-3-oxobutyrate, respectively, 2-amino-3-oxobutyrate leaves the enzyme, and NADH is finally released (22). This process is the same as that for chicken liver l-ThrDH (12) and P. cruciviae d-ThrDH (16).
Effects of inhibitors.
We examined the effects of the following compounds on the enzyme activity: metals (NiCl2, MnCl2, CoCl2, ZnCl2, BaCl2, CaCl2, HgCl2, CuCl2, MgCl2, and LiCl2), chelating reagents (EDTA, EGTA, 1,10-phenanthroline, and 2,2′-bipyridyl), and thiol reagents [p-(chloromercuri)benzoic acid, monoiodoacetate, HgCl2, and N-ethylmaleimide]. The remaining activity was determined under conditions that were standard except for the addition of each compound to the reaction mixture. The enzyme was strongly inhibited by 10 mM thiol reagents [p-(chloromercuri)benzoic acid (44%), monoiodoacetate (100%), HgCl2 (100%), and N-ethylmaleimide (48%)], and inhibition was observed with 1 mM ZnCl2 (72%) and CuCl2 (100%), whereas P. cruciviae d-ThrDH was not inhibited by thiol reagents or Cu2+ and Zn2+ ions (16). Other metal chlorides and chelating reagents had no effect on the activity of the Cytophaga enzyme. The activity of E. coli l-ThrDH was stimulated by Mn2+ (4), but the Cytophaga enzyme did not require divalent cations.
Sequencing of amino-terminal and internal regions.
Sequencing of the N-terminal region was carried out for 12 cycles yielding identifiable residues. The amino acid sequence of the N-terminal region was MNPKILIIGAXG. Peptides formed by trypsin digestion were separated and sequenced. The partial N-terminal sequences of three tryptic peptides were STPPGGGTTDYAVDI, HIPEFT, and EVVNALD.
Cloning and sequence analysis.
The degenerate oligonucleotides, 5′-GCCATGAAYCCDAARAT-3′ and 5′-ATRTCIACTGCRTARTC-3′, were designed from the N-terminal and internal sequences and used as forward and reverse PCR primers, respectively. An amplified DNA fragment (about 550 bp) was cloned into a pT7 Blue T-Vector, and nucleotides were sequenced. The results showed an open reading frame lacking initiation and termination codons. The overlapping segments, upstream and downstream regions of about 750 and 500 bp, respectively, were amplified by a genome-walking PCR method, and nucleotides were sequenced to obtain the entire gene, but the termination codon was not found. Furthermore, when genome-walking PCR was performed, the resulting 1-kb overlapping segment was found to contain a termination codon. The entire open reading frame is accordingly composed of 939 bases and codes for a protein of 312 amino acid residues with an estimated molecular weight of 35,384. The deduced amino acid sequence was used for searching the GenBank and protein databases with the BLAST program. We found significant similarities between the sequence of the Cytophaga enzyme and amino acid sequences of UDP-glucose 4-epimerases from Staphylococcus aureus (identity, 50%) and Thermoplasma volcanium (identity, 43%). The N-terminal sequence of l-ThrDH of porcine is similar to that of the Cytophaga enzyme, but except for its molecular weight and subunit structure (21), enzymological properties of the porcine enzyme have not been investigated. The amino acid sequence of the Cytophaga enzyme was not similar to those of E. coli l-ThrDH, which belongs to the medium-chain alcohol dehydrogenase family, and P. cruciviae d-ThrDH, which belongs to the 3-hydroxyacid dehydrogenase superfamily. The important amino acid residues reported for horse liver alcohol dehydrogenase, which belongs to the medium-chain alcohol dehydrogenase family (namely, six cysteines at positions 46, 96, 99, 103, 111, and 132 and Ser 48, His 51, and His 67 [position numbers are those in the horse liver alcohol dehydrogenase sequence] [7, 9]), are highly conserved in the E. coli l-ThrDH (data not shown), but the corresponding amino acid residues were not found in the Cytophaga l-ThrDH. The cysteine residue at position 38 of E. coli l-ThrDH is located in the active-site region based on its selective S carboxymethylation reaction with iodoacetic acid (10), but a corresponding cysteine residue was not found in Cytophaga l-ThrDH. The enzymes that belong to the short-chain dehydrogenase-reductase superfamily contain two characteristic signature motifs. The first of these is a YXXXK motif, and the other is a GXXGXXG motif. The amino acid sequence of Cytophaga l-ThrDH was aligned with those of UDP-glucose 4-epimerases, which belong to the short-chain dehydrogenase-reductase superfamily. The motifs are conserved in the Cytophaga enzyme, although this enzyme shows very low identities with human (identity, 18.5%) and E. coli (identity, 21.6%) epimerases (Fig. 3). This indicates that the Cytophaga enzyme belongs to the short-chain dehydrogenase-reductase superfamily. The catalytically essential amino acid residues were estimated by comparative analysis of primary structures. The essential residues of UDP-glucose 4-epimerases from E. coli and human are well conserved in the Cytophaga enzyme. The two conserved amino acid residues of the E. coli enzyme, Tyr 149, which occurs in the YXXXK motif, and Ser 124, which correspond to Tyr 143 and Ser 117 of the Cytophaga enzyme, respectively, were reported to play catalytically important roles (23). The GXXGXXG motif, which is located near the cofactor-binding pocket, is also conserved in the N-terminal regions of the eight sequences. The five residues of the human enzyme, namely, Asp 33, Asn 37, Asp 66, Tyr 157, and Lys 161, are directly involved in nucleotide binding (24), and these amino acid residues are completely conserved in the Cytophaga enzyme (Fig. 3). However, the key amino acid residues responsible for binding of the substrate, UDP-galactose, in the active site of the human enzyme (Asn 187, Asn 207, Asn 224, Phe 226, Arg 239, Arg 300, and Asp 303) do not occur in the Cytophaga enzyme.
FIG. 3.
Multiple alignment of Cytophaga l-ThrDH (Cyt L-TheDH) and UDP-glucose 4-epimerases. Sources of UDP-glucose 4-epimerases of which individual sequences are shown are designated as follows (accession numbers of the corresponding nucleotide sequences are given in parentheses): Hum, human (1EK6A); Eco, E. coli (NP415280); Mha, Mannheimia haemolytica (Q59678); Bsu, Bacillus subtilis (P55180); Sth, Streptococcus thermophilus (P21977); Abr, Azospirillum brasilense (Q59083); and Cgl, Corynebacterium glutamicum (Q45291). Identical and similar residues in more than 70% of the sequences are shaded in black and gray, respectively. Gaps (-) are introduced to obtain maximal matching. The GXXGXXG and YXXXK motifs are underlined. The asterisks show key residues, including Ser 118 and Tyr 143 (position numbers are those in the Cytophaga enzyme sequence). The arrows indicate the residues involved in the binding of the coenzyme.
We found that Cytophaga l-ThrDH did not show epimerase activity and, vice versa, that l-threonine and UDP-galactose did not inhibit UDP-glucose 4-epimerase and Cytophaga l-ThrDH, respectively.
DISCUSSION
A psychrophilic bacterium, Cytophaga sp. strain KUC-1, that abundantly produces a NAD+-dependent l-ThrDH was isolated from Antarctic seawater, and the enzyme was purified. The enzyme is composed of four identical subunits with a molecular weight of 35,000 and contains no metals. In this respect, it is distinct from any other l-ThrDHs so far reported (Table 3).
TABLE 3.
Comparison of structural properties of l-ThrDHs
| Source | Subunit structure | Mr per subunit | Metala | Reference |
|---|---|---|---|---|
| Cytophaga sp. strain KUC-1 | Homotetramer | 35,000 | None | This work |
| C. sticklandii | Homodimer | 33,000 | ND | 26 |
| E. coli K-12 | Homotetramer | 35,000 | Zn (1 g-atom/ subunit) | 5, 10 |
| Porcine liver | Homodimer | 37,000 | ND | 11 |
| Chicken liver | Monomer | 88,000 | ND | 1 |
| Goat liver | Monomer | 89,000 | ND | 21 |
ND, not determined.
The kinetic parameters of the enzyme were highly influenced by temperature. The temperature at which the lowest Km for l-threonine was found was about 20°C, which is close to the optimum growth temperature of Cytophaga cells. Thus, the enzyme is catalytically efficient even at low temperatures and is less thermostable than other amino acid dehydrogenases (3, 18, 19), including d-ThrDH (16), though the thermostabilities of l-ThrDHs from other sources have never been examined. This enzyme has probably evolved molecularly to adapt to cold conditions and thus lost thermostability. Other l-ThrDHs have not been studied in detail, and further comparative studies are needed to characterize the Cytophaga enzyme, particularly its structure and molecular evolution.
The substrate specificity of Cytophaga l-ThrDH is similar to those of the C. sticklandii (26) and chicken (1) enzymes but higher than that of the E. coli enzyme (Table 1) (4, 5). The Cytophaga enzyme recognizes strictly the stereostructures of α- and β-carbons of a substrate, whereas the E. coli enzyme acts on either d-threonine or d-allo-threonine. l-Serine is not a substrate of the Cytophaga enzyme: the β-methyl group is required. The methyl group of l-threonine can be replaced by the ethyl group but not by phenyl and thienyl groups, whereas an electrophilic group such as phenyl or thienyl at the β-position of d-threonine increases the reactivity with P. cruciviae d-ThrDH (16). These facts suggest that volume or electron attractive force of a functional group at the β-carbon plays an important role in the binding of the substrate with the enzyme and in the reactivity. Although the enzyme catalyzes the oxidation of a β-hydroxyl group of l-threonine, both the α-carboxy and amino groups of l-threonine are essential because R-(−)-hydroxybutylate and R-(−)-butanol are inert.
The amino acid sequence of Cytophaga l-ThrDH shows significant similarities to that of UDP-glucose 4-epimerase, which belongs to the short-chain dehydrogenase-reductase superfamily, but not to those of E. coli l-ThrDH, which is a member of the medium-chain alcohol dehydrogenase family, and P. cruciviae d-ThrDH, which belongs to the 3-hydroxyacid dehydrogenase superfamily. The reactions of Cytophaga l-ThrDH, E. coli l-ThrDH, and P. cruciviae d-ThrDH probably proceed through different mechanisms. The amino acid sequence of the Cytophaga enzyme was compared with those of human and E. coli UDP-glucose 4-epimerases, which also belong to the short-chain dehydrogenase-reductase superfamily, and important amino acid residues of human and E. coli UDP-glucose 4-epimerases were identified by three-dimensional structure analysis (23, 24). The catalytically important amino acid residues of the E. coli and human epimerases are significantly conserved in the Cytophaga l-ThrDH, except for the residues participating in substrate binding. The two strictly conserved amino acid residues of the E. coli UDP-glucose 4-epimerase are Tyr 149 and Ser 124, which correspond to Tyr 143 and Ser 117 of Cytophaga l-ThrDH, respectively, and these residues were reported to play important roles: Tyr 149 of E. coli UDP-glucose 4-epimerase interacts directly with the 4′-hydroxyl group of the glycosyl moiety as the active side-base, and Ser 124 acts as a proton shuttle between the sugar moiety and the presumed phenolic side chain of Tyr 149 (23). The reaction mechanism of Cytophaga l-ThrDH is probably similar to that of the UDP-glucose 4-epimerase and can be regarded as a half reaction of the epimerase reaction. However, the reaction mechanism of Cytophaga l-ThrDH is different from that of E. coli l-ThrDH because the amino acid residues involved in the catalysis of the E. coli enzyme are not at all conserved in the Cytophaga enzyme. Furthermore, the reaction mechanism of P. cruciviae d-ThrDH does not resemble that of the Cytophaga enzyme, either, although these enzymes catalyze similar reactions; this is because the TXXXK motif does not exist in the sequence of the P. cruciviae enzyme (data not shown). As a matter of course, the reaction mechanism of the Cytophaga enzyme is different from those of other amino acid dehydrogenases because l-ThrDH catalyzes β-dehydrogenation and other amino acid dehydrogenases catalyze α-deamination. Thus, the Cytophaga enzyme does not share the same structural motifs with E. coli l-ThrDH, P. cruciviae d-ThrDH, and other amino acid dehydrogenases and belongs to a structural family that is different from that of these enzymes. The Cytophaga enzyme is absorbing in these respects.
We determined the UDP-glucose 4-epimerase activity of Cytophaga l-ThrDH because the catalytically important amino acid residues of the UDP-glucose 4-epimerases are conserved in the Cytophaga l-ThrDH. However, Cytophaga l-ThrDH did not show epimerase activity, and UDP-glucose 4-epimerase did not catalyze the l-ThrDH reaction. l-Threonine and UDP-galactose did not inhibit UDP-glucose 4-epimerase or Cytophaga l-ThrDH, respectively. Accordingly, both enzymes are similar to each other in the sense that their amino acid residues are involved in enzyme catalysis but not to the extent that they share a common catalytic activity, and a substrate of one enzyme does not inhibit the other enzyme. This suggestion is supported by the fact that the amino acid residues participating in the substrate recognition by the UDP-glucose 4-epimerase are not found in the Cytophaga l-ThrDH.
This is probably the first example of the structural similarity of l-ThrDH and UDP-glucose 4-epimerase, though we have no information about their evolutionary relationship. We are now studying the three-dimensional structure of the enzyme to show the relationship between the enzyme structure and characteristics.
Acknowledgments
This work was supported in part by a research grant from the Japan Foundation of Applied Enzymology, by a research grant from the Kansai University High Technology Research Center, and by the Science Research Promotion Fund from the Japan Private School Promotion Foundation.
REFERENCES
- 1.Aoyama, Y., and Y. Motokawa. 1981. l-Threonine dehydrogenase of chicken liver. Purification, characterization, and physiological significance. J. Biol. Chem. 256:12367-12373. [PubMed] [Google Scholar]
- 2.Aronson, B. D., R. L. Somerville, B. R. Epperly, and E. E. Dekker. 1989. The primary structure of Escherichia coli l-threonine dehydrogenase. J. Biol. Chem. 264:5226-5232. [PubMed] [Google Scholar]
- 3.Asano, Y., A. Nakazawa, and K. Endo. 1987. Novel phenylalanine dehydrogenases from Sporosarcina ureae and Bacillus sphaericus. Purification and characterization. J. Biol. Chem. 262:10346-10354. [PubMed] [Google Scholar]
- 4.Boylan, S. A., and E. E. Dekker. 1978. l-Threonine dehydrogenase of Escherichia coli K-12. Biochem. Biophys. Res. Commun. 85:190-197. [DOI] [PubMed] [Google Scholar]
- 5.Boylan, S. A., and E. E. Dekker. 1981. L-threonine dehydrogenase. Purification and properties of the homogeneous enzyme from Escherichia coli K-12. J. Biol. Chem. 256:1809-1815. [PubMed] [Google Scholar]
- 6.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 7.Branden, C. I., H. Jornvall, H. Eklund, and B. Furugren. 1975. Enzymes, 3rd ed., p. 104-190. Academic Press, New York, N.Y.
- 8.DeLuna, A., A. Avendano, L. Riego, and A. Gonzalez. 2001. NADP-glutamate dehydrogenase isoenzymes of Saccharomyces cerevisiae. Purification, kinetic properties, and physiological roles. J. Biol. Chem. 276:43775-43783. [DOI] [PubMed] [Google Scholar]
- 9.Eklund, H., J. P. Samama, and T. A. Jones. 1984. Crystallographic investigations of nicotinamide adenine dinucleotide binding to horse liver alcohol dehydrogenase. Biochemistry 23:5982-5996. [DOI] [PubMed] [Google Scholar]
- 10.Epperly, B. R., and E. E. Dekker. 1991. L-threonine dehydrogenase from Escherichia coli. Identification of an active site cysteine residue and metal ion studies. J. Biol. Chem. 266:6086-6092. [PubMed] [Google Scholar]
- 11.Kao, Y. C., and L. Davis. 1994. Purification and structural characterization of porcine L-threonine dehydrogenase. Protein Expr. Purif. 5:423-431. [DOI] [PubMed] [Google Scholar]
- 12.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 13.Maresin, R., and F. Schinner. 1999. Biotechnological applications of cold-adapted organisms. Springer-Verlag, Berlin, Germany.
- 14.McGilvray, D., and J. G. Morris. 1969. Utilization of L-threonine by a species of Arthrobacter. A novel catabolic role for “aminoacetone synthase.” Biochem. J. 112:657-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Misono, H., Y. Shinagawa, S. Nagata, and S. Nagasaki. 1987. Occurrence of D-threonine dehydrogenase in Pseudomonas cruciviae. Agric. Biol. Chem. 51:1467-1469. [Google Scholar]
- 16.Misono, H., I. Kato, K. Packdibamrung, S. Nagata, and S. Nagasaki. 1993. NADP(+)-dependent d-threonine dehydrogenase from Pseudomonas cruciviae IFO 12047. Appl. Environ. Microbiol. 59:2963-2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morita, R. Y. 1975. Psychrophilic bacteria. Bacteriol. Rev. 39:144-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohashima, T., and K. Soda. 1979. Purification and properties of alanine dehydrogenase from Bacillus sphaericus. Eur. J. Biochem. 100:29-30. [DOI] [PubMed] [Google Scholar]
- 19.Ohshima, T., H. Misono, and K. Soda. 1978. Properties of crystalline leucine dehydrogenase from Bacillus sphaericus. J. Biol. Chem. 253:5719-5725. [PubMed] [Google Scholar]
- 20.Oikawa, T., K. Yamanaka, T. Kazuoka, N. Kanzawa, and K. Soda. 2001. Psychrophilic valine dehydrogenase of the Antarctic psychrophile, Cytophaga sp. KUC-1: purification, molecular characterization and expression. Eur. J. Biochem. 268:4375-4383. [DOI] [PubMed] [Google Scholar]
- 21.Ray, M., and S. Ray. 1985. l-Threonine dehydrogenase from goat liver. Feedback inhibition by methylglyoxal. J. Biol. Chem. 260:5913-5918. [PubMed] [Google Scholar]
- 22.Segal, I. H. 1975. Enzyme kinetics: behavior and analysis of rapid equilibrium and steady-state enzyme systems, p. 767-783. John Wiley & Sons, New York, N.Y.
- 23.Thoden, J. B., P. A. Frey, and H. M. Holden. 1996. High-resolution X-ray structure of UDP-galactose 4-epimerase complexed with UDP-phenol. Protein Sci. 5:2149-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thoden, J. B., T. M. Wohlers, J. L. Fridovich-Keil, and H. M. Holden. 2000. Crystallographic evidence for Tyr 157 functioning as the active site base in human UDP-galactose 4-epimerase. Biochemistry 39:5691-5701. [DOI] [PubMed] [Google Scholar]
- 25.Tulchin, N., L. Ornstein, and B. J. Davis. 1976. A microgel system for disc electrophoresis. Anal. Biochem. 72:485-490. [DOI] [PubMed] [Google Scholar]
- 26.Wagner, M., and J. R. Andreesen. 1995. Purification and characterization of threonine dehydrogenase from Clostridium sticklandii. Arch. Microbiol. 163:286-290. [DOI] [PubMed] [Google Scholar]