Abstract
We previously described a 15-kb genetic cluster consisting of 11 open reading frames (cps2A to cps2K) of Enterococcus faecalis FA2-2 that is responsible for the production of the serotype 2 capsular polysaccharide. By using transcriptional fusions to a promoterless lacZ gene, we identified two independent promoters related to the expression of the polysaccharide. Both transcription initiation sites were mapped by primer extension. Reverse transcription-PCR (RT-PCR) demonstrated the transcriptional linkage of genes present in both transcripts. Real-time RT-PCR quantification of transcripts revealed maximum transcription during log phase growth, an observation confirmed by promoter fusion studies. The heterologous expression of this pathway in Escherichia coli caused reactivity with E. faecalis type 2 antiserum, thus demonstrating the essential role of this pathway in the synthesis of the type-specific polysaccharide.
Enterococcus faecalis has emerged as a leading cause of nosocomial bacteremia, surgical site infection, and urinary tract infection (29). The development of multiple antibiotic resistances in enterococci in recent years compromises existing therapies, highlighting the need for a better understanding of the pathogenicity of the organism and the development of new treatment strategies (13). Only a few traits are known to contribute to the pathogenesis of enterococcal infection (23).
In recent years, several papers have examined the role that bacterial polysaccharides play in the biology of enterococcal infection (2, 3, 12, 15, 16, 37, 40). Bottone et al. (3) described several E. faecalis clinical isolates isolated from urinary tract infections which were highly encapsulated. Huebner et al. (16) reported detection of a capsule consisting of a glucose-glycerol phosphate polymer present in a portion of E. faecalis (33%) and Enterococcus faecium (28%) isolates, which protected these organisms from phagocytic killing in the absence of opsonic antibodies. More recently, Huebner et al. (15) showed the efficacy of the purified capsule as a serotype vaccine candidate. Mice passively immunized with anticapsular antibody demonstrated enhanced clearance of the organism. Xu et al. (39, 40) described a polysaccharide operon in E. faecalis strain OG1RF, which when expressed in Escherichia coli conferred reactivity with sera from patients with enterococcal endocarditis. The limitation of these findings was that the same antisera could not detect a polysaccharide antigen on the surface of the same E. faecalis strain. More recently, Teng et al. (37) identified a cell wall polysaccharide from E. faecalis strain OG1RF reactive with patient serum, but only after liberating the polymer from the cell wall by lysozyme digestion. This is in contrast to the capsular polysaccharide from E. faecalis, which is readily agglutinated by using intact cells and serotype-specific antibodies (12).
We previously reported an operon in E. faecalis that codes for the biosynthesis of the serotype 2 capsular polysaccharide (12). Insertional inactivation of genes within this pathway resulted in a loss of reactivity with type 2 antisera. Moreover, these mutants had significantly reduced ability to persist at sites of infection. It was further shown that human polymorphonuclear leukocytes more readily phagocytosed a mutant defective in expression of the serotype polysaccharide, a hallmark of bacterial encapsulation. This capsule determinant was found to be conserved in a number of clinical isolates, including the vancomycin-resistant strain V583 (31), from which the genome sequence is being derived (L. Bannerjei, personal communication).
Because of its role in the pathogenesis of enterococcal infection and its potential value in deriving alternative therapeutic approaches for an agent increasingly refractory to antibiotics, it was of interest to analyze the underlying genetics of E. faecalis serotype 2 capsular polysaccharide expression.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
All relevant bacterial strains and plasmid constructions used in this study are listed in Table 1. Escherichia coli XL1-Blue was used for plasmid constructions. E. coli clones were cultivated on Luria-Bertani (LB) broth (32). E. faecalis strains were cultured in 5 to 10 ml of Todd-Hewitt broth (THB; Difco Laboratories, Detroit, Mich.) with various concentrations of glucose. For serum and urine expression studies, E. faecalis strains were grown in 50 ml of nonhemolyzed, heat-treated, pooled rabbit serum (Pel-Freez, Ark.) or 50 ml of sterile-filtered pooled human urine. Plasmid pTCV-lac bearing a promoterless lacZ gene was used in making reporter fusions (28). Plasmid pAT28 (38) was used as a shuttle vector for the heterologous expression of the E. faecalis serotype 2 capsular polysaccharide in E. coli. Antibiotics (Sigma, St. Louis, Mo.) used for selection of recombinant clones in E. coli included spectinomycin (150 μg/ml) and kanamycin (100 μg/ml). For selection in E. faecalis, rifampin (25 μg/ml), fusidic acid (10 μg/ml), spectinomycin (500 μg/ml), and kanamycin (1 mg/ml) were used. Isopropyl-β-d-thiogalactopyranoside (IPTG; 0.5 mM) and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal; 80 μg/ml) were used for induction and detection of β-galactosidase activity. All restriction enzymes and molecular cloning reagents were purchased from Life Technologies (Gaithersburg, Md.) and were used according to the manufacturer's instructions.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strain | ||
| E. coli XL-1 Blue | recA1 lacZM15 Tn10 Tetr | Stratagene |
| E. faecalis FA2-2 | Plasmid-free derivative of JH2, rifampinr, fusidic acidr | 5 |
| E. faecalis OG1RF | Plasmid-free derivative of OG1, rifampinr, fusidic acidr | 24 |
| Plasmid | ||
| pTCV-lac | Shuttle vector for promoter fusion studies | 28 |
| pCPS100a | 1.7-kb Sau3 A fragment containing the cpsC promoter | This study |
| pCPS110a | 2.6-kb MfeI fragment containing the cpsC promoter | This study |
| pCPSAa | 374-bp EcoRI-BamHI fragment containing the cpsA promoter (obtained by PCR with primers cpsA1 and cpsA2) | This study |
| pCPSABa | 1,968-bp EcoRI-BamHI fragment containing the cpsA promoter through the cpsB gene (obtained by PCR with primers cpsA1 and cpsB1) | This study |
| pCPSABTa | 2,086-bp EcoRI-BamHI fragment containing cpsA promoter through a transcriptional terminator 3′ to cpsB (obtained by PCR with primers cpsA1 and cpsB2) | This study |
| pCPSA-Ca | 2,459-bp EcoRI-BamHI fragment containing cpsA promoter to the translation start of cpsC (obtained by PCR with primers cpsA1 and cpsC5) | This study |
| pCPSC1a | 447-bp EcoRI-BamHI fragment containing cpsAB terminator to the translation start of cpsC (obtained by PCR with primers cpsC1 and cpsC5) | This study |
| pCPSC2a | 390-bp EcoRI-BamHI fragment containing cpsC promoter including regulatory region to the translation start of cpsC (obtained by PCR with primers cpsC2 and cpsC5) | This study |
| PCPSC3a | 241-bp EcoRI-BamHI fragment containing cpsC promoter minus regulatory region to the translation start of cpsC (obtained by PCR with primers cpsC3 and cpsC5) | This study |
| pCPSC4a | 172-bp EcoRI-BamHI fragment containing region 3′ to putative cpsC promoter to the translation start of cpsC (obtained by PCR with primers cpsC4 and cpsC5) | This study |
| pAT28 | Shuttle vector, spectinomycin resistance | 38 |
| pCPSCKb | 12,775-bp NheI-XmaI fragment containing cpsC to cpsK (primers Cps1 and Cps2) | This study |
These plasmids are derivatives of pTCV-lac, which contain the inserts described in the text for use in promoter fusion studies.
This plasmid is a derivative of pAT28, which contains the insert described in the text for use in expression studies.
For culturing E. faecalis under logarithmic and stationary-phase growth, conditions reported by Shepard and Gilmore (35) were employed. Briefly, an overnight culture was diluted 1:100 into fresh prewarmed medium and was cultured for an additional 3 to 4 h at 37°C without shaking. For serum and urine cultures, a single colony was diluted 1:50,000 into either prewarmed serum or urine and was cultured an additional 12 (serum) or 18 (urine) h at 37°C without shaking. For stationary-phase growth conditions, bacteria from a single colony were inoculated into fresh aliquots of the respective medium and were allowed to grow for 24 h at 37°C without shaking. Bacterial cell density was monitored at an optical density of 600 nm (OD600), and direct plate counts were performed by using track dilution (17).
Construction of promoter-β-galactosidase fusions in pTCV-lac.
Primers listed in Table 2 were used to amplify regions of the E. faecalis FA2-2 genome encoding type 2 capsule biosynthesis for fusion to a promoterless lacZ reporter. E. faecalis FA2-2 chromosomal DNA, prepared as described by Pospiech and Neumann (27), was used as a template in PCRs with a TaKaRa LA Taq kit (Panvera Corp., Madison, Wis.). Primers CAPl and CAPr were used to amplify a 14,849-bp fragment encompassing the cps determinant. Briefly, 500 ng of DNA template was amplified by using the supplied enzyme kit according to the manufacturer's recommendations. After the initial hot start denaturation step in which the polymerase was added (94°C, 5 min), the following parameters were used for 35 cycles of PCR: 98°C (40 s), 68°C (15 min). Upon completion of the cycles, a final extension at 72°C (10 min) was performed, and the reaction mixture was stored at 4°C. The amplified PCR product was digested to completion with either Sau3AI or MfeI, generating 35 and 14 restriction fragments, respectively. The restriction fragments were cloned into either the BamHI (for Sau3AI fragments) or EcoRI (for MfeI fragments) site immediately 5′ to the promoterless lacZ gene in pTCV-lac. Reporter fusions were propagated in E. coli, and the randomness of fragment insertion was assessed by colony PCR (39) by using primers Vlac1 and Vlac2 (28) to amplify across the cloning site. E. coli clones were pooled, and plasmid DNA was isolated by using a Wizard mini-prep kit (Promega, Madison, Wis.). Plasmid DNA was subsequently introduced into E. faecalis by electroporation (7), and the resulting transformants were plated onto brain heart infusion (BHI) agar with kanamycin and X-Gal for the detection of β-galactosidase expression. Additional promoter-fusion constructs were generated by cloning amplification products (shown in Fig. 1 and Table 1) into pTCV-lac. The inserts for plasmids pCPS1, pCPS2, pCPS3, and pCPS4 were obtained by PCR with primers Cps1, Cps2, Cps3, and Cps4, respectively, and the common reverse primer, Cps5. The inserts for plasmids pCPSA, pCPSAB, pCPSABT, and pCPSA-C were obtained by PCR with the common forward primer, CpsA1, and primers CpsA2, CpsB1, CpsB2, and Cps5, respectively. Amplification products were restricted with EcoRI and BamHI and were directionally cloned into similarly digested pTCV-lac. Plasmids were propagated in E. coli before introduction into E. faecalis by electroporation.
TABLE 2.
Oligonucleotide primers used in this studya
| Primer name and function | Sequence (5′ to 3′) |
|---|---|
| Promoter fusion constructs | |
| CAP1 | GGAACGGCGCTATTGTTCCAGTC |
| CAPr | GTTACATATAATCAACAAATTTACTTCTG |
| CpsA1 | GAGAGAATTCCGCTATTGTTCCAGTCAATGAAT (EcoRI) |
| CpsA2 | GAGAGGATCCCATCTTTTTCCCTCCATTCCA (BamHI) |
| CpsB1 | GAGAGGATCCAAGTGCATAATTGGGAAAAC (BamHI) |
| CpsB2 | GAGAGGATCCCAAGGTACCTAAACTAACCC (BamHI) |
| CpsC1 | GAGAGAATTCCGTCAAATAAGCAAAAATCCAG (EcoRI) |
| CpsC2 | GAGAGAATTCGTTAGTTTAGGTACCTTGTGAG (EcoRI) |
| CpsC3 | GAGAGAATTCCGAATAACACTGGAATGTTTATTTC (EcoRI) |
| CpsC4 | GAGAGAATTCGAAGAAAAGCAAAATAATGTATTTTACATGC (EcoRI) |
| CpsC5 | GAGAGGATCCCATTGAATTAATCCTTTACATTAG (BamHI) |
| Primer extension | |
| CpsAPE | GTGCTGCGGAATTTGGCCTTC |
| CpsCPE | GAATAACTCTCACCACGATTGAAC |
| Expression constructs | |
| Cps1 | GAGAGCTAGCGGGTTAGTTTAGGTACCTTGTGAG (NheI) |
| Cps2 | GAGACCCGGGGTTACATATAATCAACAAATTTACTTCTG (XmaI) |
Underlined nucleotides indicate restriction cleavage sites not present in the template sequence, which were employed to facilitate cloning. The corresponding restriction enzyme is shown in parentheses to the right of the primer sequence.
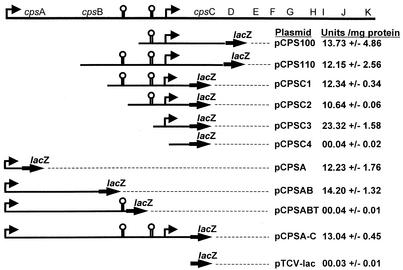
FIG. 1.
Promoter fusion analysis by using a lacZ reporter. Elevated arrows depict points of transcription initiation, and the hairpin structures show regions of dyad symmetry. Corresponding β-galactosidase activity (mean ± standard deviation) expressed from each fusion construction is indicated
β-Galactosidase activity assay.
Assays were performed according to the method of Miller (21) with modifications. Following growth in the designated culture conditions, cells were harvested by centrifugation. Cell pellets were washed once with 1.5 ml of Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, β-mercaptoethanol [pH 7.0]) and then were resuspended in 1.5 ml of Z buffer. The cell suspension was added to a 2.0-ml screw cap tube containing a 0.5-ml volume of 0.1-mm-diameter zirconia beads (BioSpec Products, Bartlesville, Okla.). The cells were disrupted in a mini-beadbeater (BioSpec Products) for 60 s at 5,000 rpm. Tubes were very briefly centrifuged to pellet beads and insoluble cell debris. The aqueous layer was removed and was further clarified by centrifugation (12,000 × g, 10 min). One milliliter of the aqueous layer was used in the β-galactosidase assay, and the remaining sample was assayed for total protein by using a Coomassie Plus Protein Assay (Pierce, Rockford, Ill.). To quantify β-galactosidase activity in the clarified supernate, 200 μl of substrate (4 mg of o-nitrophenyl-β-d-galactoside [ONPG]/ml, 0.1 M phosphate buffer [pH 7.0]) was added to 1.0 ml of the supernatant, and the reaction was allowed to develop for 30 min at room temperature. The reaction was stopped by the addition of 0.5 ml of 1 M Na2CO3. Color change was quantified by using a microtiter plate reader at 405 nm. Samples were assayed in triplicate, and values were expressed as arbitrary units/milligram of total protein.
RNA manipulations, primer extension, and reverse transcription-PCR (RT-PCR).
RNA was extracted by the method of Shepard and Gilmore (36). For bacteria grown in urine, it was necessary to first wash the cell pellet twice in cold phosphate-buffered saline (PBS; pH 7.4) for efficient RNA isolation.
For primer extension analysis, E. faecalis strain FA2-2 was grown to mid-log phase (OD600 of 0.5) in THB supplemented with 1% glucose prior to RNA isolation. Primer extension analysis was performed by using an avian myeloblastosis virus reverse transcriptase primer extension kit as recommended by the supplier (Promega). Briefly, 50 μg of total RNA was incubated with 32P end-labeled primers CpsAPE or CpsCPE (Table 2). Each primer is complementary to a region 5′ to the start of translation for either cpsA or cpsC, respectively. Reaction mixtures were incubated for 60 min at 50°C prior to the addition of stop buffer. A sequencing reaction with M13 control DNA was run as a molecular size ladder by using the Sequenase kit supplied by the manufacturer (USB, Cleveland, Ohio). Samples were denatured and resolved on an 8% polyacrylamide sequencing gel (Life Technologies, Gaithersburg, Md.), dried, and autoradiographed.
RT-PCR was performed as described previously (36) by using a Retroscript kit (Ambion, Austin, Tex.) according to the manufacturer's recommendation with the RT-PCR primer pairs shown in Table 3 and total RNA isolated from cultures grown in THB to mid-log phase (OD600 of 0.5).
TABLE 3.
Primer pairs used in this study
| Primer pair function | Primer pair sequence (5′ to 3′)
|
|||
|---|---|---|---|---|
| Forward
|
Reverse
|
|||
| Name | Sequence | Name | Sequence | |
| RT-PCR gene linkage analysis | CpsA-f | CGTGATCCAGAATTGTTGATTCG | CpsB-r | CCTAACAACAACATTACAGTGACA |
| CpsB-f | GATGCTCGTATTGACGGTTCTGT | CpsC-r | TATGGCTTGATGTATACTATTCTCT | |
| CpsC-f | CAGGAACAACGTTTATATGTGCA | CpsD-r | CTTGCAATTGCTCAATCCCAATGG | |
| CpsD-f | ATTCCCAGCAGTTCGCATGAAT | CpsE-r | CAATGCTACATAATCGTCACTTTC | |
| CpsE-f | GTCGTTCGACAAAACCAAGGCT | CpsF-r | TATCTTTCTTTGGCGCCAGAGTGT | |
| CpsF-f | TTACGGTGATTGCTTCTGAAGC | CpsG-r | CAACCATTCCAGGGATTCCACATT | |
| CpsG-f | AAGGATGCACCCACTAACACATC | CpsH-r | CGTTTGCCATCCAATGTCCACAC | |
| CpsH-f | GTGTGGACATTGGATGGCAAACG | CpsI-r | GCACCATAGTCATGGACATTAA | |
| CpsI-f | AGAAGCCTATTATCCTGTTAATG | CpsJ-r | AAGCTTAGCACGCATACCGCTA | |
| CpsJ-f | AGAATCTAGGCAGCAGGAGGAAGTT | CpsK-r | TAGAGACCATGCCAACTTGCATA | |
| CpsK-f | TATCGTAGGAGCTCATTTACATG | Chp-r | CAACTGGCTGATTTCCTGCTTGTA | |
| Real-time RT-PCR | CpsAf-RT | GTTAAAGTCAATGTAATGGGCTACC | CpsAr-RT | CCCATAATTCAAAGCAAAGTTCAA |
| CpsBf-RT | ATTGTCTGGGCAACGGATATT | CpsBr-RT | ACGGCTGATAAAATACCACCA | |
| CpsCf-RT | GAGAATGTTCCTTTCGTCAAACG | CpsCr-RT | GCCACGTCTGTATATAGATTGTTG | |
| CpsIf-RT | AATGGCTACATCAATCAAGTAGTTG | CpsIr-RT | TTGCTTGTGCTTCAGCTGGT | |
| CpsKf-RT | GTCTAATTAATTCAACTATTGCTGCAT | CpsKr-RT | CCAAGACGGTAAATTCTTTGTAC | |
| Chpf-RT | GAGGCAGGCATTCCAATAA | Chpr-RT | GCCGTCAATTACCAACGTAT | |
| rRNAf-RT | GTAGTCCACAGCTTCGGTAATATGT | rRNAr-RT | AACTAGGATGTTGGCTTAGAAGCA | |
Real-time quantitative RT-PCR.
Total RNA was isolated from E. faecalis grown to mid-log or stationary phase in rabbit serum, pooled human urine, or THB supplemented with various concentrations of glucose (1 to 5%). RNA samples were quantified spectrophotometrically, and the concentration was adjusted to 10 ng/μl in diethyl pyrocarbonate-treated water. Ten nanograms of RNA was then used to generate specific cDNA by RT with a 2.5 μM concentration of the real-time RT-PCR reverse primers (Table 3) and the TaqMan Reverse Transcription Reagents kit (Applied Biosystems, Foster City, Calif.). Five microliters from each 50-μl RT reaction mixture was used in subsequent PCR amplification with 300 nM concentrations of the appropriate forward primer, 50 nM concentrations of the corresponding reverse primer, and 25 μl of the SYBR Green PCR Master Mix (Applied Biosystems) in a total reaction volume of 50 μl, according to the manufacturer's recommendations. Change in fluorescence emission was detected and analyzed with an ABI Prism 7700 Sequence Detection System (Applied Biosystems). For quantitative analysis of specific gene expression and as an internal control, RNA concentrations were normalized to yield identical amplification curves for E. faecalis 23S rRNA in each RNA preparation. Additional controls included omission of the reverse transcriptase to measure the extent of residual genomic DNA contamination and use of a no-template control for each primer pair to measure interference from primer dimer formation.
The value used for comparison and quantitation was the threshold cycle (CT), defined as the cycle number at which the fluorescence emission exceeds an arbitrarily set baseline or threshold level (34). This threshold level reflects a midpoint in the linear range of amplification. During linear amplification, the amount of amplified target is directly proportional to the input amount of target. Thus, the higher the initial amount of specific template in each reaction mixture, the fewer the cycles that are required to exceed the threshold value. Because each cycle represents a doubling during the linear phase of amplification, the data was transformed from a logarithmic to linear scale by using the formula x = 2−Ct (34). Expression levels for each gene in each environment are presented as fold induction relative to expression levels determined for RNA isolated from stationary-phase cultures in THB. Mean values were calculated from data obtained by amplifying three independent RNA preparations.
Cloning of the cps pathway.
The plasmid shuttle vector pAT28 (38) was restricted with XmaI and XbaI. The cps pathway, including the cpsC to cpsK genes, was amplified by PCR from E. faecalis strain FA2-2 with primers Cps1 and Cps2 and the TaKaRa LA PCR kit (Panvera). The resulting 12.8-kb PCR product was cleaved with XmaI and NheI to produce compatible restriction ends for cloning into pAT28. To achieve stability in E. coli, transformants were initially cultivated at room temperature and were selected for on LB agar plates with spectinomycin (150 μg/ml), X-Gal, and IPTG. LacZ negative transformants were further screened by colony PCR by using primers specific for the insert (CpsIf-RT and CpsJr-RT) as well as by restriction digest mapping.
Colony immunoblot.
Immunoscreening was performed by a standard method (32) with slight modifications. E. coli and E. faecalis clones were inoculated onto LB or BHI agar plates and were incubated overnight at 37°C. Colonies were lifted onto nylon membranes (Amersham Pharmacia Biotech, Buckinghamshire, United Kingdom) and were exposed to chloroform vapor for 20 min. Bacterial colonies were lysed by incubation in lysis buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 5 mM MgCl2, 50 U of DNase/ml, and 0.5 mg of lysozyme/ml) for 1 h at 37°C. Membranes were washed twice in PBS (pH 7.4) to remove residual cell debris. Membranes were then incubated with primary antisera (E. faecalis type 2 antisera raised in rabbits, kindly provided by S. Maekawa, Sapporo, Japan) diluted 1:1,000 in PBS with 1% bovine serum albumin for 1 h at 37°C. Following additional PBS washes, membranes were incubated with secondary antibody (goat anti-rabbit antibody conjugated to alkaline phosphatase) diluted 1:5,000 in PBS with 1% bovine serum albumin for 1 h at 37°C. After PBS washes, membranes were incubated with the substrate (nitroblue tetrazolium-5-bromo-4-chloro-3-indolylphosphate tablets obtained from Sigma and dissolved in 10 ml of distilled water) to develop the blot.
RESULTS
Identification of promoters within the cps determinant.
To detect promoter activity within the region of the E. faecalis strain FA2-2 chromosome carrying the cps pathway, we constructed fusions to a promoterless lacZ reporter gene in pTCV-lac. A PCR fragment spanning the cpsA-cpsK junction was restricted with either Sau3AI or MfeI to generate 35 and 14 restriction fragments, respectively. These fragments were randomly inserted upstream of the promoterless lacZ reporter in pTCV-lac, linearized to yield compatible ends (BamHI or EcoRI). For the Sau3AI fragments, 6 of 330 E. faecalis clones gave rise to blue colonies on BHI agar X-Gal plates, indicative of promoter activity. DNA sequencing of the inserts revealed that all six clones, typified by pCPS100, contained an identical 1,744-bp Sau3AI fragment that included the region 304 nucleotides 5′ to the start codon for cpsC through 204 nucleotides 3′ to the start codon of cpsD (Fig. 1). The use of MfeI restriction fragments for promoter fusions resulted in five blue colonies from among 45 E. faecalis clones screened. Sequencing of these five clones, typified by pCPS110, showed an identical 2,604-bp MfeI fragment that overlapped the 1,744-bp Sau3AI fragment and included the region 73 nucleotides 3′ to the translation start of cpsB through 168 nucleotides past the start codon of cpsD (Fig. 1). No additional restriction fragments possessed detectable promoter activity, as assessed by the blue-colony phenotype, localizing promoter activity to a point 5′ to cpsC.
To further delineate the location of this promoter, additional promoter fusions were generated at staggered points 5′ to cpsC (Fig. 1 and Table 1). Plasmids pCPSC1, pCPSC2, pCPSC3, and pCPSC4 were introduced into E. faecalis by electroporation, and the resulting transformants were assessed for promoter activity as reflected by reporter β-galactosidase activity. The results showed that plasmids pCPSC1, pCPSC2, and pCPSC3 produced from 10.6 to 23.3 U of β-galactosidase activity per mg of protein, whereas the β-galactosidase activity expressed from pCPSC4 was less than 0.1 U/mg of protein, comparable to the basal level expressed by the pTCV-lac vector with no insert (Fig. 1). Plasmid pCPSC3 produced LacZ activity that was nearly double that of either pCPSC1 or pCPSC2 (Fig. 1). Examination of the DNA present in the smallest inserts conferring promoter activity (pCPS3) but absent from pCPS4, which lacked detectable promoter activity, revealed that promoter activity required sequences contained within the 174 nucleotides upstream of the inferred translation start of cpsC. This region contained a potential −10 sequence (5′ TATACT 3′) that nearly matched the consensus sequence for Bacillus (22) and E. coli (14) promoters. Spaced 17 nucleotides upstream of the −10 sequence was a near-consensus −35 sequence (5′ TTGACG 3′).
As the pCPSC3 promoter activity was twice that of pCPSC1 or pCPSC2, we examined the sequence present in pCPSC1 and pCPSC2 for regulatory elements that may serve to control capsule expression. Interestingly, two regions of dyad symmetry were identified upstream of the cpsC promoter. The first region of dyad symmetry (5′ TGAACATTCCAGTGTTATTCGATTGTTAAAAATTAAATTATAACAATCGAATAACACTGGAATGTTTA3′) located 14 nucleotides upstream of the putative −35 sequence is capable of folding into a stem-loop with a free energy of −35.7 kcals/mol. A second region of dyad symmetry (5′AGCAAAAATCCAGAAGAGAGACTCTTCTGGATTTTTGCU3′) located 194 nucleotides upstream of the −35 sequence was present only in pCPS1 and possessed a potential free energy of −21.6 kcals/mol. As the promoter activities of pCPS1 and pCPS2 were similar (Fig. 1), and because the 5′-most region of dyad symmetry was followed immediately by a poly(T) tract, we postulated that this region may serve as a transcriptional terminator for upstream genes. To examine this prospect, we first localized the next promoter 5′ to that mediating cpsC expression.
To detect promoter activity further upstream of cpsC, we cloned in pTCV-lac the 1.97 kb immediately 5′ to the region carrying promoter activity mediating cpsC expression. The resulting clone, pCPSAB, mediated expression of 14.2 U of β-galactosidase activity/mg of protein. Nucleotide sequence information for this region suggested that promoter activity could emanate from an apparent noncoding region 5′ to the two open reading frames (ORFs) that precede cpsC. To directly test this noncoding region for promoter activity, a second clone (pCPSA) was made sharing the same 5′ end as pCPSAB, but it was truncated to 374 bp. As shown in Fig. 1, pCPSA and pCPSAB possess nearly equivalent promoter activities. Inclusion of the region of dyad symmetry immediately 3′ to the two upstream ORFs (pCPSABT) returns the transcription of β-galactosidase to basal levels, confirming the hypothesis that this region of dyad symmetry serves as a transcriptional terminator for the upstream transcript. The two ORFs cotranscribed from this upstream promoter were termed cpsA and cpsB.
To examine the effect that transcription from the upstream cpsAB promoter may exert on the downstream cpsC promoter, we constructed pCPSA-C (Fig. 1). Plasmid pCPSA-C contains a 2.5-kb insert spanning the region 374 nucleotides upstream of the translational start of cpsA to the start codon of cpsC. As shown in Fig. 1, no obvious effect was seen when the promoter activity of pCPSA-C was compared to that of pCPSC1 or pCPSC2, suggesting that the promoter driving transcription for the cpsAB transcript exerts little effect on the promoter for the cpsC transcript.
Mapping of the transcription initiation sites.
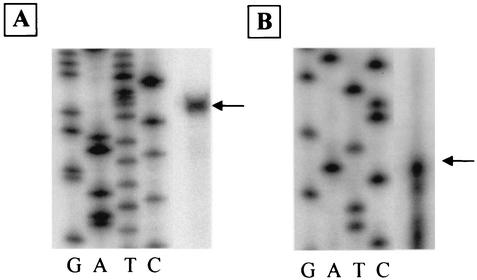
To more closely map the transcription initiation site for both the cpsA and cpsC promoters, we used primer extension analysis. As shown in Fig. 2, a band corresponding to a G residue located 29 nucleotides upstream of the ATG codon of the cpsA gene was detected. This finding localized the cpsA promoter to a point 35 nucleotides 3′ to a putative promoter of sequence TAGGCA, followed 18 bp by TATAAT. A band corresponding to a C located 160 nucleotides 5′ to the translational start of cpsC was detected (Fig. 2). This finding localized the start of cpsC transcription to a point 43 nucleotides 3′ to a sequence containing TTGACG followed 17 bp by TATACT.
FIG. 2.
Primer extension mapping of the transcription initiation site for the cpsA (A) and cpsC (B) transcripts. The M13 sequencing ladder for size determination is shown for each transcript. (A) The band identified corresponds to a G residue 29 bases upstream of the inferred cpsA start codon in the E. faecalis FA2-2 sequence. (B) The band identified corresponds to a C residue 160 bases upstream of the inferred cpsC start codon in the E. faecalis FA2-2 sequence.
Transcriptional linkage of genes in the cps determinant.
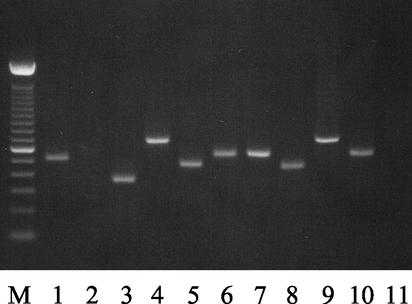
To confirm the transcriptional linkage of genes within the cps pathway as predicted by promoter fusion studies, RT-PCR was employed with primer pairs spanning the junction between each of the ORFs throughout this region. RNA isolated from mid-log THB cultures was reverse transcribed by using each of the reverse primers for RT-PCR as indicated in Table 3. The corresponding forward and reverse primers were then used in 30 cycles of PCR. Parallel reactions minus reverse transcriptase, to control for residual genomic contamination in RNA preparations, failed to yield an amplification product. DNA template controls to ensure PCR fidelity for each primer pair uniformly yielded the expected-size PCR product (data not shown). As shown in Fig. 3, a product of the expected size was seen for the RT-PCR spanning the cpsA to cpsB junction, which confirmed the prediction that cpsA and cpsB are transcriptionally linked. However, RT-PCR across the cpsB and -C intergenic region did not yield a PCR product (Fig. 3), confirming the prediction that cpsB and -C are not transcriptionally linked. RT-PCR across the remaining intergenic regions from cpsC to cpsK uniformly yielded predicted PCR products, thus demonstrating the transcriptional linkage of cpsC to cpsK (Fig. 3). Finally, the primer pair spanning the junction between cpsK and a downstream ORF, designated chp for its sequence similarity to conserved hypothetical proteins, did not produce an RT-PCR product, confirming that this downstream gene is not transcriptionally linked to those within the cps gene block.
FIG. 3.
RT-PCR analysis of transcriptional linkage of the ORFs within the cps determinant. Primer pairs specific to each intergenic junction were used to amplify across these junctions by RT-PCR. The base pairs in parentheses are the predicted sizes for the PCR products. Lane 1, RT-PCR product of the cpsAB junction (495 bp); lane 2, product of the cpsBC junction (697 bp); lane 3, product of the cpsCD junction (339 bp); lane 4, product of the cpsDE junction (645 bp); lane 5, product of the cpsEF junction (444 bp); lane 6, product of the cpsFG junction (521 bp); lane 7, product of the cpsGH junction (535 bp); lane 8, product of the cpsHI junction (431 bp); lane 9, product of the cpsIJ junction (679 bp); lane 10, product of the cpsJK junction (540 bp); and lane 11, the product of the cpsK to chp junction (807 bp). The 100-bp molecular size ladder is shown to the left (M).
Regulation of promoter activity by environmental factors.
We used plasmid pCPSA-C to examine the effect that environmental stimuli may have on expression of the cpsC-cpsK transcript. This plasmid was designed to contain the cpsC-cpsK promoter, insulated from vector sequences by the upstream cpsAB promoter and terminator, providing a native context in which to study expression.
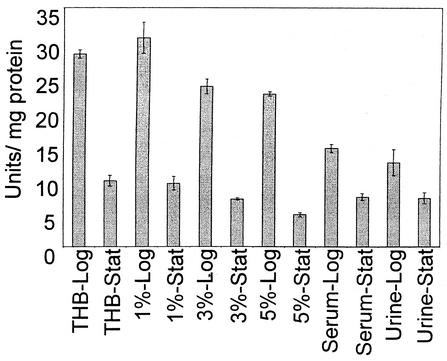
As E. faecalis is a leading cause of nosocomial urinary tract infections and bacteremia (29), we chose to examine promoter activity from bacteria grown in urine and serum. Additionally, glucose has been used routinely to supplement culture media when growing enterococcal cultures for isolating cell wall polysaccharides (16, 26). We therefore examined whether additional glucose levels had an effect on promoter activity by growing E. faecalis in THB alone (0.2% glucose) or supplemented with an additional 1, 3, or 5% glucose. Finally, it has been shown that the growth rate of the bacterial cell dramatically affects the expression of capsular polysaccharides in other systems (25, 30). Therefore, we investigated the effect that bacterial growth phase exerted on promoter activity by comparing LacZ activity from bacteria harvested in mid-log phase to that from stationary phase.
The results shown in Fig. 4 demonstrate that promoter activity, as reflected by β-galactosidase activity, is enhanced two- to threefold when cultures are grown to mid-log phase compared to the promoter activity of cultures grown to stationary phase. LacZ activity expressed from the cpsC-cpsK promoter appeared to be unaffected by the addition of glucose to THB (Fig. 4). Differences observed between the levels of β-galactosidase activity from log phase cultures grown in THB supplemented with 1 or 3% glucose were not significantly different (P = 0.34 and P = 0.08, respectively) from the levels of activity expressed in unsupplemented THB. A significant difference was observed between log phase THB culture activities and that expressed in a culture supplemented with 5% glucose (P = 0.02), but this small difference may have little biological consequence. Finally, growth in serum and urine produced a significant drop in promoter activity relative to that in THB (P < 0.01) (Fig. 4).
FIG. 4.
Effect of environmental condition on expression from the cpsC-cpsK promoter fused to the β-galactosidase reporter in pCPSA-C. Abbreviations along the X axis refer to the growth conditions under which the assays were performed. THB-Log refers to organisms grown to mid-log phase in THB. THB-Stat, stationary-phase cultures in THB; 1%-Log, log cultures grown in THB with 1% glucose; 1%-Stat, stationary-phase cultures in THB with 1% glucose; similarly, 3%-Log, 3%-Stat, 5%-Log, and 5%-Stat refer to organisms grown to log or stationary phase in THB with glucose supplementation to the percentages indicated. Serum-Log and Serum-Stat, as well as Urine-Log and Urine-Stat, refer to growth in serum or urine to log or stationary phase. Data are plotted as the means of three independent tests with error bars representing the standard deviations.
Quantitative real-time RT-PCR.
To independently and directly assess transcription of cps genes in various environmental conditions, we employed quantitative real-time RT-PCR.
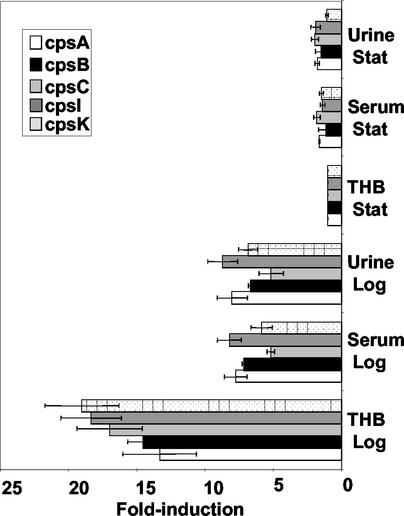
Data in Fig. 5 shows the induction of select cps genes in various environments relative to the expression of these genes in THB cultures. The results confirm that expression of cps genes is tied to growth. cps genes (cpsA, -B, -C, -I, and -K) are maximally transcribed in logarithmic phase, generally showing a greater than 10-fold induction in message levels over levels occurring in stationary phase (Fig. 5). Compared to expression in THB, cps-specific message levels were reduced in bacteria grown to mid-log phase in urine or serum (Fig. 5).
FIG. 5.
Quantitative real-time RT-PCR analysis. Effect of growth in urine or serum on gene expression in log and stationary phase relative to expression in stationary-phase THB cultures. Values represent the means from three independent RT-PCR experiments from independent RNA preparations with error bars representing standard deviation.
Heterologous expression of the cps determinant in E. coli.
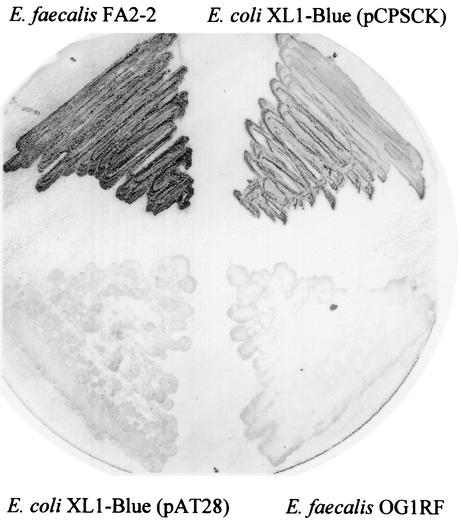
To demonstrate that the cpsC-cpsK transcript provided all the gene products necessary to produce the type 2 capsular polysaccharide, we expressed the polysaccharide determinant in a heterologous host. Employing the shuttle vector pAT28 (38), we constructed plasmid pCPSCK, which contains a 12,775-bp insert generated by PCR, which spans the region from the cpsC promoter through to the stop codon of cpsK. Colony immunoblots were performed to detect E. faecalis polysaccharide expression by this heterologous host. E. coli XL1-Blue harboring plasmid pAT28 or pCPSCK was streaked onto BHI agar plates along with E. faecalis strain FA2-2 to serve as a positive control and E. faecalis strain OG1RF to serve as a negative control, and they were incubated at 37°C for 24 h. After transfer to nylon membranes, carbohydrate was detected with E. faecalis type 2 antisera (Fig. 6). The results showed that E. coli expressing pCPSCK was reactive along with the E. faecalis strain FA2-2, whereas E. faecalis strain OG1RF and E. coli containing only the pAT28 vector were not reactive. The fact that E. coli harboring pCPSCK was reactive with type 2 antisera suggests that the gene products of cpsC to cpsK are sufficient for the production of the E. faecalis capsular polysaccharide in E. coli. It is likely that E. coli possesses complementary functions for the products of the cpsA and cpsB genes, as these genes were not required for expression in the heterologous host. What contribution, if any, that the gene products of cpsA, an undecaprenyl pyrophosphate synthetase (uppS) homolog, and cpsB, a phosphatidate cytidylyltransferase homolog, play in capsular polysaccharide biosynthesis will be difficult to assess, as they appear to be essential for cell viability (1). Our previous attempts to inactivate these genes in E. faecalis proved unsuccessful (12), indicative of their essential nature.
FIG. 6.
Colony immunoblot of E. faecalis strains FA2-2 and OG1RF along with E. coli strain XL1-Blue harboring plasmids pAT28 or pCPSCK. Immunoblotting was performed with E. faecalis type 2 antiserum as described in the text.
DISCUSSION
We previously reported a genetic determinant specifying synthesis of the type 2 polysaccharide capsule in E. faecalis (12). This ∼15-kb putative determinant consists of 11 ORFs, all transcribed in the same direction. Mutants in this pathway were unable to synthesize the capsular antigen. A polar mutation in the cpsI gene could not be complemented in trans by cpsI alone but could be complemented in trans with a plasmid expressing cpsI to cpsK, suggesting that these genes are transcriptionally linked. Additional mutations in cpsC and cpsK also resulted in loss of the polysaccharide capsule. A mutation in an ORF 3′ to cpsK was unaffected in its ability to produce the carbohydrate antigen. These observations indicated that the region from cpsC to cpsK contributed to the production of the type 2 capsule. The contribution of cpsA and cpsB to the production of the polysaccharide was not clear from this study, as attempts to generate mutants in these genes did not yield viable bacteria indicating that these genes are essential to cell survival. However, based on their sequence similarity to known cell wall precursors (18), this prospect seemed likely.
In the present work, we examined the transcriptional organization of the cps determinant, including ORFs cpsA to cpsK, by using promoter fusions to a lacZ reporter gene in pTCV-lac and by primer extension analysis. By these complementary approaches, the transcriptional start sites for two independent promoters were mapped. The first promoter, controlling expression of the cpsA and -B genes, produces a transcript that begins with a G nucleotide 29 residues 5′ to the inferred translational start of cpsA. Sequences immediately 5′ to this point possess a consensus −10 sequence spaced 18 nucleotides from a near-consensus −35 sequence (four out of six positions) (14, 22). The second promoter, which controls expression of the cpsC to -K genes, produces a transcript that begins with a cytosine 160 residues 5′ from the inferred cpsC start codon. Sequences immediately 5′ to this point also contain a near-consensus −35 sequence spaced 17 nucleotides from a near-canonical −10 sequence. 5′ to the cpsC promoter are two regions of dyad symmetry. The first region appears to serve a regulatory role in cpsC to cpsK gene expression, as its deletion caused a twofold increase in promoter activity from the cpsC promoter. The precise role of this potential regulatory element awaits further experimentation. However, its proximity to the −35 sequence may influence the interaction of RNA polymerase in this region or local folding. While the role of the first region of dyad symmetry 5′ to the cpsC promoter remains open to debate, the second region of dyad symmetry further upstream was found to function as the transcriptional terminator for the cpsAB transcript.
The prediction of two independent cps transcripts from promoter fusion studies was confirmed by RT-PCR analysis. The first transcript includes cpsA and -B, while the second larger transcript encompasses cpsC to -K. Production of a large polycistronic message is a common organizational motif in the synthesis of bacterial polysaccharides (4, 33).
With the exception of the E. faecalis peptide pheromone system (8), and more recently the regulation of enterococcal cytolysin (11), relatively little is known regarding gene regulation in enterococci, particularly in ecologies relevant to infection. Earlier reports (9, 10) suggested that enterococci vary carbohydrate expression in response to serum, but these observations have not been further extended. In this study, we examined regulation of the type 2 polysaccharide determinant under a variety of environmental conditions by transcriptional reporter fusion and by real-time RT-PCR. We found that cps gene expression was primarily growth-phase dependent, with maximal expression occurring in the mid-exponential phase irrespective of the growth medium. Growth in serum or urine caused a modest reduction in reporter enzyme activity, as well as in cps-specific mRNA levels. Growth rates in these two biological environments were lower than those in THB, which may account for the reduced expression of cps genes relative to the internal 23S rRNA control. This is consistent with the observation that in all environments, cps expression is maximal in periods of rapid cell division (i.e., log phase). It is noteworthy that in a recent report, Shepard and Gilmore (35) found that all of the putative E. faecalis virulence-related genes were down regulated as cells entered stationary phase when cultured in both serum and urine, consistent with the observations reported here. What role these gene expression changes play with respect to phenotype and the overall biology of the organism awaits further investigation.
The synthesis of bacterial polysaccharides by gram-positive bacteria appears to be a highly regulated process (6, 19, 20, 30). Growth-phase-dependent regulation of capsular polysaccharides in Streptococcus pyogenes (6, 19) and Streptococcus agalactiae (30) occurs with maximal expression in mid-log phase. Paoletti et al. (25) showed that group B streptococci synthesize about sixfold more type III capsular polysaccharide when grown in a controlled chemostat with a doubling time every 48 min compared to that of cultures limited to doubling times every 11 h.
It is interesting that recently reported cases (3) of mucoid variants of E. faecalis were isolated from patients with chronic urinary tract infection. What role chronic infection at this site or exposure to factors in urine or serum may have in selecting for variants remains unclear. It is conceivable that ecological cues not tested in the present study also affect expression of the cps determinant. On occasion it has been observed that enterococci in intimate association with host cells appear mucoid (L. E. Hancock, unpublished data). The biological significance of a linkage between growth rate and E. faecalis capsular polysaccharide expression is the subject of ongoing study.
REFERENCES
- 1.Apfel, C. M., B. Takacs, M. Fountoulakis, M. Stieger, and W. Keck. 1999. Use of genomics to identify bacterial undecaprenyl pyrophosphate synthetase: cloning, expression, and characterization of the essential uppS gene. J. Bacteriol. 181:483-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bottone, E. J. 1999. Encapsulated Enterococcus faecalis: role of encapsulation in persistence in mouse peritoneum in absence of mouse lethality. Diagn. Microbiol. Infect. Dis. 33:65-68. [DOI] [PubMed] [Google Scholar]
- 3.Bottone, E. J., L. Patel, P. Patel, and T. Robin. 1998. Mucoid encapsulated Enterococcus faecalis: an emerging morphotype isolated from patients with urinary tract infections. Diagn. Microbiol. Infect. Dis. 31:429-430. [DOI] [PubMed] [Google Scholar]
- 4.Chaffin, D. O., S. B. Beres, H. H. Yim, and C. E. Rubens. 2000. The serotype of type Ia and III group B streptococci is determined by the polymerase gene within the polycistronic capsule operon. J. Bacteriol. 182:4466-4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clewell, D. B., P. K. Tomich, M. C. Gawron-Burke, A. E. Franke, Y. Yagi, and F. Y. An. 1982. Mapping of Streptococcus faecalis plasmids pAD1 and pAD2 and studies relating to transposition of Tn917. J. Bacteriol. 152:1220-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crater, D. L., and I. van de Rijn. 1995. Hyaluronic acid synthesis operon (has) expression in group A streptococci. J. Biol. Chem. 270:18452-18458. [DOI] [PubMed] [Google Scholar]
- 7.Cruz-Rodz, A. L., and M. S. Gilmore. 1990. High efficiency introduction of plasmid DNA into glycine treated Enterococcus faecalis by electroporation. Mol. Gen. Genet. 224:152-154. [DOI] [PubMed] [Google Scholar]
- 8.Dunny, G. M., and B. A. Leonard. 1997. Cell-cell communication in gram-positive bacteria. Annu. Rev. Microbiol. 51:527-564. [DOI] [PubMed] [Google Scholar]
- 9.Guzman, C. A., C. Pruzzo, and L. Calegari. 1991. Enterococcus faecalis: specific and non-specific interactions with human polymorphonuclear leukocytes. FEMS Microbiol. Lett. 68:157-162. [DOI] [PubMed] [Google Scholar]
- 10.Guzman, C. A., C. Pruzzo, M. Plate, M. C. Guardati, and L. Calegari. 1991. Serum dependent expression of Enterococcus faecalis adhesins involved in the colonization of heart cells. Microb. Pathog. 11:399-409. [DOI] [PubMed] [Google Scholar]
- 11.Haas, W., B. D. Shepard, and M. S. Gilmore. 2002. Two-component regulator of Enterococcus faecalis cytolysin responds to quorum-sensing autoinduction. Nature 415:84-87. [DOI] [PubMed] [Google Scholar]
- 12.Hancock, L. E., and M. S. Gilmore. 2002. The capsular polysaccharide of Enterococcus faecalis and its relationship to other polysaccharides in the cell wall. Proc. Natl. Acad. Sci. USA 99:1574-1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hancock, L. E., and M. S. Gilmore. 2000. Pathogenicity of enterococci, p. 251-258. In V. Fischetti, R. Novick, J. Ferretti, D. Portnoy, and J. Rood (ed.), Gram-positive pathogens. ASM Press, Washington, D.C.
- 14.Hawley, D. K., and W. R. McClure. 1983. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 11:2237-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huebner, J., A. Quaas, W. A. Krueger, D. A. Goldmann, and G. B. Pier. 2000. Prophylactic and therapeutic efficacy of antibodies to a capsular polysaccharide shared among vancomycin-sensitive and -resistant enterococci. Infect. Immun. 68:4631-4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huebner, J., Y. Wang, W. A. Krueger, L. C. Madoff, G. Martirosian, S. Boisot, D. A. Goldmann, D. L. Kasper, A. O. Tzianabos, and G. B. Pier. 1999. Isolation and chemical characterization of a capsular polysaccharide antigen shared by clinical isolates of Enterococcus faecalis and vancomycin-resistant Enterococcus faecium. Infect. Immun. 67:1213-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jett, B. D., K. L. Hatter, M. M. Huycke, and M. S. Gilmore. 1997. A simplified agar plate method for quantifying viable bacteria. BioTechniques 23:648-650. [DOI] [PubMed] [Google Scholar]
- 18.Kato, J., S. Fujisaki, K. Nakajima, Y. Nishimura, M. Sato, and A. Nakano. 1999. The Escherichia coli homologue of yeast RER2, a key enzyme of dolichol synthesis, is essential for carrier lipid formation in bacterial cell wall synthesis. J. Bacteriol. 181:2733-2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leonard, B. A., M. Woischnik, and A. Podbielski. 1998. Production of stabilized virulence factor-negative variants by group A streptococci during stationary phase. Infect. Immun. 66:3841-3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKenney, D., K. L. Pouliot, Y. Wang, V. Murthy, M. Ulrich, G. Doring, J. C. Lee, D. A. Goldmann, and G. B. Pier. 1999. Broadly protective vaccine for Staphylococcus aureus based on an in vivo-expressed antigen. Science 284:1523-1527. [DOI] [PubMed] [Google Scholar]
- 21.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 22.Moran, C. P., Jr., N. Lang, S. F. LeGrice, G. Lee, M. Stephens, A. L. Sonenshein, J. Pero, and R. Losick. 1982. Nucleotide sequences that signal the initiation of transcription and translation in Bacillus subtilis. Mol. Gen. Genet. 186:339-346. [DOI] [PubMed] [Google Scholar]
- 23.Mundy, L. M., D. F. Sahm, and M. Gilmore. 2000. Relationship between enterococcal virulence and antimicrobial resistance. Clin. Microbiol. Rev. 13:513-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murray, B. F., K. V. Singh, R. P. Ross, J. D. Heath, G. M. Dunny, and G. M. Weinstock. 1993. Generation of restriction map of Enterococcus faecalis OG1 and investigation of growth requirements and regions encoding biosynthetic function. J. Bacteriol. 175:5216-5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paoletti, L. C., R. A. Ross, and K. D. Johnson. 1996. Cell growth rate regulates expression of group B Streptococcus type III capsular polysaccharide. Infect. Immun. 64:1220-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pazur, J. H. 1982. β-D-glucose 1-phosphate: a structural unit and an immunological determinant of a glycan from streptococcal cell walls. J. Biol. Chem. 257:589-591. [PubMed] [Google Scholar]
- 27.Pospiech, A., and B. Neumann. 1995. A versatile quick-prep of genomic DNA from gram-positive bacteria. Trends Genet. 11:217-218. [DOI] [PubMed] [Google Scholar]
- 28.Poyart, C., and P. Trieu-Cuot. 1997. A broad-host-range mobilizable shuttle vector for the construction of transcriptional fusions to beta-galactosidase in gram-positive bacteria. FEMS Microbiol. Lett. 156:193-198. [DOI] [PubMed] [Google Scholar]
- 29.Richards, M. J., J. R. Edwards, D. H. Culver, and R. P. Gaynes. 2000. Nosocomial infections in combined medical-surgical intensive care units in the United States. Infect. Control. Hosp. Epidemiol. 21:510-515. [DOI] [PubMed] [Google Scholar]
- 30.Ross, R. A., L. C. Madoff, and L. C. Paoletti. 1999. Regulation of cell component production by growth rate in the group B Streptococcus. J. Bacteriol. 181:5389-5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sahm, D. F., J. Kissinger, M. S. Gilmore, P. R. Murray, R. Mulder, J. Solliday, and B. Clarke. 1989. In vitro susceptibility studies of vancomycin-resistant Enterococcus faecalis. Antimicrob. Agents Chemother. 33:1588-1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory Manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 33.Sau, S., J. Sun, and C. Y. Lee. 1997. Molecular characterization and transcriptional analysis of type 8 capsule genes in Staphylococcus aureus. J. Bacteriol. 179:1614-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmittgen, T. D., B. A. Zakrajsek, A. G. Mills, V. Gorn, M. J. Singer, and M. W. Reed. 2000. Quantitative reverse transcription-polymerase chain reaction to study mRNA decay: comparison of endpoint and real-time methods. Anal. Biochem. 285:194-204. [DOI] [PubMed] [Google Scholar]
- 35.Shepard, B. D., and M. S. Gilmore. 2002. Differential expression of virulence-related genes in Enterococcus faecalis in response to biological cues in serum and urine. Infect. Immun. 70:4344-4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shepard, B. D., and M. S. Gilmore. 1999. Identification of aerobically and anaerobically induced genes in Enterococcus faecalis by random arbitrarily primed PCR. Appl. Environ. Microbiol. 65:1470-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Teng, F., K. D. Jacques-Palaz, G. M. Weinstock, and B. E. Murray. 2002. Evidence that the enterococcal polysaccharide antigen gene (epa) cluster is widespread in Enterococcus faecalis and influences resistance to phagocytic killing of E. faecalis. Infect. Immun. 70:2010-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trieu-Cuot, P., C. Carlier, C. Poyart-Salmeron, and P. Courvalin. 1990. A pair of mobilizable shuttle vectors conferring resistance to spectinomycin for molecular cloning in Escherichia coli and in gram-positive bacteria. Nucleic Acids Res. 18:4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu, Y., B. E. Murray, and G. M. Weinstock. 1998. A cluster of genes involved in polysaccharide biosynthesis from Enterococcus faecalis OG1RF. Infect. Immun. 66:4313-4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu, Y., K. V. Singh, X. Qin, B. E. Murray, and G. M. Weinstock. 2000. Analysis of a gene cluster of Enterococcus faecalis involved in polysaccharide biosynthesis. Infect. Immun. 68:815-823. [DOI] [PMC free article] [PubMed] [Google Scholar]