Abstract
Lysobacter enzymogenes strain N4-7 produces multiple biochemically distinct extracellular β-1,3-glucanase activities. The gluA, gluB, and gluC genes, encoding enzymes with β-1,3-glucanase activity, were identified by a reverse-genetics approach following internal amino acid sequence determination of β-1,3-glucanase-active proteins partially purified from culture filtrates of strain N4-7. Analysis of gluA and gluC gene products indicates that they are members of family 16 glycoside hydrolases that have significant sequence identity to each other throughout the catalytic domain but that differ structurally by the presence of a family 6 carbohydrate-binding domain within the gluC product. Analysis of the gluB gene product indicates that it is a member of family 64 glycoside hydrolases. Expression of each gene in Escherichia coli resulted in the production of proteins with β-1,3-glucanase activity. Biochemical analyses of the recombinant enzymes indicate that GluA and GluC exhibit maximal activity at pH 4.5 and 45°C and that GluB is most active between pH 4.5 and 5.0 at 41°C. Activity of recombinant proteins against various β-1,3 glucan substrates indicates that GluA and GluC are most active against linear β-1,3 glucans, while GluB is most active against the insoluble β-1,3 glucan substrate zymosan A. These data suggest that the contribution of β-1,3-glucanases to the biocontrol activity of L. enzymogenes may be due to complementary activities of these enzymes in the hydrolysis of β-1,3 glucans from fungal cell walls.
Members of the genus Lysobacter typically are found in soil and water habitats and are characterized by gliding motility and the ability to lyse other microorganisms, including fungi and nematodes (4). One species within this group, Lysobacter enzymogenes, produces an impressive repertoire of extracellular degradative enzyme activities. These include chitinase, glucanase, and protease activities, which can degrade components found in fungal cell walls and which are presumed to contribute to the lytic activity of L. enzymogenes. Despite these described traits, relatively little is known about the hydrolytic enzymes that L. enzymogenes produces or the contributing role for each enzyme class in the lytic activity of L. enzymogenes. To date, only proteases (7, 33, 40) from the species have been characterized at the molecular and biochemical levels.
β-1,3-Glucanases, which hydrolyze glucan polymers containing β-1,3 linkages (3, 39) from a number of microbial species, have been characterized. Proposed physiological functions for these enzymes vary depending on the source of enzyme. Among bacteria, many β-1,3-glucanases have been studied for purposes of fungal and yeast cell wall degradation (see, e.g., references 2, 9, 17, 27, 28, 31, and 41). However, despite obvious associations with fungal antagonism, few β-1,3-glucanases from bacteria with demonstrated biological control activity toward fungal plant pathogens have been characterized. The bacterial biocontrol strain N4-7 was isolated originally as a biocontrol agent for plant diseases (20) and was classified recently as L. enzymogenes (35). Production of lytic enzymes, including β-1,3-glucanases, was thought to contribute significantly to the biocontrol activity of this strain. Since initial characterization of extracellular enzyme activities of strain N4-7 indicated the production of a number of different β-1,3-glucanase activities (16), we hypothesized that these activities are encoded by multiple genes, rather than arising as multiple enzyme species from a single gene. We were interested in identifying genes encoding β-1,3-glucanase activity in strain N4-7 as a first step in defining the genetic elements responsible for extracellular enzyme production in this strain and establishing the role of these enzymes in the biocontrol activity of strain N4-7. Due to the complexity of β-1,3-glucanase activity produced by this bacterium, which precluded mutagenesis and screening for loss of enzyme activity, individual genes were identified following internal peptide sequence analysis of active proteins partially purified from strain N4-7 culture filtrates. In addition, biochemical analysis of individual β-1,3-glucanase-active proteins was initiated by expression in Escherichia coli of each β-1,3-glucanase activity-encoding gene.
MATERIALS AND METHODS
Bacterial strains and media.
Bacterial strains and plasmids used in this study are listed in Table 1. L. enzymogenes strain N4-7 was grown at 30°C with shaking in medium 813, composed of 2.5 g of Casamino Acids, 3 g of K2HPO4, 1.2 g of NaH2PO4, 1 g of NH4Cl, 0.3 g of MgSO4 · 7H2O, 0.15 g of KCl, 10 mg of CaCl2, and 2.5 mg of FeSO4 per liter. Yeast cell walls were prepared by autoclaving fresh baker's yeast (Saccharomyces cerevisiae) dissolved in distilled H2O, followed by centrifuging (5,000 × g, 10 min). Pelleted cell solids were washed three times with distilled H2O, sterilized by autoclaving, and added (1% [wt/vol]) as a carbon source to medium 813, creating medium 813Y. E. coli cultures were grown in Luria-Bertani medium (Difco, Detroit, Mich.) supplemented with ampicillin (100 μg ml−1) or tetracycline (25 μg ml−1) when appropriate.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| L. enzymogenes N4-7 | Wild-type biocontrol bacterium | 20 |
| E. coli | ||
| DH5α | Host strain for general cloning; supE44 ΔlacU169φ80lacZΔM15 hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | |
| HB101 | Host strain for genomic library; F−thi-1 hsdS20 (rB− mB−) supE44 recA13 ara-14 leuB6 proA2 lacY1 rpsL20 (Strr) xyl-5 mtl-1 | |
| TOP10 | Host strain for PBAD expression vectors; F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(araA-leu)7697 galU galK rpsL endA1 nupG | Invitrogen |
| BL21 (DE3) | Host strain for T7 expression vectors; F−dcm ompT hsdS (rB− mB−) galλ(DE3) | Stratagene |
| Plasmids | ||
| pUC119 | Multicopy cloning vector; Apr | 38 |
| pBluescript SK(+) | Multicopy cloning vector; Apr | Stratagene |
| pLAFR3 | Cosmid cloning vector; Tcr | 34 |
| pBAD/myc-HisA | Multiple cloning site downstream from PBAD promoter; Apr | Invitrogen |
| pET3C | T7 expression vector; BamHI site in reading frame 3 relative to T7 gene 10 initiation codon; Apr | Stratagene |
| pNG2 | pLAFR3 cosmid library clones containing gluA | This study |
| pNG6 | pLAFR3 cosmid library clones containing gluB | This study |
| pNG9 | pLAFR3 cosmid library clones containing gluC | This study |
| pNG21 | 3.2-kb PstI gluA fragment of pNG2 in pUC119; ORF oriented downstream of lac promoter | This study |
| pNG602 | 2.0-kb NotI gluB fragment of pNG6 in pBluescript SK(+); ORF oriented downstream of lac promoter | This study |
| pNG94 | 3.2-kb PstI gluC fragment of pNG9 in pBluescript SK(+); ORF oriented downstream of lac promoter | This study |
| pNG2503 | Coding region of gluA cloned in pET3C | This study |
| pNG6027 | Coding region of gluB cloned in pBAD/myc-HisA | This study |
| pNG9615 | Coding region of gluC cloned in pBAD/myc-HisA | This study |
Native polyacrylamide gel electrophoresis (PAGE).
Strain N4-7 was grown for 2 days in medium 813Y. Extracellular proteins were concentrated from 5 ml of culture filtrate by adsorption onto a 1-ml phenyl-Sepharose 6 Fast Flow (high-sub) column (Amersham Pharmacia Biotech, Piscataway, N.J.), followed by elution with 50% acetonitrile in 20 mM Tris-HCl, pH 6.8. The eluate was vacuum dried and resuspended in 200 μl of electrophoresis buffer (25 mM histidine, 30 mM MOPS [morpholinepropanesulfonic acid] [pH 6.6]). Samples (5 μl) were loaded onto 6% acrylamide gels containing 0.4% laminarin and electrophoresed in the same buffer at 10 mA on ice for 4 h. β-1,3-Glucanase activity was detected by incubating gels at 37°C for 30 min and staining with 2,3,5-triphenyltetrazolium chloride (25).
Partial purification of β-1,3-glucanase-active proteins.
Culture filtrate (600 ml) from strain N4-7 grown for 2 days in medium 813Y was loaded onto an 80-ml phenyl-Sepharose 6 Fast Flow (high-sub) column equilibrated with 20 mM bis-Tris, pH 6.7. Proteins were eluted with a linear gradient from 0 to 50% acetonitrile in 20 mM bis-Tris, pH 6.7, over 800 ml at a flow rate of 5 ml min−1. Fractions (8 ml) were assayed for β-1,3-glucanase activity by measuring reducing sugars (6) produced from laminarin as described previously (42). Units of activity are defined as micromoles of reducing sugar equivalents produced per minute, relative to glucose standards. Active fractions were pooled and applied to an 80-ml Q Sepharose high-performance (Amersham Pharmacia Biotech) column equilibrated with 20 mM bis-Tris, pH 6.7. Proteins were eluted with a linear gradient of 0 to 0.5 M NaCl in 20 mM bis-Tris, pH 6.7, over 800 ml at a flow rate of 6 ml min−1. Active fractions were pooled and concentrated into 1 ml of 20 mM bis-Tris, pH 6.7, by ultrafiltration using Amicon PM10 disks (10,000-molecular-weight-cutoff polyethersulfone membranes; Millipore, Bedford, Mass.). Samples were separated by sodium dodecyl sulfate (SDS)-10% PAGE and visualized by staining with Coomassie brilliant blue R-250. Three protein bands (I-1, I-2, and II) that corresponded to bands of β-1,3-glucanase activity on native gels were excised from SDS-PAGE gels and subjected to trypsin digestion and internal amino acid sequence analysis (Protein Chemistry Laboratory, University of Texas Medical Branch, Galveston, Tex., for bands I-2 and II and the Harvard Microchemistry Facility, Harvard University, Cambridge, Mass., for band I-1).
Identification and analysis of genes encoding β-1,3-glucanase activities.
Oligonucleotide primer sequences are listed in Table 2. Oligonucleotide glu31 was designed by reverse translation of identical internal peptide sequences obtained from activity bands I-1 and I-2 and corresponded to the first seven residues of the sequenced protein fragment. Oligonucleotide glu43 was designed by reverse translation of an internal peptide sequence obtained from activity band II and corresponded to the first 13 amino acid residues, omitting residue G10, which did not affect DNA hybridization results. Oligonucleotide probes were 5′ end labeled with digoxigenin (DIG). A genomic library of strain N4-7 DNA was constructed in cosmid vector pLAFR3 (34) and maintained in E. coli HB101 cells as individual clones. Cosmid clones containing DNA predicted to encode β-1,3-glucanase-active proteins were identified by Southern hybridization of the DIG-labeled oligonucleotide to DNA isolated from the genomic library. Hybridizations were performed at 50 (glu31 probe) or 65°C (glu43 probe), and membranes were washed under high-stringency conditions according to the manufacturer's recommendations (Genius nonradioactive labeling and detection kit; Roche Diagnostics, Indianapolis, Ind.). Oligonucleotide-hybridizing fragments were subcloned into pUC119 (38) or pBluescript SK(+) (Stratagene, La Jolla, Calif.) and sequenced on an ABI 373 DNA automated sequencer (Applied Biosystems, Foster City, Calif.). Sequences were assembled with Lasergene sequence analysis software (DNASTAR, Madison, Wis.). Open reading frames (ORFs) encoding proteins with β-1,3-glucanase activity were identified by similarity to known bacterial β-1,3-glucanase genes by using the BLAST (Basic Local Alignment Search Tool) at the National Center for Biotechnology Information (NCBI) (1) website (http://www.ncbi.nlm.nih.gov/BLAST). Glycoside families were determined by using the CAZy(ModO) Coutinho and Henrissat (1999) Carbohydrate-Active Enzymes server (http://afmb.cnrs-mrs.fr/∼cazy/CAZY/index.html).
TABLE 2.
Oligonucleotide primer sequences used for Southern hybridization probes and PCR amplificationa
| Primer | Sequence (5′-3′) | Restriction site |
|---|---|---|
| glu43 | TTCAACCTCGCCCTGCTCAACGCCAGCC AGAACACCGC | |
| glu31 | ACCTACGGCACCATCCACTG | |
| NGAm | CCCGGATCCAGAGCTGGCAGCTGGTCT | BamHI |
| NGAup | GGCGGATCCGAACGCGCGGCTTACTG | BamHI |
| N43m | CCCGGATCCGCCACCCCGGCGCGCTTC AACC | BamHI |
| N43up | GCCGGATCCTCAGCGCAGCGCCCGCAA TGT | BamHI |
| NGCm | CGCCTCGAGGCAGAACTGGCAATTGGT | XhoI |
| NGCup | GCGCTCGAGGAACCCGAGTGTTCAGAT | XhoI |
Underlining designates restriction sites. All primers were synthesized by Integrated DNA Technologies, Coralville, Iowa, or Genosys Biotechnologies, Inc., The Woodlands, Tex.
Protein domains were initially profiled by using CD-Search against the Conserved Domain Database (CDD) with the RPS-BLAST program (NCBI) and also by using the Protein Domain database BLAST (http://prodes.toulouse.inra.fr/prodom/2002.1/html/form.php). After conserved domains in our protein sequences were identified and delimited, NCBI BLAST was used to find homologous sequences not yet deposited in the CDD. DIALIGN 2.1 (24) was used to align the domain sequences, and the Genetics Computer Group (GCG; Madison, Wis.) program Gap and the SeqLab interface for the Wisconsin Package, version 9.1, were used to analyze domain sequences and make manual adjustments. Ambiguous regions of uncertain alignment were excluded from the analysis. MrBayes, version 1.11, a Bayesian phylogenetic inference program (J. P. Huelsenbeck, MrBayes: Bayesian inference of phylogeny, Department of Biology, University of Rochester, Rochester, N.Y., 2000 [distributed by the author]) was used for maximum-likelihood analysis to find the most likely tree for each domain alignment and to derive branch support. PAUP (D. L. Swofford, PAUP: phylogenetic analysis using parsimony [and other methods], version 8, Sinauer Associates, Sunderland, Mass., 1999) was used to determine partition frequencies of consensus trees. A Markov chain Monte Carlo (four-chain) analysis was run for 102,000 generations by using the PAM250 (250 substitutions per 100 residues) mutation matrix of Jones et al. (20). Trees were sampled every 100 generations, yielding 1,020 trees. Plots of tree number versus −ln likelihood determined that the first 20 trees were not asymptotic, and they were discarded, leaving 1,000 likely trees. For each analysis, the most likely trees (tree with lowest −ln score) were selected for presentation in Fig. 2 and 3.
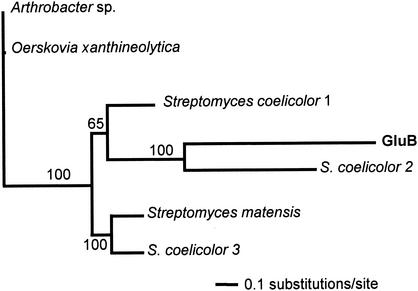
FIG. 2.
Maximum-likelihood tree for GluB and family 64 glycoside hydrolases. Analysis was performed as described in Materials and Methods with the following enzymes (GenBank accession numbers in parentheses): Arthrobacter sp. β-1,3-glucanase (BAA04892); O. xanthineolytica β-1,3-glucanase (AAA25520); S. coelicolor 1 putative glycosyl hydrolase (CAC16456); S. coelicolor 2 putative glycosyl hydrolase (CAB69688); S. coelicolor 3 putative secreted sugar hydrolase (CAC16439); S. matensis laminaripentaose-producing β-1,3-glucanase (BAA34349). Branch partition frequencies from a consensus tree of 1,000 likely trees are shown above nodes.
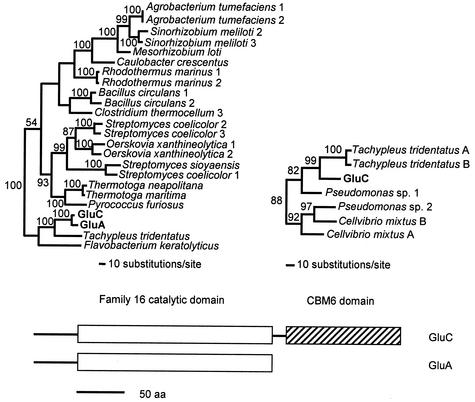
FIG. 3.
Structural organization and phylogenetic trees of GluA and GluC domains. Relevant clades of maximum-likelihood trees for the family 16 catalytic domain (open box) and CBM6 domain (striped box), with branch partition frequencies from a consensus tree of 1,000 likely trees near nodes, are shown. For the catalytic domain, the tree depicts the following (GenBank accession numbers in parentheses): Agrobacterium tumefaciens 1 endo-1,3-1,4-β-glycanase (NP_357049), Agrobacterium tumefaciens 2 endo-1,3-1,4-β-glycanase (NP_534060), Bacillus circulans 1 glucan endo-1,3-β-glucosidase A1 (P23903), Bacillus circulans 2 glucan endo-1,3-β-d-glucosidase (JN0772), Caulobacter crescentus β-glucanase (NP_419199), Clostridium thermocellum 3 endo-1,3(4)-β-glucanase (T18265), GluA from L. enzymogenes β-1,3-glucanase (AY157838), GluC from L. enzymogenes β-1,3-glucanase (AY157840), “F. keratolyticus” endo-β-galactosidase (AF083896), Mesorhizobium loti endo-1,3-1,4-β-glycanase (NP_103892), O. xanthineolytica 1 β-1,3-glucanase (AAC44371), O. xanthineolytica 2 β-1,3-glucanase (AAC38290), Pyrococcus furiosus endo-β-1,3-glucanase (AAC25554), Rhodothermus marinus 1 endo-β-1,3-1,4-glucanase (P45798), Rhodothermus marinus 2 laminarinase (AAC69707), Sinorhizobium meliloti 2 endo-1,3-1,4-β-glycanase (CAB38101), Sinorhizobium meliloti 3 endo-1,3-1,4-β-glycanase (O33680), S. coelicolor 1 probable secreted glucosidase (T35164), S. coelicolor 2 putative secreted hydrolase (CAC14352), S. coelicolor 3 putative secreted hydrolase (CAC16455), S. sioyaensis endo-1,3-β-glucanase (AAF31438), T. tridentatus α-subunit of coagulation factor G (A49878), Thermotoga neapolitana laminarinase (CAA88008), and Thermotoga maritima laminarinase (NP_227840). For the CBM6 domain, the tree depicts the following (GenBank accession numbers, followed by residue region, in parentheses): C. mixtus A cellulase (AAB61462; 359 to 484), C. mixtus B cellulase (AAB61462; 492 to 620), GluC from L. enzymogenes β-1,3-glucanase (AY157840; 263 to 382), Pseudomonas sp. strain 1 chitinase (BAB79259; 414 to 533), Pseudomonas sp. strain 2 cellulase (BAB79288; 345 to 464), T. tridentatus A α-subunit of coagulation factor G (A49878; 414 to 534), and T. tridentatus B α-subunit of coagulation factor G (A49878; 551 to 672).
Expression of genes encoding β-1,3-glucanase activities in E. coli.
All primers constructed for PCR amplification are listed in Table 2. The region of gluA predicted to encode the mature form of GluA (bases 68 to 776 relative to the start codon) was amplified from pNG25 with primers NGAm and NGAup, designed to incorporate 5′ BamHI restriction sites at both ends of the amplified product. The amplified gluA fragment was digested with BamHI and cloned into the same site of the T7 expression vector pET3C (Stratagene), resulting in pNG2503, and transformed into E. coli strain BL21(DE3). The region of gluB predicted to encode the mature form of GluB (bases 79 to 1188 relative to the start codon) was amplified from pNG602 with primers N43m and N43up, incorporating 5′ BamHI restriction sites. The amplified gluB fragment was digested with BamHI and cloned into the BglII site of the arabinose-inducible expression vector pBAD/myc-HisA (Invitrogen, Carlsbad, Calif.), resulting in pNG6027, and was transformed into E. coli strain TOP10. The region of gluC predicted to encode the mature form of GluC (bases 66 to 1164 relative to the start codon) was amplified from pNG96 with primers NGCm and NGCup, incorporating 5′ XhoI restriction sites. The amplified gluC fragment was digested with XhoI and cloned into the XhoI site of pBAD/myc-HisA, resulting in pNG9615, and was transformed into E. coli strain TOP10. Expression of recombinant proteins from clones in BL21(DE3) and TOP10 was induced in mid-exponential phase (optical density at 600 nm = 0.5) by the addition of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) or 0.02% arabinose, respectively, and incubation for an additional 2 to 4 h. Sonic lysates were prepared by centrifuging 5 ml of induced cultures, resuspending cell pellets in 1 ml of 20 mM Tris-HCl, pH 6.8, and sonicating on ice with four 30-s pulses by using a Fisher Sonic Dismembrator. Lysates were concentrated with a 1-ml phenyl-Sepharose column as described above. β-1,3-Glucanase activities were detected by native PAGE, performed as described above, with a 43 mM imidazole-30 mM HEPES, pH 7.4, buffer system.
Purification of recombinant enzymes.
Expression of β-1,3-glucanase-active proteins in 1-liter cultures of E. coli TOP10 or BL21(DE3) grown in Luria-Bertani medium was induced as described above. Cells were pelleted by centrifugation and resuspended in 20 ml of 20 mM Tris-HCl, pH 7.4. Cells were lysed, and cell debris was removed by centrifugation (26,900 × g, 30 min, 4°C). For each enzyme, the supernatant was loaded onto a 100-ml phenyl-Sepharose 6 Fast Flow (high sub) column and eluted as described for native proteins. Recombinant GluB was further purified by pooling β-1,3-glucanase-active fractions, adjusting pH to 6.2 with HCl, and loading them onto a 100-ml SP Sepharose high-performance (Amersham Pharmacia Biotech) column. Proteins were eluted with a linear gradient from 0 to 0.5 M NaCl in 20 mM bis-Tris, pH 6.0, over 1,000 ml at a flow rate of 5 ml min−1. Recombinant GluC was further purified after phenyl-Sepharose chromatography by pooling β-1,3-glucanase-active fractions and loading onto a 100-ml Q Sepharose high-performance (Amersham Pharmacia Biotech) column. Proteins were eluted with a linear gradient from 0 to 0.5 M NaCl in 20 mM bis-Tris, pH 6.5, over 2,000 ml at a flow rate of 5 ml min−1. Active fractions from each purification were pooled and concentrated into 4 ml of 1 mM Tris-HCl, pH 7.5, by ultrafiltration as described above. Each concentrated enzyme was mixed with 4 ml of storage buffer (100 mM Tris-HCl [pH 7.4], 200 mM NaCl, 0.2 mM EDTA, 1 mM dithiothreitol, 2% Triton X-100)-8 ml of glycerol and stored at −20°C. At each purification step, β-1,3-glucanase activity relative to total protein was determined (by measuring A280 and converting to milligrams of protein by using the calculated extinction coefficient of each protein: GluA, 0.336 mg ml−1 A280 unit−1; GluB, 1.008 mg ml−1 A280 unit−1; GluC, 0.332 mg ml−1 A280 unit−1 [1-cm path length]).
Biochemical analyses of recombinant enzymes.
The optimal pH for recombinant β-1,3-glucanase activity was determined by diluting purified enzymes 1:100 in 10 μl of 50 mM sodium acetate (pH 4.0 to 5.5), 50 mM bis-Tris (pH 6.0 to 7.0), 50 mM Tris-HCl (pH 7.5 to 9.0), or 50 mM glycine (pH 10.0) and measuring reducing sugars produced from laminarin after 30 min at 37°C. Subsequent experiments were performed with enzymes diluted 1:100 in 50 mM sodium acetate (pH 4.5). The optimal reaction temperature for recombinant β-1,3-glucanase activity was determined by measuring reducing sugars produced from laminarin by 10 μl of diluted enzyme after 30 min at 25 to 45°C. The substrate range of recombinant β-1,3-glucanase activity was determined by measuring reducing sugars produced after incubating 10 μl of diluted enzyme with laminarin (soluble β-1,3 glucan), curdlan (insoluble β-1,3 glucan), pachyman (insoluble β-1,3 glucan), zymosan A (insoluble Saccharomyces cerevisiae cell walls), amylose (soluble α-1,4 glucan), or carboxymethylcellulose (soluble β-1,4 glucan) for 30 min at 45 (GluA and GluC) or 41°C (GluB).
Nucleotide sequence accession numbers.
Nucleotide sequence data were deposited in the GenBank database under accession numbers AY157838 (gluA), AY157839 (gluB), and AY157840 (gluC).
RESULTS
Partial purification of β-1,3-glucanase activities.
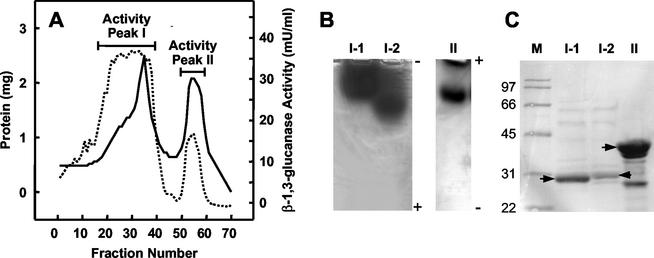
To characterize β-1,3-glucanase activity produced by L. enzymogenes strain N4-7, culture filtrates were subjected to hydrophobic interaction chromatography, which resulted in the separation of two peaks of β-1,3-glucanase activity (Fig. 1A). Further purification of activity peak I by anion-exchange chromatography resulted in the separation of two active fractions (I-1 and I-2) that resolved differently by native PAGE running toward the anode (Fig. 1B). In contrast, β-1,3-glucanase activity in peak II resolved as a single band on native gels run toward the cathode (Fig. 1B). SDS-PAGE analysis of fractions I-1, I-2, and II showed the sizes of the major proteins in the active fractions to be approximately 30, 31, and 43 kDa, respectively (Fig. 1C), which corresponded to similar-size fragments of activity bands cut from native gels run on SDS-PAGE gel (data not shown).
FIG. 1.
Partial purification of β-1,3-glucanase activities from culture filtrate of strain N4-7. (A) Extracellular proteins from strain N4-7 were resolved by phenyl-Sepharose chromatography (solid line), and fractions were assayed for β-1,3-glucanase activity (dotted line). (B) β-1,3-Glucanase activities purified from phenyl-Sepharose activity peaks I and II. Lanes I-1 and I-2, anodic native PAGE of active fractions following separation of peak I by anion exchange chromatography; lane II, cathodic native PAGE of peak II. (C) SDS-PAGE of β-1,3-glucanase-active fractions after chromatographic separations. Lane M, protein molecular mass standards in kilodaltons. All other lanes correspond to those in panel B. Arrows, major bands extracted for protein sequencing.
Identification of the 43-kDa β-1,3-glucanase-encoding ORF.
The partial amino acid sequence of one fragment of the trypsin-digested 43-kDa protein obtained from activity peak II was determined to be FNLALLNASGQNTAY. This sequence was used to design oligonucleotide probe glu43 by reverse translation. Three overlapping cosmid clones were identified from a genomic library of strain N4-7 by using glu43 as a probe in Southern hybridizations. The hybridizing region was localized to a 2.0-kb NotI fragment on cosmid pNG6, which was subcloned into pBluescript SK(+) to yield pNG602. Nucleotide sequence analysis of this fragment revealed a 1,188-bp ORF, designated gluB. The deduced 395-amino-acid protein encoded by gluB contained the exact 15-amino-acid sequence obtained for the 43-kDa protein beginning at residue 32. A signal peptide sequence of 26 residues at the N terminus of the predicted gluB gene product was detected. The size of the mature protein is predicted to be 39.8 kDa, with a pI of 8.14. A 23-bp G+C-rich sequence that includes a 10-bp inverted repeat indicative of a transcription termination signal was identified immediately downstream of the ORF.
Identification of gluA and gluC ORFs.
Amino acid sequencing of fragments obtained by trypsin digestion of the 30- and 31-kDa proteins (activity peaks I-1 and I-2, respectively) yielded the identical sequence, TYGTIHWR, suggesting that the two proteins were translational products of the same gene or two separate genes that had high sequence similarity to each other. The oligonucleotide probe glu31, constructed by reverse translation of the internal amino acid sequence, was used as a probe in a Southern hybridization to N4-7 genomic DNA, which revealed two hybridizing EcoRI fragments of 0.5 and 1.3 kb.
The screening of the N4-7 genomic library identified two overlapping cosmid clones that contained a 1.3-kb EcoRI hybridizing fragment. This fragment was localized on a 3.2-kb PstI fragment contained in cosmid pNG9, which was subcloned into pBluescript SK(+) to give plasmid pNG96. Sequence analysis of the fragment indicated the presence of a 1,152-bp ORF, which was designated gluC. The deduced amino acid sequence encoded by gluC revealed the presence of the sequence TYGTIHWR, beginning at residue 150; this sequence was identical to the internal amino acid sequences obtained from isolated proteins I-1 and I-2.
Three additional overlapping cosmid clones that contained a 0.5-kb EcoRI hybridizing fragment were identified. The hybridizing fragment was localized to a 3.2-kb PstI fragment on cosmid pNG2. This fragment, unrelated to the similar-size fragment containing gluC, was subcloned into pBluescript SK(+), yielding pNG21. Sequence analysis of this fragment detected a 765-bp ORF, designated gluA. The deduced amino acid sequence encoded by gluA revealed the sequence SYGTIHWR, beginning at residue 150, which differed from the sequence used to generate the glu31 oligonucleotide probe only by the first residue.
Similar to what was found for GluB, signal peptide sequences 22 residues in length were found at the beginning of the deduced translation products for GluA and GluC. The molecular mass of the mature GluA protein was predicted as 26.9 kDa, with a pI of 6.71, compared to a predicted molecular mass of 40.1 kDa, with a pI of 4.98, for the mature GluC protein. Stem-loop structures downstream of both ORFs were identified. For gluA, the stem-loop consisted of a 35-bp G+C-rich sequence including a 15-bp inverted repeat located 17-bp downstream from the stop codon. A 30-bp G+C-rich sequence including a 13-bp inverted repeat was identified 85 bp downstream of the gluC ORF.
Structural organization and phylogenetic analysis of glu genes.
Searches of databases revealed no conserved domains within the gluB gene product; however, a BLAST database search of the gluB gene product indicated that it has significant similarity (27 to 32% identical over the entire gene product in pairwise comparisons) to β-1,3-glucanases comprising family 64 glycoside hydrolases. Phylogenetic analysis indicated that GluB forms a distinct clade with putative glycoside hydrolases originating from Streptomyces coelicolor and a β-1,3-glucanase from Streptomyces matensis and is more distantly related to β-1,3-glucanases from Arthrobacter sp. and Oerskovia xanthineolytica (Fig. 2).
Analysis of the predicted proteins for gluA and gluC gene products indicated that the two have significant sequence identity (86% by pairwise analysis), through GluA in its entirety and through the first 251 residues of GluC. An RPS-BLAST search of the CDD revealed that both proteins contain catalytic domains between residues 46 and 249 belonging to family 16 glycoside hydrolases (Fig. 3). The sequence EIDIME, which includes the catalytic glutamate residues of the active site conserved within family 16 glycoside hydrolases (14, 15, 19, 26) was identified between residues 137 to 142 for both proteins. An RPS-BLAST search of GluC revealed that, in addition to the catalytic domain, there was a C-terminal region between residues 263 and 382 with sequence similarity to family 6 carbohydrate-binding module (CBM6) domains (Fig. 3). The two domains are separated by a short sequence rich in Gly, Pro, Ser, and Thr residues, which typically comprise domain linker regions (36).
Phylogenetic analyses of GluA and GluC sequences indicate that the catalytic domains form a clade with the α-subunit of coagulation factor G, which contains a glucan-binding domain from the horseshoe crab, Tachypleus tridentatus (30), as well as with a keratan sulfate endo-1,4-β-galactosidase from “Flavobacterium keratolyticus,” which has demonstrated broad activity toward substrates with β-1,3 linkages (22) (Fig. 3). These proteins cluster within a much larger clade, which is formed with other bacterial proteins with enzymatic activities toward β-1,3-glycan linkages. In contrast to those for the catalytic domain, sequences related to CBM6 of GluC occur in a variety of hydrolytic enzymes that originate from various organisms (Fig. 3). The CBM6 of GluC clusters in a clade with two domains found in the glucan-binding α-subunit of coagulation factor G from T. tridentatus (30), as well as a cellulase originating from Cellvibrio mixtus and a chitinase and cellulase from two Pseudomonas spp. (Fig. 3).
Expression of recombinant β-1,3-glucanase-active proteins in E. coli.
β-1,3-Glucanase activity from gluA, gluB, or gluC was not detected in cosmid library clones (pNG2, pNG6, and pNG9, respectively) or subclones (pNG21, pNG602, and pNG94, respectively). Therefore, translational fusions to the predicted mature protein encoded by each gene were constructed with the expression vector pBAD/myc-HisA or pET3C as described in Materials and Methods, resulting in detectable activity in E. coli lysates.
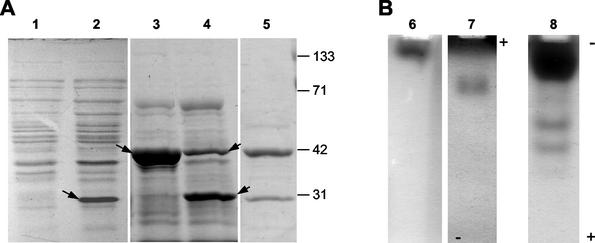
Expression of gluA in E. coli BL21(DE3) harboring pNG2503 resulted in the production of a predominant protein of approximately 31 kDa in crude cell lysates (Fig. 4A, lane 2); this protein showed β-1,3-glucanase activity when separated by cathodic native PAGE at pH 7.4 (Fig. 4B, lane 6). This activity was not observed during initial characterization of L. enzymogenes culture filtrates but was later detected in culture filtrates when native gels were run at the higher pH (data not shown). The majority of the overexpressed GluA protein in IPTG-induced cultures was produced in the form of inactive inclusion bodies, which hampered purification of recombinant GluA to homogeneity. Nevertheless, sufficient amounts of active GluA were recovered following hydrophobic interaction chromatography for further analysis (Table 3).
FIG. 4.
SDS-PAGE (A) and β-1,3-glucanase activity on native PAGE gels (B) of recombinant glucanases from E. coli lysates. Lanes: 1, pET3C vector control (crude lysate); 2 and 6, GluA from pNG2503 (crude lysate); 3 and 7, GluB from pNG6027 (phenyl-Sepharose-concentrated lysate); 4 and 8, GluC from pNG9615 (phenyl-Sepharose-concentrated lysate); 5, L. enzymogenes culture filtrate. Arrows, prominent proteins in lysates.
TABLE 3.
Summary of purification of recombinant β-1,3-glucanase-active proteins
| Purification step | Total protein (mg) | Total activity (U) | Sp act (U/mg) | Purification (fold) | Yield (%) |
|---|---|---|---|---|---|
| GluA | |||||
| Culture filtrate | 420 | 39 | 0.09 | 1 | 100 |
| Phenyl-Sepharose | 1.2 | 10 | 8.3 | 92 | 26 |
| GluB | |||||
| Culture filtrate | 1,300 | 53 | 0.04 | 1 | 100 |
| Phenyl-Sepharose | 30 | 20 | 0.7 | 17 | 38 |
| SP Sepharose | 5.9 | 19 | 3.2 | 80 | 36 |
| GluC | |||||
| Culture filtrate | 380 | 46 | 0.12 | 1 | 100 |
| Phenyl-Sepharose | 19 | 41 | 2.2 | 18 | 89 |
| Q Sepharose | 0.13 | 9 | 69 | 575 | 20 |
Expression of gluB in E. coli TOP10 harboring pNG6027 and concentration of cell lysate by using phenyl-Sepharose resulted in cell lysates containing a predominant protein of approximately 41 kDa (Fig. 4A, lane 3) and β-1,3-glucanase activity on cathodic native PAGE gel (Fig. 4B, lane 7). Both the size of the predominant protein and the native mobility of the glucanase activity corresponded closely with those of activity peak II (Fig. 1), isolated from L. enzymogenes culture filtrate. Fractionation of activity by hydrophobic interaction and cation-exchange chromatography resulted in 80-fold purification of GluB (Table 3).
Expression of gluC in E. coli TOP10 harboring pNG9615 and concentration of cell lysate using phenyl-Sepharose resulted in multiple β-1,3-glucanase activities on native PAGE gel (Fig. 4B, lane 8), suggesting the occurrence of posttranslational processing. SDS-PAGE analysis of the cell lysate indicated the presence of a predominant 31-kDa protein (Fig. 4A, lane 4) similar in size to the predominant proteins in activity peak I from culture filtrate of L. enzymogenes (Fig. 1C). The 31-kDa protein size is substantially smaller than the predicted size for the mature gluC gene product. However, a protein of 42 kDa, close to the predicted size of 40 kDa, was also detected in the lysate, further supporting the possibility that posttranslational processing of the gluC gene product had occurred. Recombinant GluC activity was fractionated by hydrophobic interaction and anion-exchange chromatography, resulting in 575-fold purification relative to culture lysates (Table 3).
Biochemical characteristics of recombinant β-1,3-glucanase activities.
Laminarin hydrolysis was used to determine relative optimal β-1,3-glucanase activity for each enzyme in terms of reaction pH and temperature. Activity was highest for all three enzymes in the range of pH 4.0 to 7.0 (data not shown). Activity of both GluA and GluC was optimal at pH 4.5; activity of GluB was optimal between pH 4.5 and 5.0. At pH 4.5, the temperature optima for β-1,3-glucanase activities of GluA and GluC were both found to be approximately 45°C, and that for GluB was approximately 41°C (data not shown).
The activity ranges of recombinant β-1,3-glucanases against different glucan substrates relative to activity against laminarin were determined (Table 4). GluB was more active against zymosan A, a commercial preparation of partially purified yeast cell walls containing branched glucan chains, than linear β-1,3 glucans. Among linear-chain substrates, soluble laminarin was hydrolyzed by GluB more effectively than the insoluble β-1,3 glucans curdlan and pachyman. In contrast, GluA and GluC were more active against the linear β-1,3 glucans curdlan and laminarin than against zymosan A. The reason for the relatively low activity of all three enzymes against pachyman is unclear, but it may be due to poor dispersal of the substrate in the assay tubes or to the presence of β-1,6-linked branches found within molecules of the substrate (21), which prevents accessibility of cleavable β-1,3 linkages. GluA, GluB, and GluC did not show appreciable levels of activity toward the substrates carboxymethylcellulose (β-1,4-glucan) or amylose (α-1,4-glucan), further supporting the hypothesis that these enzymes act primarily as β-1,3-glucanases and are separate from other enzymatic activities expressed by L. enzymogenes (4).
TABLE 4.
Substrate specificity of recombinant β-1,3-glucanase-active proteins
| Substrate | Main linkage and properties | Relative activity (%)a of:
|
||
|---|---|---|---|---|
| GluAb | GluBc | GluCd | ||
| Laminarin | β-1,3; linear, soluble | 100 ± 4.1 | 100 ± 5.3 | 100 ± 7.9 |
| Curdlan | β-1,3; linear, insoluble | 114 ± 7.3 | 56 ± 2.3 | 132 ± 0.7 |
| Pachyman | β-1,3; with β-1,6 branches, insoluble | 14 ± 0.3 | 29 ± 1.3 | 17 ± 1.0 |
| Zymosan A | β-1,3; yeast cell walls, insoluble | 47 ± 3.1 | 148 ± 1.4 | 62 ± 3.6 |
| CM-cellulosee | β-1,4; linear, soluble | 3.9 ± 4.6 | 3.2 ± 2.5 | 2.6 ± 1.0 |
| Amylose | α-1,4; linear, soluble | 1.6 ± 0.7 | 1.5 ± 2.5 | 1.4 ± 2.9 |
Activity ± standard error of triplicate samples relative to laminarin.
100% activity = 6.8 U of GluA protein mg−1.
100% activity = 2.1 U of GluB protein mg−1.
100% activity = 55 U of GluC protein mg−1.
CM-cellulose, carboxymethyl cellulose.
DISCUSSION
Our studies indicate that three genes in L. enzymogenes strain N4-7 encode β-1,3-glucanase activities belonging to two different families of glycoside hydrolases. Analysis of the gluB gene showed that it encodes a protein product consisting of a single catalytic domain related to those of family 64 glycoside hydrolases (Fig. 2), a family which currently consists of enzymes originating from fungal and prokaryotic origin (see the Carbohydrate-Active Enzymes server: http://afmb.cnrs-mrs.fr/∼cazy/CAZY/index.html). Currently, there are relatively few enzymes belonging to family 64 that have been characterized at the molecular and biochemical levels; GluB represents the only member of prokaryotic origin with known β-1,3-glucanase activity that does not originate from Actinomycetales. In contrast, both gluA and gluC encode proteins whose catalytic domains have 86% sequence identity and that are related to family 16 glycoside hydrolases (Fig. 3). Family 16 comprises β-1,3-glucanases, β-1,3-1,4-glucanases, lichenases, xyloglucan endotransferases, κ-carrageenases, and agarases, originating from bacteria, fungi, and plants. The predicted amino acid sequence of GluC contains a C-terminal region similar to the CBM6 domain present in a number of bacterial enzymes, including β-1,3-glucanases, β-1,3-1,4-glucanases, chitinases, and xylanases (Carbohydrate-Active Enzymes server). The diversity observed among enzyme type and source organisms is a signature indicative of domain shuffling in the evolution of glycoside hydrolases (11).
Expression of gluC in both L. enzymogenes and E. coli resulted in the production of multiple β-1,3-glucanase-active species. Initial purification of 30- and 31-kDa proteins from L. enzymogenes culture filtrate, both containing the same internal amino acid sequence, suggested that the full-length 40.1-kDa product of gluC is posttranslationally processed into biochemically distinct species during expression in L. enzymogenes (Fig. 1). Similarly, expression of gluC in E. coli resulted in the production of proteins of full-length and processed sizes, correlated with multiple β-1,3-glucanase activities (Fig. 4). The processing of GluC may occur by an autocatalytic process or by a process common to both bacteria. Conversely, the smaller forms of GluC may simply be stable degradation products of the full-length protein. Our results indicate that these smaller forms of GluC are likely missing the CBM6 domain. It is possible that the intact 40.1-kDa GluC protein is produced by L. enzymogenes under other environmental conditions and may contribute to greater activity differences between GluA and GluC. For example, Hong et al. (17) recently showed that a CBM6 domain from a β-1,3-glucanase from Streptomyces sioyaensis binds to various insoluble β-1,3-glucans and likely enhances the enzymatic activity toward these substrates.
Based on analysis of the cloned gluA gene and recombinant enzyme, it is likely that, in its initial isolation from L. enzymogenes filtrate, GluA copurified with GluC during hydrophobic interaction chromatography but was not detected on native gels run under the conditions described. Subsequent experiments with native gels at pH 7.4 rather than 6.6 resulted in detection of β-1,3-glucanase activity corresponding to GluA from both L. enzymogenes and E. coli (Fig. 4).
Recombinant GluB demonstrated higher relative activity toward the branched-chain β-1,3 glucan substrate zymosan A than toward linear β-1,3 glucan substrates. In contrast, recombinant GluA and GluC both demonstrated higher relative activity toward the linear β-1,3 glucan substrates curdlan and laminarin than toward zymosan A. From these observations, we speculate that GluB may function in an early step in the hydrolysis of complex β-1,3 glucans present in fungal cell walls, liberating shorter oligosaccharides to serve as substrates for the GluA and GluC β-1,3-glucanases secreted by strain N4-7. The activities of all three enzymes were optimal around pH 4.5 to 5.0, lower than pH optima reported for most other β-1,3-glucanases of prokaryotic origin (see, e.g., references 2, 5, 9, 13, 27, and 37). This is consistent with acidic environments surrounding fungal hyphae (12), the suspected site of action for those enzymes produced by L. enzymogenes.
Among prokaryotes known to produce multiple β-1,3-glucanase systems, L. enzymogenes has similarities to Oerskovia xanthineolytica (recently reclassified as Cellulosimicrobium cellulans) (29) and Streptomyces coelicolor. All three species contain genomes of relatively high G+C content, are soil-borne microbes that demonstrate antagonistic properties toward fungi, and contain both family 64 and family 16 enzymes. Similarities between the β-1,3-glucanase systems of O. xanthineolytica and L. enzymogenes extend to structural organization of the enzymes. GluC and GluA are structurally organized similarly to the βglII and βglIIA isoenzymes from O. xanthineolytica strain LL G109; βglII and βglIIA have significant amino acid identity throughout their family 16 catalytic domains, and βglII contains a substrate-binding domain located at the C terminal that is lacking in βglIIA (8). Furthermore, a family 64 β-1,3-glucanase from O. xanthineolytica strain 73-14 has been characterized (31). In contrast to β-1,3-glucanases from L. enzymogenes, both the family 64 enzyme and βglII from O. xanthineolytica contain ricin-type binding domains (8, 31), which have been demonstrated to be instrumental for yeast cell lysis activity of enzymes (32, 33).
The role of bacterial β-1,3-glucanases in antagonism and biological control of fungal plant pathogens has been poorly defined in comparison to that of bacterial chitinases. Fridlender et al. (10) demonstrated biocontrol of Rhizoctonia solani by an isolate of Burkholderia cepacia and showed hyphal damage presumed to be due solely to the production of β-1,3-glucanase activity. Lim et al. (23) showed that growth inhibition and cell wall lysis of Fusarium solani by Pseudomonas stutzeri was caused by a combination of extracellular β-1,3-glucanase and chitinase activities. However, these studies offer limited comparisons to L. enzymogenes, in that the number and types of β-1,3-glucanase activities produced by these bacteria have not been shown and biochemical and genetic analyses of individual β-1,3-glucanases have not been performed. To understand the relationship of extracellular enzyme production, lytic activity, and the biocontrol phenotype of L. enzymogenes, it became important to define the components of the extracellular enzyme complement of L. enzymogenes strain N4-7. Results presented in this study are intended as a first step in the elucidation of the roles of individual β-1,3-glucanase-active proteins in the biocontrol activity of strain N4-7. The identification of each β-1,3-glucanase activity-encoding gene in strain N4-7 will be instrumental in determining the overall contribution of each enzyme to biocontrol by means of site-directed mutagenesis for the sequential inactivation of each gene. Furthermore, additional analysis of the action patterns of substrate hydrolysis by individual enzymes and combinations of enzymes will aid in establishing synergistic effects of β-1,3-glucanase activities and mechanisms involved in fungal biocontrol by these and related bacteria.
Acknowledgments
This work was supported in part with funding by EcoSoil Systems, Inc., and The New Jersey Agricultural Experiment Station.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aono, R., M. Hammura, M. Yamamoto, and T. Asano. 1995. Isolation of extracellular 28- and 42-kilodalton β-1,3-glucanases and comparison of three β-1,3-glucanases produced by Bacillus circulans IAM1165. Appl. Environ. Microbiol. 61:122-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bielecki, S., and E. Galas. 1991. Microbial β-glucanases different from cellulases. Crit. Rev. Biotechnol. 10:275-304. [DOI] [PubMed] [Google Scholar]
- 4.Christensen, P., and F. D. Cook. 1978. Lysobacter, a new genus of nonfruiting, gliding bacteria with high base ratio. Int. J. Syst. Bacteriol. 28:367-393. [Google Scholar]
- 5.Doi, K., and A. Doi. 1986. Cloning and expression in Escherichia coli of the gene for an Arthrobacter β-1,3-glucanase. J. Bacteriol. 168:1272-1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dygert, S., L. H. Li, D. Florida, and J. A. Thoma. 1965. Determination of reducing sugar with improved precision. Anal. Biochem. 13:367-374. [DOI] [PubMed] [Google Scholar]
- 7.Epstein, D. M., and P. C. Wensink. 1988. The α-lytic protease gene of Lysobacter enzymogenes. J. Biol. Chem. 263:16586-16590. [PubMed] [Google Scholar]
- 8.Ferrer, P., T. Halkier, L. Hedegaard, D. Savva, I. Diers, and J. A. Asenjo. 1996. Nucleotide sequence of a β-1,3-glucanase isoenzyme IIA gene of Oerskovia xanthineolytica LLG109 (Cellulomonas cellulans) and initial characterization of the recombinant enzyme expressed in Bacillus subtilis. J. Bacteriol. 178:4751-4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fiske, M. J., K. L. Tobey-Fincher, and R. L. Fuchs. 1990. Cloning of two genes from Bacillus circulans WL-12 which encode 1,3-β-glucanase activity. J. Gen. Microbiol. 136:2377-2383. [DOI] [PubMed] [Google Scholar]
- 10.Fridlender, M., J. Inbar, and I. Chet. 1993. Biological control of soilborne plant pathogens by a β-1,3 glucanase-producing Pseudomonas cepacia. Soil Biol. Biochem. 25:1211-1221. [Google Scholar]
- 11.Gilkes, N. R., B. Henrissat, D. G. Kilburn, R. C. Miller, Jr., and R. A. J. Warren. 1991. Domains in microbial β-1,4-glycanases: sequence conservation, function and enzyme families. Microbiol. Rev. 55:303-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Griffen, D. H. 1994. Fungal physiology, 2nd ed. Wiley-Liss, New York, N.Y.
- 13.Gueguen, Y., W. G. B. Voorhorst, J. van der Oost, and W. M. de Vos. 1997. Molecular and biochemical characterization of an endo-β-1,3-glucanase of the hyperthermophilic archaeon Pyrococcus furiosus. J. Biol. Chem. 272:31258-31264. [DOI] [PubMed] [Google Scholar]
- 14.Hahn, M., O. Olsen, O. Politz, R. Borriss, and U. Heinemann. 1995. Crystal structure and site-directed mutagenesis of Bacillus macerans endo-1,3-1,4-β-glucanase. J. Biol. Chem. 270:3081-3088. [DOI] [PubMed] [Google Scholar]
- 15.Hoj, P. B., R. Condron, J. C. Traeger, J. C. McAuliffe, and B. A. Stone. 1992. Identification of glutamic acid 105 at the active site of Bacillus amyloliquefaciens 1,3-1,4-β-D-glucan 4-glucanohydrolase using epoxide-based inhibitors. J. Biol. Chem. 267:25059-25066. [PubMed] [Google Scholar]
- 16.Holtman, M. A. 1998. Ph.D. thesis. Rutgers, The State University of New Jersey, New Brunswick.
- 17.Hong, T.-Y., C.-W. Cheng, J.-W. Huang, and M. Meng. 2002. Isolation and biochemical characterization of an endo-1,3-β-glucanase from Streptomyces sioyaensis containing a C-terminal family 6 carbohydrate-binding module that binds to 1,3-β-glucan. Microbiology 148:1151-1159. [DOI] [PubMed] [Google Scholar]
- 18.Jones, D. T., W. R. Taylor, and J. M. Thornton. 1992. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8:275-282. [DOI] [PubMed] [Google Scholar]
- 19.Juncosa, M., J. Pons, T. Dot, E. Querol, and A. Planas. 1994. Identification of active site carboxylic residues in Bacillus licheniformis 1,3-1,4-β-D-glucan 4-glucanohydrolase by site-directed mutagenesis. J. Biol. Chem. 269:14530-14535. [PubMed] [Google Scholar]
- 20.Kobayashi, D. Y., and N. E.-H. El-Barrad. 1996. Selection of bacterial antagonists using enrichment cultures for the control of summer patch disease in Kentucky bluegrass. Curr. Microbiol. 32:106-110. [Google Scholar]
- 21.Lee, I.-Y. 2002. Curdlan, p. 135-154. In E. Vandamme, S. De Baets, and A. Steinbüchel (ed.), Biopolymers, vol. 5. Wiley, Hoboken, N.J.
- 22.Leng, L., A. Zhu, Z. Zhang, R. Hurst, and J. Goldstein. 1998. Cloning, functional expression and purification of endo-β-galactosidase from Flavobacterium keratolyticus. Gene 222:187-194. [DOI] [PubMed] [Google Scholar]
- 23.Lim, H.-S., Y.-S. Kim, and S.-D. Kim. 1991. Pseudomonas stutzeri YPL-1 genetic transformation and antifungal mechanism against Fusarium solani, an agent of plant root rot. Appl. Environ. Microbiol. 57:510-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morgenstern, B. 1999. DIALIGN 2: improvement of the segment-to-segment approach to multiple sequence alignment. Bioinformatics 15:211-218. [DOI] [PubMed] [Google Scholar]
- 25.Pan, S.-Q., X.-S. Ye, and J. Kuc. 1989. Direct detection of β-1,3-glucanase isozymes on polyacrylamide electrophoresis and isoelectrofocusing gels. Anal. Biochem. 182:136-140. [DOI] [PubMed] [Google Scholar]
- 26.Planas, A., M. Juncosa, J. Lloberas, and E. Querol. 1992. Essential catalytic role of Glu134 in endo-β-1,3-1,4-D-glucan 4-glucanohydrolase from B. licheniformis as determined by site-directed mutagenesis. FEBS Lett. 308:141-145. [DOI] [PubMed] [Google Scholar]
- 27.Saeki, K., J. Iwata, S. Yamazaki, Y. Watanabe, and Y. Tamai. 1994. Purification and characterization of a yeast lytic β-1,3-glucanase from Oerskovia xanthineolytica TK-1. J. Ferment. Bioeng. 78:407-412. [Google Scholar]
- 28.Sakellaris, H., J. M. Pemberton, and J. M. Manners. 1990. Genes from Cellvibrio mixtus encoding β-1,3 endoglucanase. Appl. Environ. Microbiol. 56:3204-3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schumann, P., N. Weiss, and E. Stackerbrandt. 2001. Reclassification of Cellomonas cellulans (Stackebrandt and Keddie 1986) as Cellulosimicrobium cellulans gen. nov., comb. nov. Int. J. Syst. E vol. Microbiol. 51:1007-1010. [DOI] [PubMed] [Google Scholar]
- 30.Seki, N., T. Muta, T. Oda, D. Iwaki, K. Kuma, T. Miyata, and S. Iwanaga. 1994. Horseshoe crab (1,3)-β-D-glucan-sensitive coagulation factor G. J. Biol. Chem. 269:1370-1374. [PubMed] [Google Scholar]
- 31.Shen, S.-H., P. Chretien, L. Bastien, and S. N. Slilaty. 1991. Primary sequence of the glucanase gene from Oerskovia xanthineolytica. J. Biol. Chem. 266:1058-1063. [PubMed] [Google Scholar]
- 32.Shimoi, H., Y. Iimura, T. Obata, and M. Tadenuma. 1992. Molecular structure of Rarobacter faecitabidus protease I. J. Biol. Chem. 267:25189-25195. [PubMed] [Google Scholar]
- 33.Silen, J. L., C. N. McGrath, K. R. Smith, and D. A. Agard. 1988. Molecular analysis of the gene encoding lytic protease: evidence for a preproenzyme. Gene 69:237-244. [DOI] [PubMed] [Google Scholar]
- 34.Staskawicz, B., D. Dahlbeck, N. Keen, and C. Napoli. 1987. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J. Bacteriol. 169:5789-5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sullivan, R. F., M. A. Holtman, G. J. Zylstra, J. F. White, and D. Y. Kobayashi. 2003. Taxonomic positioning of two biological control agents for plant diseases as Lysobacter enzymogenes based on phylogenetic analysis of 16S rDNA, fatty acid composition and phenotypic characteristics. J. Appl. Microbiol. 94:1079-1086. [DOI] [PubMed] [Google Scholar]
- 36.Tomme, P., R. A. J. Warren, and N. R. Gilkes. 1995. Cellulose hydrolysis by bacteria and fungi. Adv. Microb. Physiol. 37:1-81. [DOI] [PubMed] [Google Scholar]
- 37.Ventom, A. M., and J. A. Asenjo. 1991. Characterization of yeast lytic enzymes from Oerskovia xanthineolytica LL-G109. Enzyme Microb. Technol. 13:71-75. [Google Scholar]
- 38.Vieira, J., and J. Messing. 1987. Production of single-stranded plasmid DNA. Methods Enzymol. 153:3-11. [DOI] [PubMed] [Google Scholar]
- 39.Warren, R. A. J. 1996. Microbial hydrolysis of polysaccharides. Annu. Rev. Microbiol. 50:183-212. [DOI] [PubMed] [Google Scholar]
- 40.Wright, D. S., L. D. Graham, and P. A. Jennings. 1998. Cloning of a Lysobacter enzymogenes gene that encodes an arginyl endopeptidase (endoproteinase Arg-C). Biochim. Biophys. Acta 1443:369-374. [DOI] [PubMed] [Google Scholar]
- 41.Yahata, N., T. Watanabe, Y. Nakamura, Y. Yamamoto, S. Kamimiya, and H. Tanaka. 1990. Structure of the gene encoding β-1,3-glucanase A1 of Bacillus circulans WL-12. Gene 86:113-117. [DOI] [PubMed] [Google Scholar]
- 42.Zheng, Y., and C. A. Wozniak. 1997. Adaptation of a β-1,3-glucanase assay to microplate format. BioTechniques 22:922-926. [DOI] [PubMed] [Google Scholar]