RNA polymerase II (RNAPII) is the enzyme responsible for synthesis of all mRNA in higher cells. As the central component of the eukaryotic transcription machinery, RNAPII is the final target of regulatory pathways that are ultimately responsible for cellular development, differentiation, and metabolic control. Publication of the high-resolution structure of yeast RNAPII (1, 2) and of a transcribing RNAPII/DNA/RNA complex (3) identified key structural elements central to many of the enzyme's functions and suggested mechanisms for RNA chain elongation and translocation of the polymerase along DNA (2). Although a possible mechanism for interaction of RNAPII with promoter DNA was also proposed, absent from the x-ray structures were subunits Rpb4 and Rpb7, which form a dissociable complex in yeast and are required for initiation.
The problem posed for crystallization of the complete initiation-competent RNAPII by the presence of substoichiometric amounts of the Rpb4/Rpb7 complex has now been overcome, and reports by Armache et al. (4) and Bushnell and Kornberg (5) in this issue of PNAS describe the structure of RNAPII, including all 12 component subunits. The x-ray structures of the initiation-competent RNAPII are in agreement with each other and with a previously published 18-Å resolution structure calculated by cryoelectron microscopy (cryo-EM) and image analysis of single RNAPII particles (6). Although the cryo-EM structure of the 12-subunit RNAPII was correct, the proposed mode of interaction between the Rpb4/Rpb7 heterodimer and the 10-subunit (core) RNAPII [based on fitting the x-ray structure of the Archaeal homolog of the eukaryoytic Rpb4/Rpb7 complex (7) to the EM reconstruction] was flawed. A correct fit of the Archaeal Rpb4/Rpb7 homolog to the x-ray structures of the complete RNAPII confirms the location of the Rpb4/Rpb7 heterodimer with respect to the core RNAPII and establishes that contact of the heterodimer with the core RNAPII is mediated by Rpb7. The significance of the structure of the complete initiation-competent RNAPII goes well beyond determining the mode of interaction between the Rpb4/Rpb7 heterodimer and the core RNAPII. In fact, the most interesting information derived from the x-ray and cryo-EM structures regards the conformation of RNAPII and its implications for the initiation mechanism (Fig. 1).
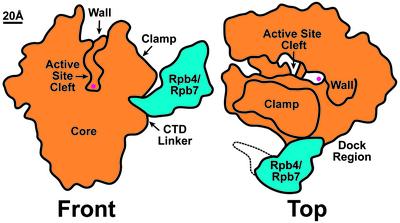
Fig. 1.
Structure of the 12-subunit initiation-competent RNAPII. Two schematic views show the core (10-subunit) enzyme and its mode of interaction with the dissociable Rpb4/Rpb7 complex, which is required for transcription initiation. (The dotted line marks the position of partially ordered density localized in the cryo-EM reconstruction but not in the x-ray map.) The presence of Rpb4/Rpb7 locks the mobile “clamp” domain in a conformation that results in a narrow active site cleft, blocked at the upstream end by the “wall” domain. This arrangement of the cleft effectively prevents double-stranded promoter DNA from reaching the active site (marked by the red circle). The DNA strands separate when the transcription bubble is formed, allowing the template strand to reach the active site. The Rpb4/Rpb7 heterodimer is likely to play additional roles during initiation, because it extends the effective area of the “dock” region involved in interaction with additional components of the preinitiation complex.
The x-ray structure of the core RNAPII revealed that the active site of the enzyme is located at the bottom of a deep cleft, blocked at the upstream end by a domain appropriately named the “wall.” One side of the active site cleft is formed by a domain determined to adopt different conformations in two crystal forms of polymerase (1, 2) and in the first 3D structure of core RNAPII calculated by 2D electron crystallography (8). This mobile domain, named the “clamp,” was proposed to control access by double-stranded promoter DNA to the active site. The clamp would adopt the open conformation detected in one of the 3D crystal forms (2) to allow straight double-stranded promoter DNA to lie along the bottom of the active site cleft, clear the wall, and reach the RNAPII active site. In a subsequent step of the initiation mechanism, the clamp would switch to a closed state and effectively “clamp” the DNA in place, thereby explaining the processivity of the polymerase.
The mechanism just described was based on structural characterization of the core RNAPII, a form of the enzyme defective for initiation. Therefore, it seemed appropriate to examine the conformation of the clamp and its role in controlling access of DNA to the active site by using a technique that did not require crystallization of the enzyme. Cryo-EM analysis of the complete initiation-competent RNAPII revealed that in single RNAPII particles in solution, the position of the clamp varies between closed and collapsed conformations, which are separated only by a small energy barrier (6). That the clamp does not adopt an open conformation in the complete RNAPII led to the seemingly controversial proposition that double-stranded DNA would never reach the bottom of the active site cleft, but rather that formation of the transcription bubble mediated by the action of general transcription factor TFIIH would allow DNA to “melt” into the cleft and reach the active site (6).
The x-ray structure of the initiation-competent RNAPII now makes it apparent that the clamp cannot swing open when the Rpb4/Rpb7 heterodimer is present. The N-terminal domain of Rpb7 acts as a wedge that, when inserted in a conserved pocket formed by the combined contribution of RNAPII subunits Rpb1, Rpb2, and Rpb6, locks the clamp in a conformation that most closely resembles that observed in the transcribing RNAPII/DNA/RNA complex (3). Five flexible domains that connect the clamp to the remainder of the polymerase, named the “switches,” are not disordered as they were in the structure of core RNAPII but well ordered, as observed for the transcribing RNAPII complex. As discussed by Armache et al. (4), the ordered switches might in fact form an extended binding site for the template DNA strand.
If promoter binding and initiation occur with a closed clamp, as is now clear, how does the template DNA strand reach the active site of the polymerase? Armache et al. (4) contemplate a scenario in which Rpb4/Rpb7 must dissociate from the enzyme to allow the clamp to open and allow entry of double-stranded DNA into a wide active site cleft and are later recruited to complete the initiation process after promoter binding. However, as pointed out by the authors, this would conflict with the observation that Rpb4/Rpb7 homologs in RNAPIIs from most other organisms are not known to dissociate from the enzymes. A similar dilemma has come up in the field of bacterial transcription, where the x-ray structure of the core polymerase shows the clamp in an open conformation (9), but the structure of the RNA polymerase holoenzyme, the form of the enzyme relevant for initiation that includes the core polymerase plus the σ factor, shows a closed clamp (10, 11). It now seems clear that, in both bacteria and eukaryotes, the conformation of RNA polymerase that is relevant for initiation is the same as that observed in the transcribing yeast RNAPII/DNA/RNA complex, and that promoter melting must be required for the template DNA strand to reach the RNAPII active site. This possibility was considered by one of the groups reporting on the bacterial holoenzyme structure (11) and is supported by the x-ray structure of a bacterial holoenzyme/DNA complex, in which promoter DNA binds along the top of the clamp and the wall. A similar mode of interaction with promoter DNA was first suggested for RNAPII when the cryo-EM structure of the complete enzyme was reported (6), and the structures reported by Armache et al. (4) and Bushnell and Kornberg (5) lead to the same conclusion.
The conformation of RNAPII relevant for promoter binding and initiation is also the conformation of the enzyme that will interact with the general transcription factors to form the preinitiation complex (12). The approximate location for binding of transcription factors TBP and TFIIB to RNAPII can be anticipated based on the location of the RNAPII active site, the topology of eukaryotic promoters, and structural information about eukaryotic and bacterial polymerase/DNA complexes (3, 13–15). It encompasses the previously identified “dock” region on the upstream face of the RNAPII (2). As pointed out by both Armache et al. (4) and Bushnell and Kornberg (5), the position of the Rpb4/Rpb7 heterodimer increases the area of the dock region, thereby suggesting a role of Rpb4/Rpb7 in assembly of the preinitiation complex. Indeed, the role of Rpb4/Rpb7 in preinitiation complex assembly is likely to be even more significant. MS analysis of immunopurified yeast macromolecular complexes has indicated that Rpb7 interacts with Tfg1, the largest subunit of transcription factor IIF (16). The Rpb4/Rpb7 heterodimer might play a multifaceted role in initiation, locking the active site area of RNAPII into the right conformation and functioning as scaffolding for addition of further components of the preinitiation complex.
The structure of the preinitiation complex is more than the sum of the structure of its parts.
What other roles might the Rpb4/Rpb7 heterodimer play in the preinitiation complex? Biochemical studies of Rpb4/Rpb7 (17) and structural characterization of the corresponding Archaeal complex (7) pointed to the existence of single-stranded nucleic acid-binding sites in Rpb7. The cryo-EM reconstruction of the 12-subunit polymerase placed the Rpb4/Rpb7 complex near the proposed RNA exit groove in RNAPII and adjacent to the linker to the C-terminal domain (CTD) of the largest RNAPII subunit, known to be involved in recruitment of complexes involved in RNA processing (18–20). This location immediately suggested a role for Rpb4/Rpb7 in binding the newly polymerized RNA chain and directing it toward the RNA processing machinery (6). Analysis of the x-ray structure of the 12-subunit RNAPII has led to similar propositions. Unfortunately, whereas the x-ray structure now available has much higher resolution, it still does not clarify exactly how Rpb4 and Rpb7 would interact with the nascent RNA as it exits the active site cleft of RNAPII. Roles for Rpb4/Rpb7 in interaction with the Mediator complex (proposed by Bushnell and Kornberg in ref. 5) and in recruitment of Fcp1, the CTD phosphatase that, through its effect on the phosphorylation state of the CTD, functions in recycling of RNAPII, are interesting but remain speculative.
Comparison of the x-ray and cryo-EM structures of the 12-subunit RNAPII indicates that a significant portion of the Rpb4 density is partially disordered. This density gives rise to the extended structure that runs along the base of the clamp toward the downstream end of the RNAPII active site in the low-resolution cryo-EM reconstruction (see Fig. 1), and Bushnell and Kornberg (5) suggest that it might correspond to the central charged domain of Rpb4. Sequence analysis of several general transcription factors reveals the presence of charged, most likely unstructured domains, suggesting that components of the preinitiation complex are not rigid but instead might assemble like a puzzle formed by flexible pieces. The interfaces between components might in some cases be rather extended, as in the case of the interaction between the bacterial σ factor and core polymerase (10, 11), and some factors most likely adopt their functional conformations only when involved in synergetic interactions with other components of the preinitiation complex.
The structure of the complete initiation-competent RNAPII provides important information about the mechanism of initiation and adds to the evidence suggesting that the structure of the preinitiation complex amounts to more than the sum of the structure of its parts. Interactions between different components of the preinitiation complex might be essential to bring individual pieces into their functional conformations, which emphasizes the importance of generating structural information about the most inclusive complexes possible. Cryo-EM can be used to calculate low-resolution (10- to 25-Å) reconstructions of multicomponent assemblies under near-physiological conditions. High-resolution NMR and x-ray structures of individual pieces, assembled as suggested by the overall low-resolution structure of a complex, will then result in the structure of the entire assembly at high resolution, as required for a complete mechanistic understanding of the transcription process and its regulation.
References
- 1.Cramer, P., Bushnell, D. A., Fu, J., Gnatt, A. L., Maier-Davis, B., Thompson, N. E., Burgess, R. R., Edwards, A. M., David, P. R. & Kornberg, R. D. (2000) Science 288, 640–649. [DOI] [PubMed] [Google Scholar]
- 2.Cramer, P., Bushnell, D. A. & Kornberg, R. D. (2001) Science 292, 1863–1876. [DOI] [PubMed] [Google Scholar]
- 3.Gnatt, A. L., Cramer, P., Fu, J., Bushnell, D. A. & Kornberg, R. D. (2001) Science 292, 1876–1882. [DOI] [PubMed] [Google Scholar]
- 4.Armache, K.-J., Kettenberger, H. & Cramer, P. (2003) Proc. Natl. Acad. Sci. USA 100, 6964–6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bushnell, D. A. & Kornberg, R. D. (2003) Proc. Natl. Acad. Sci. USA 100, 6969–6973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Craighead, J., Chang, W. & Asturias, F. (2002) Structure (Cambridge, Mass.) 10, 1117–1125. [DOI] [PubMed] [Google Scholar]
- 7.Todone, F., Brick, P., Werner, F., Weinzierl, R. O. & Onesti, S. (2001) Mol. Cell 8, 1137–1143. [DOI] [PubMed] [Google Scholar]
- 8.Darst, S. A., Edwards, A. M., Kubalek, E. W. & Kornberg, R. D. (1991) Cell 66, 121–128. [DOI] [PubMed] [Google Scholar]
- 9.Zhang, G., Campbell, E. A., Minakhin, L., Richter, C., Severinov, K. & Darst, S. A. (1999) Cell 98, 811–824. [DOI] [PubMed] [Google Scholar]
- 10.Murakami, K. S., Masuda, S. & Darst, S. A. (2002) Science 296, 1280–1284. [DOI] [PubMed] [Google Scholar]
- 11.Vassylyev, D. G., Sekine, S., Laptenko, O., Lee, J., Vassylyeva, M. N., Borukhov, S. & Yokoyama, S. (2002) Nature 417, 712–719. [DOI] [PubMed] [Google Scholar]
- 12.Woychik, N. A. & Hampsey, M. (2002) Cell 108, 453–463. [DOI] [PubMed] [Google Scholar]
- 13.Korzheva, N., Mustaev, A., Kozlov, M., Malhotra, A., Nikiforov, V., Goldfarb, A. & Darst, S. A. (2000) Science 289, 619–625. [DOI] [PubMed] [Google Scholar]
- 14.Murakami, K. S., Masuda, S., Campbell, E. A., Muzzin, O. & Darst, S. A. (2002) Science 296, 1285–1290. [DOI] [PubMed] [Google Scholar]
- 15.Mekler, V., Kortkhonjia, E., Mukhopadhyay, J., Knight, J., Revyakin, A., Kapanidis, A. N., Niu, W., Ebright, Y. W., Levy, R. & Ebright, R. H. (2002) Cell 108, 599–614. [DOI] [PubMed] [Google Scholar]
- 16.Gavin, A. C., Bosche, M., Krause, R., Grandi, P., Marzioch, M., Bauer, A., Schultz, J., Rick, J. M., Michon, A. M., Cruciat, C. M., et al. (2002) Nature 415, 141–147. [DOI] [PubMed] [Google Scholar]
- 17.Orlicky, S. M., Tran, P. T., Sayre, M. H. & Edwards, A. M. (2001) J. Biol. Chem. 276, 10097–10102. [DOI] [PubMed] [Google Scholar]
- 18.Furuichi, Y. & Shatkin, A. J. (2000) Adv. Virus Res. 55, 135–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shuman, S. (2001) Prog. Nucleic Acid Res. Mol. Biol. 66, 1–40. [DOI] [PubMed] [Google Scholar]
- 20.Proudfoot, N. J., Furger, A. & Dye, M. J. (2002) Cell 108, 501–512. [DOI] [PubMed] [Google Scholar]