Species are part of the common coinage of biology. Taxonomists name them, developmental biologists deconstruct them, physiologists compare them, ecologists count them, conservation biologists conserve them, and evolutionary biologists study their multiplication and extinction. It may be that only the individual is a more important biological unit than the species. It is thus no surprise that Darwin named his great work On the Origin of Species, nor is it a surprise that many evolutionary biologists today concentrate their efforts on understanding species formation, or speciation. Riitta Savolainen and Kari Vepsäläinen (1), in a study in this issue of PNAS, have contributed significantly to a long-standing debate in evolutionary biology, over whether geographic isolation is a necessary first step in speciation, in a study of social parasites in Myrmica ants (2). The resolution of this argument has important consequences beyond evolutionary biology. If species do not have to wait for the next geological event to speciate, then speciation could conceivably be very rapid. This, in turn, means that the process might be conducive to study in real time. We might expect to see speciation in response to humanity's incessant modification of the globe. Anthropogenic change might drive not only such well known adaptations as the evolution of insecticide, herbicide, and antibiotic resistance, but also, for example, speciation of herbivorous insects adapting to plants moved to new parts of the world.
To fully understand Savolainen and Vepsäläinen's results, some history is in order. In the 144 years since publication of On the Origin of Species, enormous progress has been made in illuminating the things we need to know to understand speciation (2). We now know a lot about geographic isolation and the biogeography of closely related species. We know the kinds of features that prevent mating between closely related species or, in other words, that produce “reproductive isolation.” We understand that many close species differ in ecology, and we appreciate the consequences of lack of ecological differences. We know a great deal about how to count differences between the genomes of closely related species, and we are finally, if still dimly, seeing some of the specific genes that underlie reproductive isolation (3).
But fundamental questions remain unanswered. A critical question is how reproductive isolation could evolve in the face of gene flow; random mating within a population and gene flow between neighboring populations are enormously powerful homogenizing forces. How could selection tear a single population into two reproductively isolated species? For Darwin, who had an over-riding faith in the power of natural selection, this was not a difficulty. But with the mathematical sophistication that the new science of population genetics brought to the “modern synthesis” of the 1930s (4) came an understanding of just how strong selection had to be to overcome gene flow. The solution that Ernst Mayr, Theodosius Dobzhansky, and others (5) proposed is that geographic barriers could break the chain of gene flow between populations. If two populations exchange no genes because of geographic isolation, then these populations can evolve completely independently, adapting to new challenges until genetic reproductive isolation evolves as an evolutionary byproduct. The two new species that have evolved in geographic isolation (or allopatry) can later coexist in the same area (in sympatry). Geographic (or allopatric) speciation is such an obvious solution to the problem of evolving reproductive isolation, and the evidence for it so great (5), that it became the only accepted mode of speciation (excepting speciation via polyploidy and a few other similar mechanisms, which must occur in sympatry).
We might expect to see speciation in response to humanity's modification of the globe.
But a few evolutionary biologists, such as Guy Bush (6), started in the 1960s to argue that sympatric speciation was not limited to a handful of special cases, but was quite common. Bush was driven to his stance by the observation that one could find many groups of closely related, sympatric species that use different ecological niches. Conspicuous among these groups are parasites of animals and plants, which are often highly specialized ecologically and frequently mate on the host, a factor that potentially links any adaptation to a new host with a reduction in gene flow between the new and ancestral populations.
Another group of organisms that has attracted undying speculation about sympatric speciation are the social parasites in the Hymenoptera (7). Social parasites of the wasps, ants, and bees have fascinating natural histories. Of the usual social insect divisions into female reproductives (queens), males, and non-reproductive females (workers), only two occur in social parasites: female reproductive and males (7). Workers are often totally absent. Social parasite species live by tricking workers of the host species into feeding themselves and their progeny. They are the ultimate bad houseguests. Social parasites occur in most groups of social Hymenoptera, including bumble bees, several wasp groups, and many ant groups.
How does social parasitism involve sympatric speciation? The entomologist Emery in 1909 noted that social parasites almost always parasitized close relatives, an observation now known as “Emery's rule” (7). Others in the decades since (7) have been entranced by the idea that the social parasites could evolve from their hosts. The most often proposed scenario is evolution from a state of polygyny (multiple queens) within a species that all share equally in worker production, to polygyny with some queens “cheating” to increase their fitness by producing mostly reproductives, to assortative mating among cheaters, to finally a new parasitic species reproductively isolated from its ancestor (1, 7). Such a sympatric process would produce species pairs that are sister species (nearest relatives), which should pair together in a phylogenetic tree.
But in evolutionary biology it can be a long, rocky road from scenario to proof. “Proving” sympatric speciation has been a particularly tough proposition. The only potentially simple cases involve divergence in historical time (8, 9). For example, the apple host race of the fruit fly Rhagoletis pomonella arose from the ancestral, native, hawthorn-infesting population in North America ≈ 140 years ago, and has evolved substantial reproductive isolation from the ancestral race. The catch here, however, is that reproductive isolation between the races is only partial; they are not generally accepted as ”complete“ species.
For the vast majority of cases, the hypothesis of sympatric speciation must stand or fall based on inferences about the evolutionary past. This is certainly true for hymenopteran social parasites; it is unlikely that we will see the evolution of new ones in historical time. What kinds of observations can be used to test the competing hypotheses of sympatric and allopatric speciation in the past? Fundamentally, they are (i) the types of intermediate stages seen and (ii) the phylogenetic relationships observed in the relevant populations and species.
Over many decades, Ernst Mayr has pointed out that a parasite could adapt to a new host in a geographically isolated population, evolve reproductive isolation there, and then come back into sympatry with the original form (5). Mayr's allopatric hypothesis for the existence of sympatric, specialized species thus requires allopatric intermediate populations in varying degrees of adaptation to a new host; the competing sympatric hypothesis requires sympatric “host races” in varying degrees of adaptation to new hosts (8). For social parasites, the corresponding allopatric scenario has had the parasite attaining full species status in allopatry, then becoming sympatric with the host, and then evolving into a social parasite, leaving its old free-living lifestyle behind (1, 7). The allopatric explanation thus requires that one should, if enough groups are studied, see all possible intermediate forms, such as sister species that have just come into sympatry and are partially adapted to parasitism. Although more extensive searching needs to be done, such intermediates have not been reported for Myrmica (1) or for ants in general (7). Savolainen and Vepsäläinen also point out that social parasitism and its hypothesized precursor state of polygyny are both common in the tribe to which Myrmica belongs, and that the life history of the Myrmica social parasites would facilitate preferential mating between newly originated parasites (1).
Using the powerful tools of modern phylogeny estimation based on molecular data, it is now possible to infer phylogenies with some confidence. As Savolainen and Vepsäläinen point out, a relatively simple and straightforward inference is that if the parasites and hosts each form a monophyletic group, then sympatric speciation can be cleanly rejected. That is, if one finds that the social parasites are all grouped together on one branch or clade of a phylogenetic tree, and their hosts are all grouped on another branch, then the parasites cannot have evolved from the hosts. This is, in fact, the exact finding of several studies of social parasites and their hosts in wasps and bees (see ref. 1 and references therein), in which the “strict form” of Emery's rule is violated.
In Myrmica, however, Savolainen and Vepsäläinen did find evidence for host–parasite sister species pairs. Using trees built with sequences for three mitochondrial genes, they have shown that two of three Myrmica social parasites are in fact sister species of their host species, and thus follow the strict version of Emery's rule, whereas the third apparently parasitizes several close relatives, and thus supports at least the “loose version” of Emery's rule. Altogether, Savolainen and Vepsäläinen make a good case for sympatric speciation in Myrmica from the standpoint of both intermediate stages, and from phylogeny.
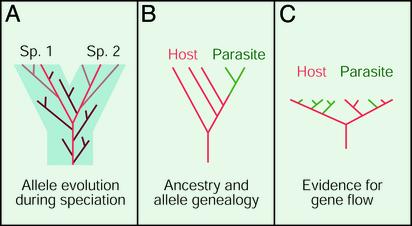
In the near future we can look forward to even more thorough tests of the allopatric and sympatric hypotheses for the evolution of social parasites in Hymenoptera. As has been appreciated ever since the pioneering work of Avise and colleagues (10) in the late 1970s, very intricate and informative genealogical relationships of haplotypes (or alleles) exist within species trees. Such a “gene tree” within a “species tree” is shown in Fig. 1A. When only one allele is sampled per species or population, as is the case for most of Savolainen and Vepsäläinen's work, then one can see only allele clades like those shown in red in Fig. 1. Such sampling is suitable for determining relationships at the species and higher level. However, other extant lineages, like those shown in light brown, could also tell us much about the history of speciation events. Fig. 1B provides a hypothetical example of how the study of gene trees, which would be possible with greater sampling than done in the present Myrmica study, could provide a powerful test of sympatric speciation. Here, all of the parasite alleles belong to one clade (are monophyletic). But the host alleles are not monophyletic. Although they all have a common ancestor, the parasite alleles are also derived from one of the clades (so that the host is said to be paraphyletic). What would such a finding tell us? It would tell us that the host was older than the parasite, which is critically important because this is what the sympatric hypothesis demands.
Fig. 1.
(A) Species tree (blue area) and allele tree (lines). Sampling just one allele per species reveals only part of what could be known about a speciation (red allele branches, or clades), whereas sampling extensively (red + light brown clades) reveals much more about timing, geographical patterns of events, etc. Dark brown extinct clades are unknowable. (B) Allele tree for a host (red) and parasite (green) that would support sympatric speciation, because the parasite is not only sister species to the host but also originated later than the host (parasite alleles originate at a later time). (C) A pattern that has been observed for mitochondrial genes implying both interspecific gene flow and rapid evolution, possibly because of “selective sweeps” of selected mutations through the species (tree labeled as in B).
But there is another complication here. As Savolainen and Vepsäläinen point out, interspecific gene flow (or more precisely passage of entire mitochondria across species lines) is a significant problem for testing the sympatric vs. allopatric hypotheses. Based on studies in Drosophila and other organisms (ref. 11 and references therein), mitochondrial gene trees from closely related species may look like those in Fig. 1C. In this case, neither of the species is monophyletic, or even paraphyletic, and moreover the sequences are quite similar. This pattern seems to be caused by a combination of interspecific gene flow and “selective sweeps,” in which a single strongly selected base position can drag an entire, nonrecombining mitochondrial sequence to high frequency. In some cases, species trees based on mitochondrial DNA may be positively misleading (11). As Savolainen and Vepsäläinen (1) discuss, the sister pairing of the English sample of the parasite Myrmica microrubra and its host M. rubra, and the corresponding pairing of host and parasite in Finland, may indicate gene flow between host and parasite. Sequencing independent nuclear genes would address this possibility.
One of the more conspicuous divisions among evolutionary biologists is between what could be called the consolidators and the challengers. Consolidators wish to bind those things about evolution which are undeniably true, such as genetic drift, natural selection, and allopatric speciation, into larger explanatory frameworks that organize as much as possible of the spectacular complexity of nature. Challengers (otherwise known as troublemakers) are irrevocably drawn to ideas that, at least in the short run, threaten consolidation. Of course, this is simplistic; a challenger with respect to sympatric speciation might be so threatened by, say, Lamarckian inheritance as to be a staunch consolidator on Darwinian selection. But there is little doubt that the most effective challengers, like Darwin himself, are driven by a core of facts that keep the challenge from being just an exercise in rhetoric, that keep the challenge vital. Sympatric speciation is a challenge to conventional thinking that will not die, because the challengers now have the support of a growing base of empirical support (ref. 8 and references therein). When the idea of sympatric speciation is combined with the great font of evolutionary ideas that arise from the study of social insects, as Savolainen and Vepsäläinen's contribution (1) makes clear, we should expect both scientific challengers and challenging research for many years to come.
See companion article on page 7169.
References
- 1.Savolainen, R. & Vepsäläinen, K. (2003) Proc. Natl. Acad. Sci. USA 100, 7169–7174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Howard, D. J. & Berlocher, S. H., eds. (1998) Endless Forms: Species and Speciation (Oxford Univ. Press, Oxford).
- 3.Ting, C.-T., Tsaur, S.-C. & Wu, C.-I. (2000) Proc. Natl. Acad. Sci. USA 97, 5313–5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berlocher, S. H. (1998) in Endless Forms: Species and Speciation, eds. Howard, D. J. & Berlocher, S. H. (Oxford Univ. Press, Oxford), pp. 3–15.
- 5.Mayr, E. (1963) Animal Species and Evolution (Harvard Univ. Press, Cambridge, MA).
- 6.Bush, G. L. (1969) Evolution (Lawrence, Kans.) 23, 237–251. [Google Scholar]
- 7.Hölldobler, B. & Wilson, E. O. (1990) The Ants (Harvard Univ. Press, Cambridge, MA).
- 8.Berlocher, S. H. & Feder, J. (2002) Annu. Rev. Entomol. 47, 773–815. [DOI] [PubMed] [Google Scholar]
- 9.Feder, J. L. (1998) in Endless Forms: Species and Speciation, eds. Howard, D. J. & Berlocher, S. H. (Oxford Univ. Press, Oxford), pp. 130–144.
- 10.Avise, J. C. (1994) Molecular Markers, Natural History, and Evolution (Chapman & Hall, New York).
- 11.Machado, C. A. & Hey, J. (2003) Proc. R. Soc. London B, 10.1098/rspb.2003.2333.