Abstract
Pathogenic microbes can devastate populations of marine plants and animals. Yet, many sessile organisms such as seaweeds and sponges suffer remarkably low levels of microbial infection, despite lacking cell-based immune systems. Antimicrobial defenses of marine organisms are largely uncharacterized, although from a small number of studies it appears that chemical defenses may improve host resistance. In this study, we asked whether the common seaweed Lobophora variegata is chemically defended against potentially deleterious microorganisms. Using bioassay-guided fractionation, we isolated and characterized a 22-membered cyclic lactone, lobophorolide (1), of presumed polyketide origin, with sub-μM activity against pathogenic and saprophytic marine fungi. Deterrent concentrations of 1 were found in 46 of 51 samples collected from 10 locations in the Bahamas over a 4-year period. Lobophorolide (1) is structurally unprecedented, yet parts of the molecule are related to tolytoxin, the scytophycins, and the swinholides, macrolides previously isolated from terrestrial cyanobacteria and from marine sponges and gastropods. Until now, compounds of this structural class have not been associated with marine macrophytes. Our findings suggest that seaweeds use targeted antimicrobial chemical defense strategies and that secondary metabolites important in the ecological interactions between marine macroorganisms and microorganisms could be a promising source of novel bioactive compounds.
Atypical milliliter of seawater contains 103 fungal cells, 106 bacteria, and 107 viruses, including pathogens that cause widespread mortalities and microbes that initiate fouling of host surfaces (1). Thus, marine plants and animals are continually exposed to high concentrations of potentially harmful microbes. Microbial pathogens cause black band disease in stony corals (2), Caribbean sea fan mortality (3), coralline lethal orange disease in coralline algae (4), green spot rotting disease in the alga Porphyra (5), red spot disease in the kelp Laminaria (6), and raisin disease in the brown alga Sargassum (7), to name just a few. In the 1930s, a slime mold wasting disease eliminated almost all of the eelgrass Zostera marina on the Atlantic coasts of North America and Europe, with continuing negative effects on soft-sediment communities, waterfowl populations, and scallop fisheries (8, 9). In the 1980s, an unidentified pathogen killed 95–99% of the herbivorous sea urchin Diadema antillarum throughout the Caribbean Sea; without these herbivores to control seaweeds, reef-building corals were rapidly overgrown (10–12). Harvell and colleagues (13, 14) proposed that the frequency of disease among marine macroorganisms has increased in recent decades and may continue to increase, because of climate change and human activities that stress hosts, introduce pathogens to new areas, and provide microbes with favorable conditions for growth.
As these examples illustrate, epidemics caused by microbes are often host-specific, but the reasons some potential hosts are resistant to infection have yet to be determined. Marine organisms are also resistant to saprophytic microbes that selectively attack senescent tissues. Given that many organisms possess nutrient-rich surfaces lacking physical barriers to microbial degradation, the mechanism of resistance could be chemical. Because microbes are more widely distributed in seawater than in air, pressures from pathogenic, parasitic, saprophytic, and fouling microbes might be greater in marine environments, a circumstance that might be expected to select for potent antimicrobial defenses. The apparently infrequent, but devastating, nature of marine epidemics suggests that evolution of resistance traits may be episodic, with occasional elimination of most susceptible individuals resulting in selection for disease resistance over evolutionary time.
We hypothesize that chemical defense could be a widespread strategy used by marine macroorganisms to deter microbial infection. In support of this, Weinberger and Friedlander (15) found that the red alga Gracilaria conferta responds to the presence of epiphytic bacteria with a burst of hydrogen peroxide and other reactive oxygen species that kill bacteria. Kim and colleagues (16, 17) reported that crude extracts of some gorgonian corals inhibit growth and germination of the sea fan pathogen Aspergillus sydowii, although defensive metabolites were not identified. Gil-Turnes et al. (18) discovered that shrimp embryos are covered by a bacterium, Alteromonas sp., that produces the broad-spectrum antifungal compound isatin and that this compound protects the embryos from a pathogenic fungus Lagenidium callinectes. This relationship represents an associational defense (19) acquired by the shrimp via its symbiosis. Other prokaryotes, particularly cyanobacteria, have been the source of numerous antifungal metabolites (20), which may provide competitive advantages to bacteria in their interactions with fungi.
In the course of exploring interactions between marine macroorganisms and microorganisms, we asked whether macroalgae, which lack cell-based inducible immune responses, enhance their fitness via chemical defenses against microbes. This hypothesis could explain why macroscopic algae are rarely infected, despite constant exposure to potentially deleterious microorganisms (e.g., ref. 7). Not surprisingly, those few diseases that are well understood affect commercially important seaweeds such as Porphyra, Laminaria, and Sargassum (21). It is likely that the microbial challenge to other marine plants is as high as for harvested species, but poorly understood. In preliminary experiments surveying 55 species of Caribbean seaweeds for antimicrobial potential (M. Puglisi, P.R.J., J.K., and W.F., unpublished work), extracts from Lobophora variegata exhibited especially potent antifungal activity against two co-occurring and broadly distributed marine fungi, the pathogenic Ascomycete Lindra thalassiae and the saprophytic Deuteromycete Dendryphiella salina. This circumtropical brown alga dominates many Caribbean reefs, growing from near the surface to depths of 88 m (22). Herein, we report the discovery of a compound responsible for its antifungal activity and present evidence that this compound provides L. variegata with an effective chemical defense against ecologically important microbes.
Materials and Methods
General Chemical Methods. Analytical HPLC and liquid chromatography (LC)-MS were performed with a Hewlett–Packard Series II 1100 liquid chromatograph with diode array UV detection and Agilent (Palo Alto, CA) 1100 electrospray ionization (ESI) mass spectrometer and a Waters 2695 Alliance HPLC system with diode array UV detection and Micromass (Manchester, U.K.) ZQ2000 ESI mass spectrometer. Semi-preparative HPLC purifications were performed with a Waters 510 pump with refractive index detection. HPLC columns used were Rainin Instruments Dynamax and Waters XTerra. Preliminary 1H and homonuclear 2D NMR experiments were performed on a 300-MHz Varian Inova spectrometer. NMR spectral data for fine-scale structure determination were acquired on a 500-MHz Varian spectrometer equipped with a nanoprobe, using the following experiments: 1H, 13C, gradient heteronuclear single-quantum correlation, gradient heteronuclear multiple-bond correlation, gradient COSY, total COSY with 80-msec mix time, total COSY with 60-msec mix time, homonuclear 2D J correlation, and rotating frame Overhauser effect spectroscopy (23). Additionally, single irradiation nuclear Overhauser effect and long-range COSY experiments were performed on a Bruker DRX 500-MHz NMR spectrometer. NMR spectra were recorded in deuteriochloroform and referenced to internal CHCl3 (1H δ 7.24, 13C δ 77.0). IR spectra were recorded on a Perkin–Elmer 1600 Series Fourier transform IR system as thin films. UV measurements were made on a Perkin–Elmer Lambda 3B spectrophotometer. Optical rotations were measured by using a Jasco (Tokyo) DIP-360 digital polarimeter with a path length of 10 cm recorded at the sodium D line. The CD spectrum was acquired on a Jasco J720 spectropolarimeter with a path length of 10 mm. High-resolution mass measurements were acquired on a Micromass quadrupole time-of-flight Ultima API mass spectrometer. X-ray data (24) were accessed from the Cambridge Structural Database and analyzed with MERCURY 1.0 software provided by the Cambridge Crystallographic Data Centre (Cambridge, U.K.). Geometry optimization was performed by molecular mechanics calculations using TITAN 6.0.6 (Wavefunction, Irvine, CA).
The Marine Alga L. variegata. The brown alga L. variegata (Dictyotaceae, Phaeophyta) was collected by scuba divers at depths of 6–30 m at several reef locations throughout the Islands of the Bahamas and from the Red Sea near Hurghada, Egypt.** Initial collections were quantified by volumetric displacement of fresh algal tissue in seawater. Subsequent collections were weighed after freeze-drying. Two algal collections were used for bioassay-guided fractionation: a combined collection of the common reef form (also called decumbent or shelf form; ref. 25) from several sites in the Bahamas (1,100 ml) and a single collection of the same form from Cay Lobos, Bahamas (1,500 ml). Smaller individual collections (including heavily fouled and unfouled plants) were made at multiple locations to measure antifungal activity and for LC-MS analysis.
Bioassays. L. variegata extracts and purified compounds were tested against four ecologically relevant marine microorganisms by using assays designed to provide a realistic approximation of in situ microbial exposure to these materials. In all cases, materials were tested at concentrations that were volumetrically equivalent to those occurring in the algae. For example, the extract obtained from 2 ml of alga was incorporated into 2 ml of media, which was then exposed to a test microorganism. The amount assayed of a pure compound was equal to that obtained from 2 ml of alga.
The two filamentous fungi used in the bioassays were D. salina and L. thalassiae. D. salina, collected in the Bahamas, was used for bioassay-guided fractionation of L. variegata extracts; L. thalassiae, originally collected in Belize, was acquired from the American Type Culture Collection (ATCC 56663). Extracts or compounds were dried under vacuum and then dissolved in 50 μl of acetone. Two milliliters of molten medium (16 g/liter granulated agar, 2 g/liter yeast extract, 2 g/liter peptone, 4 g/liter D-mannitol, 250 mg/liter of both penicillin G and streptomycin sulfate in 1 liter of natural seawater) was then added to the extract or compound, and the solution was mixed by swirling. For each sample, three 400-μl aliquots were dispensed into sterile, 24-well microtiter plates and allowed to solidify. Solvent-only control wells were prepared in the same manner. Sections of fungal hyphae (1 mm2) were then cut with a sterile needle from working plates of the test strains and inoculated into the center of each agar-filled well. The 24-well plates were kept at 25°C for 6 days, after which time the area covered by fungal growth was measured under a microscope for each well and compared with fungal coverage on control wells. Percent fungal inhibition is reported as a mean of the three subsamples for each treatment, relative to the mean coverage on three controls. When more than one replicate algal sample was tested against a given fungus, differences between treatments and controls were analyzed by using a one-tailed, paired t test on the means of three subsamples for each replicate (26). P values <0.05 were considered significant. Error bars depict ± one standard error. IC50 values were determined (in triplicate) in a similar manner except 1-ml equivalents were solubilized in acetone and serially diluted 1:1 with sterile seawater to a total volume of 500 μl. Five hundred microliters of 2× molten medium was then added to each well, the wells were inoculated, and the dilution at which fungal growth was inhibited by 50% relative to controls was determined.
Assays were also conducted with the bacterium Pseudalteromonas bacteriolytica, with a modified 96-well plate format in which a 48-h shake culture of P. bacteriolytica was diluted 1:160 with media and added to all wells in the plate. Five microliters of test solution (concentrated 40× in DMSO) was added to the top row (final volume 200 μl) and serially diluted 1:1 down the plate such that the concentration of test compound in the top row was equivalent to that obtained from 200 μl of algal tissue. Gentamicin was used as a standard, and solvent-only wells were run as a control, with bacterial growth assessed turbidimetrically. Assays using the thrausochytrid Schizochitrium aggregatum were performed as described (27), using cultures originally collected in the Bahamas. Additionally, standard microtiter plate biomedical assays were performed to determine antifungal activity against the pathogenic yeast Candida albicans and cytotoxicity against the human colon tumor cell line HCT-116. Field herbivory assays were conducted at Pickles Reef near Key Largo, FL, as reported (28), testing pure lobophorolide (1) at 2 × 10–4 percent of plant dry weight, using a one-tailed, paired t test to compare consumption rates (26).
Isolation of Lobophorolide (1) by Bioassay-Guided Fractionation. Frozen L. variegata was lyophilized and then extracted four times with methanol/dichloromethane (1:1). Crude extracts were combined, reduced in vacuo, and then subjected to solvent partitioning according to a method modified from Kupchan et al. (29). Thus, the crude extract was partitioned between hexanes and methanol/water (9:1), and the latter extract was partitioned between methanol/water (3:2) and chloroform. The methanol/water fraction was further partitioned between ethyl acetate and water, and the latter was then partitioned against n-butanol. The chloroform extract, which possessed the entire antifungal activity, was fractionated by using vacuum LC with reversed-phase silica gel (C18) using a gradient eluent system from methanol/water (1:1) to methanol to ethyl acetate. Active fractions, eluted with methanol/water (4:1 to 9:1), were pooled and then further fractionated by using two rounds of Sephadex LH-20 size exclusion chromatography eluting with methanol. A single active compound was finally purified by HPLC, first using reversed-phase HPLC (eluent methanol/water 9:1) and then multiple rounds of normal-phase HPLC (gradient eluent hexane/ethyl acetate 1:4 to ethyl acetate).
Structure Elucidation of Lobophorolide (1). Lobophorolide (1) was isolated as a white amorphous solid [0.2 mg from 1,100 ml of fresh algae or 135 g dry weight (collection from various Bahamian sites), and 0.2 mg from 1,500 ml of fresh algae or 224 g dry weight (collection from Cay Lobos, Bahamas)]. Lobophorolide (1) showed the following spectral characteristics: [α]25D –33.0° (c 0.067, CH3OH); CD (CH3OH) [θ]260 +17, ΔA260 +1.0 × 10–4; UV (CH3OH) λmax (log ε) 262 (4.0); IR (thin film) νmax 3425 (br), 2,919, 2,849, 1,737, 1,684, 1,625, 1,584, 1,455, 1,378, 1,349, 1,267, 1,196, 1,091 cm–1; 1H NMR (500 MHz, CDCl3) δ 0.81 (d, J = 7.0, Me-39), 0.84 (d, J = 7.0, Me-38), 1.01 (d, J = 6.5, Me-40), 1.14 (m, H-30a), 1.18 (d, J = 6.0, Me-42), 1.26 (m, H-26a), 1.31 (m, H-25a), 1.34 (m, H-8a), 1.40 (m, H-18a), 1.42 (m, H-25b), 1.46 (m, H-14a), 1.58 (m, H-8b), 1.60 (m, H-28a), 1.63 (m, H-14b), 1.70 (m, H-24), 1.82 (br d, J = 13.0, H-28b), 1.89 (s, Me-32), 1.91 (m, H-26b), 1.92 (m, H-12a), 1.92 (m, H-18b), 1.92 (m, H-22), 1.94 (m, H-20), 1.97 (m, H-30b), 2.00 (m, H-12b), 2.62 (d, J = 4.0, H-35a), 2.64 (d, J = 4.5, H-35b), 3.01 (dd, J = 10.0, 2.0, H-23), 3.19 (s, Me-36), 3.21 (m, H-19), 3.21 (s, Me-37), 3.28 (m, H-13), 3.32 (s, Me-41), 3.43 (s, Me-34), 3.44 (m, H-7), 3.52 (m, H-29), 3.62 (s, Me-33), 3.68 (m, H-31), 3.76 (dd, J = 10.5, 4.5, H-17), 3.87 (dd, J = 7.5, 3.0, H-15), 4.00 (m, H-27), 4.28 (dd, J = 9.0, 9.0, H-6), 4.48 (br d, J = 10.0, H-9), 5.20 (dd, J = 10.5, 1.6, H-21), 5.59 (br d, J = 10.0, H-10), 5.82 (m, H-11), 5.85 (d, J = 15.5, H-2), 5.85 (d, J = 9.7, H5), 7.54 (d, J = 16.0, H-3); 13C NMR (125 MHz, CDCl3) δ 9.2 (C-39), 9.3 (C-38), 12.8 (C-32), 18.2 (C-40), 22.0 (C-42), 24.4 (C-25), 27.5 (C-18), 29.4 (C-26), 31.4 (C-12), 33.0 (C-24), 34.8 (C-14), 35.2 (C-28), 36.2 (C-8), 37.6 (C-22), 37.8 (C-20), 39.1 (C-30), 45.2 (C-35), 53.1 (C-36), 55.7 (C-41), 57.8 (C-34), 58.6 (C-37), 60.5 (C-16), 61.0 (C-33), 64.6 (C-31), 66.8 (C-13), 70.2 (C-9), 71.1 (C-6), 71.8 (C-27), 73.5 (C-29), 74.6 (C-17), 76.1 (C-23), 76.5 (C-21), 77.2 (C-19), 78.3 (C-15), 82.4 (C-7), 118.0 (C-2), 125.5 (C-11), 129.5 (C-10), 138.0 (C-4), 138.5 (C-5), 150.5 (C-3), 168.0 (C-1); 2D NMR and nuclear Overhauser effect, see Table 2, which is published as supporting information on the PNAS web site, www.pnas.org; high-resolution quadrupole time-of-flight (QTOF) MS m/z [M+H]+ 767.4922 (calculated for C42H71O12 767.4945), [M+Na]+ 789.4761 (calculated for C42H70O12Na, 789.4765), [M+K]+ 805.4507 (calculated for C42H70O12K, 805.4504); QTOF MS-MS on 767.4945 m/z 749.4875, 735.4775, 717.4679, 703.4520, 685.4408, 667.4298, 653.4133, 635.4005, 621.3818, 603.3627, 571.3482, 553.3347; 535.3488, 521.3234, 503.3276, 485.3249, 471.2972, 437.1985, 263.2035; positive electrospray ionization-MS m/z 789.4, 767.5; electron impact MS m/z 735 (2), 419 (5), 387 (10), 263 (12), 223 (15), 183 (17), 113 (50), 81 (100).
Geographic Distribution of Lobophorolide (1) and Antifungal Activity of L. variegata Extracts. Small quantities (2–400 ml) of L. variegata were collected from all sampling sites to determine whether extracts possessed antifungal activity and lobophorolide (1). Samples were extracted three times with acetone, and the extracts were combined and reduced in vacuo. Activity against D. salina was measured as described above. The remaining material was fractionated by solvent partitioning, then by gradient reversed-phase flash column chromatography (Supelco Envi-18 cartridge), and the antifungal materials that eluted with methanol/water 9:1 were concentrated. Aliquots were analyzed by reversed-phase LC-MS with a gradient of methanol/water (0.1% acetic acid) 3:2 to 9:1 over 15 min, monitoring by UV at 262 nm. Six standard solutions of pure 1 (0.003–1.0 μg/ml) were used to establish a standard curve (R2 = 0.993) for quantification of 1 in extracts by integration of the [M+Na]+ peak at m/z 789.4. The detection limit was determined to be ≈5 × 10–6 percent by dry mass.
To test whether lobophorolide (1) could be removed from seaweed surfaces, seaweed samples (2 ml each, n = 7) were shaken with n-hexane for 60 s, and the hexane extract was filtered, dried in vacuo, and subjected to antifungal assaying as described above. The remaining alga was then extracted with acetone as described above and assayed.
Results and Discussion
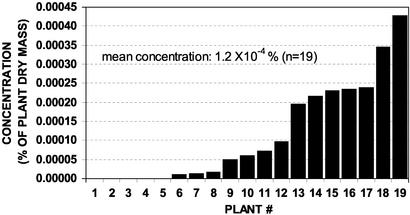
Identification of Antifungal Compound Lobophorolide (1). Bioassay-guided fractionation of L. variegata extracts using the marine Deuteromycete D. salina led to the isolation of the antifungal compound lobophorolide (1), in a yield of 0.1 and 0.2 μg/ml, or 9 × 10–5 and 2 × 10–4 percent of plant dry mass, respectively, from two different batches of L. variegata. Using LC-MS, quantification of 1 in 19 independently collected samples from multiple sites in the Bahamas revealed the mean concentration of 1 to be 1.2 ± 0.3 × 10–4 percent of plant dry mass.
A nominal molecular mass of 766 Da and a molecular formula of C42H70O12 were indicated by MS. Exact mass measurement on daughter ions produced by MS–MS indicated the successive loss of 5 units of methanol and 2 units of water, suggesting five O-methyl groups and two alcohol functionalities. Among the 42 carbon signals observed in the 13C NMR spectrum were an ester carbonyl functionality (δ 168.0) and six olefinic carbons (δ 117–151), thus, four of the eight unsaturations were from rings. Both UV (262 nm) and IR (1,737 cm–1) absorbances highlighted an α,β,γ,δ-unsaturated carbonyl system. From a total of 12 oxygen atoms, the five O-methyls, two alcohols, and two oxygens of the ester group left the likelihood of three oxygens being present in ether functionalities.
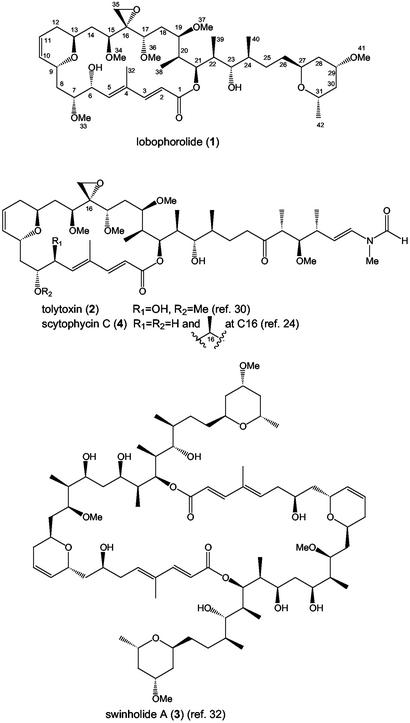
Analysis of 1H, 13C, and 2D NMR spectral data of lobophorolide (1) allowed all protons and carbons to be related and a planar structure to be proposed (Fig. 1). Comparison of these NMR features with published data for tolytoxin (2) (30) and swinholide A (3) (31–33) suggested a hybrid structure for 1, consisting of the macrolide portion of 2 and the side chain of 3 (Fig. 1). Because the NMR data for 1 (this study) and 2 (30) are in close agreement throughout the macrolide portion, we believe that 1 and 2 share the same relative configuration at all stereogenic centers in that region of the molecule. However, we suggest an R configuration at C6 for both 1 and 2 rather than the S configuration proposed for 2 by Carmeli et al. (30).†† With an optical rotation of [α]D –30°, lobophorolide (1) probably has the same absolute stereochemistry as 2 and scytophycin B ([α]D –23° and –38°, respectively), which was determined for scytophycin B by CD analysis of a synthesized derivative (24). In this study, CD analysis of 1 showed the same sign ellipticity near its λmax as 2 and the scytophycin group (24, 30).
Fig. 1.
Structure of lobophorolide (1) and related macrolides.
Lobophorolide (1) is almost certainly of polyketide origin, requiring the step-wise biosynthesis of acetyl CoA and 15 units of malonyl CoA, plus five methylations at C2 of acetate and five O-methylations. Carbons 6 and 16 appear to have been oxidized, with all other oxygens originating from C1 of acetate. The isolation of a polycyclic macrolide from L. variegata was surprising, because this class of secondary metabolite has not been previously known to occur in marine macrophytes.
Ecological Function of Lobophorolide (1). Throughout the bioassay-guided fractionation process, only lobophorolide (1) and fractions containing 1 inhibited fungal growth in laboratory assays, suggesting that 1 accounts for all of the antifungal activity in extracts of L. variegata. To date, most seaweed secondary metabolites that possess antimicrobial or antifeedant activities have been isolated at concentrations of 0.1–10% of plant dry mass (34, 35). In contrast, the natural concentration of 1 was exceedingly low, averaging 1.2 × 10–4 percent of plant dry mass. Despite these low levels, 1 showed potent activity at and below its natural concentration against two marine fungi, D. salina and L. thalassiae, isolated from Caribbean waters (Table 1, Fig. 2). D. salina is a globally distributed saprophytic marine fungus that degrades senescent brown and red algal tissue, whereas L. thalassiae is a pathogenic fungus that causes disease in a variety of marine plants but is not known to infect L. variegata (7, 36). We propose that the presence of 1 acting as a chemical defense may be the reason this seaweed is not known to be susceptible to fungal infection and disease. It is also possible that chemical defenses such as 1 help prevent the degradation of live algal tissue by saprophytic fungi such as D. salina. The biological activity of 1 is highly specific, targeting filamentous fungi while being inactive against the thrausochytrid S. aggregatum and the bacterium P. bacteriolytica, which causes red spot disease in the kelp Laminaria (6) (Table 1). Using a standard field assay (28), 1 was also ineffective as a feeding deterrent to herbivorous fishes (amount eaten: control 52 ± 5%, treatment 51 ± 6%; n = 29, P = 0.44).
Table 1. Biological activities of lobophorolide (1).
| Assay organism | IC50, μg/ml |
|---|---|
| D. salina | 0.034 |
| L. thalassiae | 0.135 |
| S. aggregatum | Not active* |
| P. bacteriolytica | Not active* |
| C. albicans | 1.3 |
| C. albicans (amphotericin-resistant strain) | 0.5 |
| Human colon tumor cell line HCT-116 | 0.03 |
Tested up to 0.2 μg/ml.
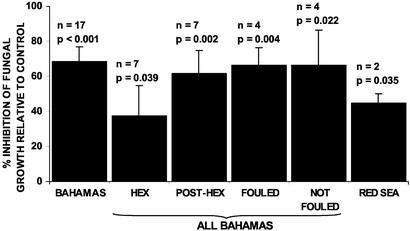
Fig. 2.
Antifungal activity of L. variegata extracts against D. salina. The activity of each extract is the mean of three subsamples. P values indicate results of one-tailed paired t test of treatments vs. controls. HEX, 60-s hexane extraction; POST-HEX, acetone extraction after HEX treatment; FOULED, samples that had relatively high levels of macroscopic fouling; NOT FOULED, samples that had relatively low levels of macroscopic fouling.
Despite the prevalence of microbes in the ocean (1) and the devastating nature of pathogen-borne epidemics (9), disease among marine plants appears to be rare (36), and thus many plants likely possess traits that make them resistant to microbial attack. Nevertheless, few examples of seaweed antimicrobial defenses are known. Jensen et al. (27) found that the seagrass Thalassia testudinum is protected from a zoosporic fungus by a flavone glycoside present at high concentration in seagrass tissues. Halogenated furanones in gland cell vesicles of the red alga Delisea pulchra prevent bacterial colonization and biofilm formation, by inhibiting bacterial quorum sensing (35). Although defensive properties against bacteria and fungi have been attributed to brown algal polyphenolics (phlorotannins) (37), the few tests conducted with purified phlorotannins have not supported this generalization (38). In this study, phlorotannins isolated from the ethyl acetate and 1-butanol fractions of L. variegata showed no antifungal activity when tested at their natural concentration (2% of plant dry mass), whereas all of the activity in the crude extracts could be attributed to a single compound, lobophorolide (1). Thus, there is mounting evidence that seaweeds use specific compounds as defenses against harmful microorganisms.
It appears that lobophorolide (1) may be found both within the tissues and on the surface of L. variegata. Hexane surface extracts [as in de Nys et al. (39)] and acetone extracts of algae performed after the hexane treatment both had significant antifungal activity (Fig. 2, HEX and POST-HEX). Because the hexane solubility of 1 is low [only 10% of 1 could be recovered from coated glass using the surface extraction technique (J.K., data not shown)], there may be a higher concentration of 1 on algal surfaces than the above experiment suggests. Because average whole plant levels of 1 were 3–10 times the concentration required to inhibit D. salina and L. thalassiae (Table 1, Fig. 3), even a small proportion of 1 on the surface could reduce fungal colonization. Perhaps more importantly, natural concentrations of 1 should prevent the penetration of fungal hyphae into healthy algal tissues and defend damaged tissues from fungal invasion. Because some pathogenic fungi, including L. thalassiae, are believed to attack primarily algal tissues that have already sustained physical damage (36), 1 should provide an effective antifungal defense.
Fig. 3.
Concentrations of lobophorolide (1) in 19 independently collected samples of L. variegata from several locations in the Bahamas, determined by LC-MS. IC50 values of 1 against D. salina and L. thalassiae (Table 1) correspond to 1 × 10–5 percent and 4 × 10–5 percent of plant dry mass, respectively.
Geographic Variation of Chemical Defenses. Thirty-two Bahamian samples of the common reef form of L. variegata exhibited antifungal activity (Fig. 2, sum of all Bahamian samples), and the presence of lobophorolide (1) was confirmed in four of the four extracts from this group that were analyzed by LC-MS. LC-MS quantification of 1 in 19 other samples from several locations in the Bahamas indicated that concentrations of 1 varied from below the detection limit of 5 × 10–6 percent to 4 × 10–4 percent of plant dry mass (Fig. 3). Overall, from a total of 51 samples collected at 10 Bahamian sites** over 4 years, 46 samples contained detectable, deterrent concentrations 1 (Fig. 3) or showed potent antifungal activity most likely caused by 1 (Fig. 2).
Extracts of two collections of L. variegata from the Red Sea exhibited antifungal activity (Fig. 2), but when this activity was pursued by chromatographic separation and LC-MS analysis, 1 was not found and the antifungal activity tracked to a different chromatographic fraction. Thus, although L. variegata from the Red Sea also appears to possess antifungal chemical defenses, the defensive compound(s) is different from that found in Caribbean L. variegata.
Relationship to Other Polyketide Macrolides. Lobophorolide (1) and tolytoxin (2) share a 22-membered cyclic lactone, whose carbon skeleton is also the basis for the 44-membered dimeric cyclic lactone of swinholide A (3) (Fig. 1). Despite the structural similarities, these compounds and their analogs were isolated from very diverse sources. Cultured terrestrial cyanobacteria of the genera Tolypothrix and Scytonema have been consistent sources of 2 and the scytophycins (30). Biosynthetic investigations using stable isotope-labeled acetate, glycine, and methionine have supported a cyanobacterial biogenesis for these compounds, with glycine as a starter unit and the successive condensation of 15 acetate units by polyketide biosynthesis (40). Swinholide A (3) and related compounds have been isolated from marine sponges of the genus Theonella from the Red Sea, Japan, and the tropical Indo-Pacific (e.g., refs. 41 and 42). Because of their structural similarity to 2 and the scytophycins, and because of the presence of large populations of filamentous cyanobacteria in Theonella spp., 3 and related compounds have been hypothesized to be cyanobacterial products (32, 42). However, Bewley et al. (43) separated sponge, cyanobacterial, and eubacterial cells of Theonella swinhoei and found 3 to be associated with a heterotrophic eubacterial fraction. This association, plus the discovery of tolytoxin-23-acetate from a cephalaspidean mollusk (44) and other macrolides isolated from sea hares and nudibranchs (e.g., ref. 45), has weakened the assertion that such metabolites are all cyanobacterial in origin. However, in some cases, cyanobacteria are at the base of food webs involving macrolide-containing mollusks, suggesting that cyanobacteria may be the biosynthetic sources of some polyketide macrolides.
Lobophorolide (1) could be the product of a microbial symbiont. Epiphytic bacteria including cyanobacteria were observed by scanning electron and light microscopy and several bacterial strains have been cultured from L. variegata surfaces, although none of these has to date yielded 1 (P.R.J. and T. Mincer, unpublished work). Specimens of L. variegata that were heavily fouled by macroscopic organisms, including algae and hydroids, and other specimens only lightly fouled all had significant antifungal activities, suggesting that 1 is not associated with the epiphyte community (Fig. 2). However, because of the low yield of 1, its structural similarity to microbial metabolites, and the fact that polyketides of this type are unprecedented from macroalgae, a microbial origin warrants further exploration. Recently, genetic sequencing revealed that a beetle chemical defense is produced by a symbiotic bacterium, using mixed polyketide and nonribosomal peptide biosynthesis, implying probable bacterial origins of closely related polyketides from marine sponges (46). Although examples are rare (e.g., ref. 18), symbionts may be an important source of antimicrobial chemical defenses for plants and animals. Regardless of its origin, 1 appears to fulfill an important ecological function as an antifungal defense for Caribbean L. variegata.
Other Biological Activities of Lobophorolide (1). Lobophorolide (1) was active in biomedical screens (Table 1). The potency of 1 against C. albicans was similar to that of the swinholides (47) but less than that of tolytoxin (2) (48). The antineoplastic (anticancer) activity of 1 was of a similar magnitude to the swinholide and tolytoxin/scytophycin classes of compounds (41, 48, 49), although this comparison is potentially confounded by the use of different human cancer cell lines. The tolytoxin/scytophycin group inhibits actin polymerization (50, 51), and because of the structural similarity of 1 and 2, it is possible that 1 acts by a similar mechanism. Although L. variegata has been the subject of several earlier chemical studies, 1 was not previously discovered, possibly because of its low natural concentration and specific biological activity. Ecologically driven studies, such as this one, which used pathogenic and saprophytic marine fungi in operationally simple assays, may be a promising strategy for uncovering novel natural products of commercial interest.
Conclusions
It is likely that many marine plants use chemical defenses against microbial pathogens, epiphytes, and saprophytes, but this hypothesis has rarely been tested. Lobophorolide (1), a polycyclic macrolide with sub-μM antifungal activity, appears to function as an antifungal chemical defense, protecting L. variegata from at least one pathogenic (L. thalassiae) and one saprophytic fungus (D. salina), while being inactive against a thrausochytrid fungus, a pathogenic bacterium, and herbivorous fishes. The structural similarity of 1 to bacterial metabolites suggests that 1 could be the product of a microbial symbiont of L. variegata, although no such symbiont has yet been identified. The specific association between 1 and Bahamian L. variegata provides an example of an abundant and broadly distributed seaweed that uses a targeted chemical strategy to prevent fungal attack. Further investigations of chemically mediated interactions between marine microbes and macroorganisms are likely to reveal novel molecules and mechanisms that enable marine plants and animals to persist despite intense microbial challenge.
Supplementary Material
Acknowledgments
We thank the captains and crew of the R/V Seward Johnson and the M/V Golden Shadow and Joseph Pawlik for the invitation to participate in research cruises, and the governments of Egypt and the Islands of the Bahamas for permission to perform research in their territorial waters. We also thank HRH Prince Khaled Bin Sultan Bin Abdul-Aziz for his generous support of our Red Sea research program. Matt Woolery assisted with extractions, and Sara Kelly, Sebastian Engel, and Melany Puglisi conducted additional assays. Assay microorganisms were kindly provided by E. B. Gareth Jones, Tomoo Sawabe, and David Porter. Algal collections were obtained with assistance from Eliane Garo, Mark Hay, Michelle Hoogstra, Chris Kauffman, Sara Kelly, Sarah Kelly, Jeremy Long, Tracy Mincer, Monica Puyana, Allan Spyere, and Kristen Whalen. Laren Tolbert, Jack Eichler, and Javier Concepcion helped with conformational analysis, and Mark Hay and Deron Burkepile provided useful comments on the manuscript. We thank the The Scripps Institution of Oceanography analytical facility for the use of UV, IR, and some NMR spectroscopic instrumentation, Leslie Gelbaum at the Georgia Institute of Technology NMR Center for nuclear Overhauser effect and long-range COSY NMR data, and the Micromass Facility (Beverly, MA) for use of mass spectrometric demonstration equipment. This research was supported by National Science Foundation Grants CHE-9807098 and CHE 01-11270 (to W.F.), a Natural Sciences and Engineering Research Council (Canada) postdoctoral fellowship (to J.K.), and an O'Donnell Fellowship from the Life Sciences Research Foundation (to J.K.).
Abbreviation: LC, liquid chromatography.
Footnotes
Bahamas locations: Andros Island Salvador Point 24°25′N, 77°45′W; Bimini Cat Cay 25°33.13′N, 79°18.13′W; Cay Lobos 22°25′N, 77°40′W; Great Egg Island 25°20.79′N, 76°53.82′W; Little San Salvador Island 24°32.68′N, 75°55.73′W; Long Island 23°38.49′N, 75°21.50′W; San Salvador Island 24°01.14′N, 74°32.68′W; Santo Domingo Cay 21°43.25′N, 75°45.50′W; Stirrup Cay 25°50.06′N, 77°54.97′W; Sweetings Cay, 26°33.65′N, 77°52.52′W; in September 1998, August 1999, August 2000, and June 2002. Red Sea locations: Elphinstone 25°19.41′N, 34°47.15′E; Gez Siyul 24°22.55′N, 35°23.03′E in February 2000.
The observed 9-Hz pseudo-triplet signal for H6 in 1 and 2 indicated either syn or anti orientations of H6 to both H5 and H7. Scytophycin C (4) is itself achiral at C6, but from its x-ray crystal structure, the pro-S H6 of 4 appears anti to H7, whereas the pro-R H6 is approximately gauche to H7, supporting locating the C6 hydroxyl group of 1 and 2 in place of the pro-R H6 of 4, resulting in an R orientation at C6 in 1 and 2. In this study, nuclear Overhauser effects observed for 1, in particular H6 to H32, H6 to H8a and H8b, H7 to H5 and H9 (see Table 2), also favor the R assignment at C6.
References
- 1.Rheinheimer, G. (1992) Aquatic Microbiology (Wiley, New York), 3rd Ed.
- 2.Taylor, D. L. (1983) Mar. Ecol. 4, 321–328. [Google Scholar]
- 3.Smith, G. W., Ives, L. D., Nagelkerken, I. A. & Ritchie, K. B. (1996) Nature 383, 487. [Google Scholar]
- 4.Littler, M. M. & Littler, D. S. (1995) Science 267, 1356–1360. [DOI] [PubMed] [Google Scholar]
- 5.Goff, L. J. & Glasgow, J. C. (1980) Pathogens of Marine Plants, Special Publication 7 (Center for Coastal Studies, University of California, Santa Cruz).
- 6.Sawabe, T., Makino, H., Tatsumi, M., Nakano, K., Tajima, K., Iqbal, M. M., Yumoto, I., Ezura, Y. & Christen, R. (1998) Int. J. Syst. Bacteriol. 48, 769–774. [DOI] [PubMed] [Google Scholar]
- 7.Kohlmeyer, J. (1971) Mar. Biol. 8, 344–350. [Google Scholar]
- 8.Rasmussen, E. (1977) in Seagrass Ecosystems, eds. McRoy, C. P. & Helfferich, C. (Dekker, New York), pp. 1–51.
- 9.Short, F. T., Muehlstein, L. K. & Porter, D. (1987) Biol. Bull. 173, 557–562. [DOI] [PubMed] [Google Scholar]
- 10.Lessios, H. A. (1988) Annu. Rev. Ecol. Syst. 19, 371–393. [Google Scholar]
- 11.Lessios, H. A. (1995) Proc. R. Soc. London Ser. B 259, 331–337. [Google Scholar]
- 12.Hughes, T. P. (1994) Science 265, 1547–1551. [DOI] [PubMed] [Google Scholar]
- 13.Harvell, C. D., Kim, K., Burkholder, J. M., Colwell, R. R., Epstein, P. R., Grimes, D. J., Hofmann, E. E., Lipp, E. K., Osterhaus, A. D. M. E., Overstreet, R. M., et al. (1999) Science 285, 1505–1510. [DOI] [PubMed] [Google Scholar]
- 14.Harvell, C. D., Mitchell, C. E., Ward, J. R., Altizer, S., Dobson, A. P., Ostfeld, R. S. & Samuel, M. D. (2002) Science 296, 2158–2162. [DOI] [PubMed] [Google Scholar]
- 15.Weinberger, F. & Friedlander, M. (2000) J. Phycol. 36, 1079–1086. [Google Scholar]
- 16.Kim, K., Harvell, C. D., Kim, P. D., Smith, G. W. & Merkel, S. M. (2000) Mar. Biol. 136, 259–267. [Google Scholar]
- 17.Kim, K., Kim, P. D., Alker, A. P. & Harvell C. D. (2000) Mar. Biol. 137, 393–401. [Google Scholar]
- 18.Gil-Turnes, M. S., Hay, M. E. & Fenical, W. (1989) Science 246, 116–118. [DOI] [PubMed] [Google Scholar]
- 19.Atsatt, P. R. & O'Dowd, D. J. (1976) Science 193, 24–29. [DOI] [PubMed] [Google Scholar]
- 20.Gerwick, W. H., Tan, L. T. & Sitachitta, N. (2001) in The Alkaloids: Chemistry and Biology, ed. Cordell, G. A. (Academic, New York), pp. 75–184. [DOI] [PubMed]
- 21.Andrews, J. H. (1976) Biol. Rev. 51, 211–253. [Google Scholar]
- 22.Littler, M. M., Littler, D. S., Blair, S. M. & Norris, J. N. (1985) Science 227, 57–59. [DOI] [PubMed] [Google Scholar]
- 23.Kupce, E., Keifer, P. A. & Delepierre, M. (2000) J. Magn. Reson. 148, 114–120. [DOI] [PubMed] [Google Scholar]
- 24.Ishibashi, M., Moore, R. E. & Patterson, G. M. L. (1986) J. Org. Chem. 51, 5300–5306. [Google Scholar]
- 25.Littler, D. S. & Littler, M. M. (2000) Caribbean Reef Plants (Offshore Graphics, Washington, DC).
- 26.Zar, J. H. (1996) Biostatistical Analysis (Prentice–Hall, Upper Saddle River, NJ), 3rd Ed.
- 27.Jensen, P. R., Jenkins, K. M., Porter, D. & Fenical, W. (1998) Appl. Environ. Microbiol. 64, 1490–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hay, M. E., Fenical, W. & Gustafson, K. (1987) Ecology 68, 1581–1591. [DOI] [PubMed] [Google Scholar]
- 29.Kupchan, S. M., Britton, R. W., Lacadie, J. A., Ziegler, M. F. & Sigel, C. W. (1975) J. Org. Chem. 40, 648–654. [DOI] [PubMed] [Google Scholar]
- 30.Carmeli, S., Moore, R. E. & Patterson, G. M. L. (1990) J. Nat. Prod. 53, 1533–1542. [DOI] [PubMed] [Google Scholar]
- 31.Carmely, S., Rotem, M. & Kashman, Y. (1986) Magn. Reson. Chem. 24, 343–349. [Google Scholar]
- 32.Kitagawa, I., Kobayashi, M., Katori, T., Yamashita, M., Tanaka, J., Doi, M. & Ishida, T. (1990) J. Am. Chem. Soc. 112, 3710–3712. [Google Scholar]
- 33.Kobayashi, M., Tanaka, J., Katori, T., Matsuura, M., Yamashita, M. & Kitagawa, I. (1990) Chem. Pharm. Bull. 38, 2409–2418. [DOI] [PubMed] [Google Scholar]
- 34.Paul, V. J. (1993) Ecological Roles of Marine Natural Products (Comstock, Ithaca, NY).
- 35.Kjelleberg, S., Steinberg, P., Givskov, M., Gram, L., Manefield, M. & de Nys, R. (1997) Aquat. Microbiol. Ecol. 13, 85–93. [Google Scholar]
- 36.Kohlmeyer, J. & Kohlmeyer, E. (1979) Marine Mycology: The Higher Fungi (Academic, New York).
- 37.Sieburth, J. M. & Conover, J. T. (1965) Nature 208, 52–53. [Google Scholar]
- 38.Ragan, M. A. & Glombitza, K. W. (1986) Prog. Phycol. Res. 4, 129–241. [Google Scholar]
- 39.de Nys, R. S., Dworjanyn, S. A. & Steinberg, P. D. (1998) Mar. Ecol. Prog. Ser. 162, 79–87. [Google Scholar]
- 40.Carmeli, S., Moore, R. E., Patterson, G. M. L. & Yoshida, W. Y. (1993) Tetrahedron Lett. 34, 5571–5574. [Google Scholar]
- 41.Tsukamoto, S., Ishibashi, M., Sasaki, T. & Kobayashi, J. (1991) J. Chem. Soc. Perkin Trans. 1 , 3185–3188.
- 42.Todd, J. S., Alvi, K. A. & Crews, P. (1992) Tetrahedron Lett. 33, 441–442. [Google Scholar]
- 43.Bewley, C. A., Holland, N. D. & Faulkner, D. J. (1996) Experientia 52, 716–722. [DOI] [PubMed] [Google Scholar]
- 44.Nakao, Y., Yoshida, W. Y., Szabo, C. M., Baker, B. J. & Scheuer, P. J. (1998) J. Org. Chem. 63, 3272–3280. [Google Scholar]
- 45.Yamada, K., Ojika, M., Ishigaki, T. & Yoshida, Y. (1993) J. Am. Chem. Soc. 115, 11020–11021. [Google Scholar]
- 46.Piel, J. (2002) Proc. Natl. Acad. Sci. USA 99, 14002–14007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sakai, R., Higa, T. & Kashman, Y. (1986) Chem. Lett. 1499–1502.
- 48.Patterson, G. M. L. & Carmeli, S. (1992) Arch. Microbiol. 157, 406–410. [DOI] [PubMed] [Google Scholar]
- 49.Doi, M., Ishida, T., Kobayashi, M. & Kitagawa, I. (1991) J. Org. Chem. 56, 3629–3632. [Google Scholar]
- 50.Patterson, G. M. L., Smith, C. D., Kimura, L. H., Britton, B. A. & Carmeli, S. (1993) Cell. Motil. Cytoskeleton 24, 39–48. [DOI] [PubMed] [Google Scholar]
- 51.Belmont, L. D., Patterson, G. M. L. & Drubin, D. G. (1999) J. Cell Sci. 112, 1325–1336. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.