Abstract
Type III secretion systems (TTSS) are important virulence factors that Gram-negative bacteria use to translocate proteins into the cytoplasm of eukaryotic host cells. Salmonellae encode two virulence-associated TTSS. The Salmonella pathogenicity island 1 (SPI1)-encoded TTSS is active on contact with host cells, whereas the Salmonella pathogenicity island 2 (SPI2)-encoded TTSS is expressed after phagocytosis of bacteria by host cells. Previously, no consensus signal sequence for translocation has been identified among TTSS effector proteins. In this work, seven proteins, termed Salmonella-translocated effectors (STE), are described that contain conserved amino acid sequences that direct translocation by TTSS in Salmonella typhimurium. STE that are coordinately regulated with SPI2 gene expression contain translocation signals that are recognized by the SPI2 but not by the SPI1 TTSS. STE that are constitutively expressed contain signals that direct translocation through both SPI1 and SPI2 TTSS. Of the seven STE examined, SspH1 and SspH2 have been previously shown to be translocated and involved in virulence; SlrP and SifA were identified as virulence factors, but were not previously known to be associated with TTSS; and SseI, SseJ, and SifB were previously unidentified. Three STE genes (sspH1, sspH2, and sseI) are located within temperate bacteriophages, suggesting a common mechanism for the dissemination of more recently evolved STE.
Virulence-associated type III secretion systems (TTSS) are complex macromolecular structures by which Gram-negative pathogens translocate effector proteins directly from the bacterial cytosol into the cytoplasm of eukaryotic cells (1). The TTSS apparatus spans the bacterial inner and outer membranes and exports proteins past the cell wall. Two classes of proteins are exported by TTSS: translocon and effector proteins. Translocon proteins are required for translocation of effectors across the eukaryotic plasma membrane and may also function as effectors themselves. Effector proteins function to alter host cell physiology and promote bacterial survival in host tissues (1).
Salmonellae encode two distinct virulence-associated TTSS within Salmonella pathogenicity islands 1 and 2 (SPI1 and SPI2) that have been implicated in different aspects of salmonellae pathogenicity (1). On contact with cultured cells, SPI1 TTSS translocates at least eight effector proteins that mediate several effects, including membrane ruffling, bacterial invasion, cell death, and transepithelial migration of neutrophils (1). Evidence suggests that these in vitro phenotypes are relevant to the ability of nontyphoidal salmonellae to cause gastroenteritis (2, 3). SPI1 is expressed in growth conditions that are consistent with the intestinal environment (4). This regulation depends on a variety of transcription factors encoded within and without SPI1 (4). In contrast, SPI2 expression is induced in phagocytosed bacteria and can be induced in vitro by conditions consistent with the phagosomal environment (i.e., growth in minimal medium with low magnesium or low pH) (5, 6). SPI2 mutants fail to replicate within macrophages and epithelial cells (7, 8), a defect that likely contributes to the avirulence of SPI2 mutants in mice (9). The mechanism by which SPI2 promotes bacterial virulence is unknown. Although few translocated effectors of SPI2 TTSS have been identified, one has been shown to promote intracellular survival by inhibition of phagosome–lysosome fusion (10).
Unlike proteins exported by the general secretory pathway, TTSS effector proteins are not proteolytically cleaved and have not previously been identified by conserved amino acid sequence motifs within their secretion/translocation signals (1). The mechanism by which TTSS recognize a diverse array of effectors with no apparent consensus signal sequence remains elusive. Data largely accumulated from the study of the TTSS-translocated Yop proteins of Yersinia indicate that the TTSS machinery recognizes effector proteins by two signals in their amino termini: a secretion signal and a translocation signal (11). Secretion signals have been mapped in several Yop proteins to sequences encoding the amino-terminal 15 amino acid residues (11). These signals are insensitive to frameshift mutations (12), suggesting that the recognized signal lies within the mRNA sequence. Translocation signals are located within the amino-terminal 50–100 amino acid residues of several Yop effectors (11, 13, 14). Chaperones specific for each Yop bind to the translocation signal and facilitate secretion and translocation of the effector (15).
SspH1, SspH2, and SlrP are recently identified Salmonella typhimurium virulence factors encoded outside SPI1 and SPI2 that contain leucine-rich repeats and similar amino- and carboxyl-terminal domains (16, 17). SspH1 contains a translocation signal recognized by both SPI1 and SPI2 TTSS, whereas SspH2 is an SPI2-restricted translocated protein. In this work, four ORFs, which likely encode Salmonella-translocated effectors (STE), have been identified on the basis of similarity to the SspH1/SspH2/SlrP amino termini.
Experimental Procedures
Bacterial and Eukaryotic Cell Line Growth Conditions.
Bacteria were grown overnight in LB with aeration. These bacteria were used for SPI2 translocation assays. For SPI1 expression and translocation assays, overnight cultures were diluted 1 to 50 or 1 to 100 in 2 ml of Luria–Bertani medium (LB) and grown 3 or 4 h with aeration. For SPI2 expression, bacteria were diluted 1 to 100 in 2 ml of N minimal medium with 0.2% glycerol and 8 μM MgCl2 and grown 16 h with aeration. RAW264.7 cells were grown as previously described (16).
DNA Techniques.
PCR and DNA sequence analyses were performed as described (16). sseI was sequenced from pEM55 (which was cloned from a size-selected genomic library) and various subclones. sifB was sequenced from DNA PCR amplified with Vent polymerase from S. typhimurium 14028s or Salmonella typhi Ty2. Sequences were confirmed with at least two independent reactions. Sequence comparisons were performed with the basic local alignment search tool (blast) (18).
Plasmid and Strain Construction.
Bacterial strains were constructed by P22HT int transduction as described (16). Primers were designed to amplify DNA sequences encoding putative secretion/translocation signals, and the resulting products were cloned into pMJH20 (16) to create various CyaA fusions. The SspH11–31,36–140CyaA (Δ32–35) fusion was created by splicing by overlap extension PCR (19). All resulting fusions have stop codons to terminate lacZ translation (cyaA is inserted into the multiple cloning site within the lacZ gene) and are expressed from their native ribosome binding sites and the lac promoter. The exception is LacZ21CyaA, which fuses the first 21 codons of the lacZ gene in pMJH20 to cyaA. sseI∷fluc was cloned in the vector pEM67 by digestion of pEM55 with EagI, blunting, digesting with EcoRI, and ligating into pGPLFR03 (16). pEM67 contains 1,616 bp of upstream sequence as well as the amino-terminal 61 codons of sseI. Primers were designed to PCR amplify sequences upstream of sseJ, sifB, sifA, and slrP to create the fluc transcriptional fusions in pEM87, 89, 88, and 86, respectively. Each vector contains sequences upstream of the methionine start codon (sseJ 990 bp, sifB 1,012 bp, sifA 367 bp, and slrP 620 bp) and sequences encoding amino-terminal residues (sseJ 147, sifB 143, sifA 140, and slrP 191 aa) that are terminated at the same stop codon within the pGPLFR03 vector. Resulting plasmids were introduced into CS019 by conjugation, and single chromosomal integration events that create gene duplications were confirmed by Southern blotting.
Enzymatic Assays.
Luciferase activity and β-galactosidase activity were determined from bacteria grown in culture or harvested from infected macrophages as described (16). Infected RAW264.7 macrophages were lysed in 1% Triton X-100, and half the sample was used to determine colony-forming units by dilutional plating, while the other half was mixed with 2× luciferase buffer (with 1% instead of 2% Triton X-100) for luciferase enzyme activity readings. fluc readings were taken for 30 s, whereas rluc readings were taken for 10 s. β-Galactosidase units were determined as described (20). RAW264.7 macrophages were infected with bacteria expressing CyaA fusion proteins at a multiplicity of infection of 10 as described (16). Infected macrophages were lysed in 0.1 M HCl and heated to 95°C for 5 min, and cAMP levels were determined by using the Direct cAMP Correlate-EIA Kit (Assay Designs, Ann Arbor, MI). cAMP levels were normalized for protein content of each sample as determined by the Bradford assay.
Results
Identification of Four Genes with Sequence Similarity to the SspH2/SspH1/SlrP Translocation Signal.
Southern blots of chromosomal DNA from several Salmonella isolates probed with DNA encoding the first 96 amino acid residues of sspH2 previously detected both sspH2 and a second hybridizing signal in several Salmonella isolates (16), suggesting that another gene contained sequences similar to the SspH2 amino terminus. DNA including this gene was cloned from a size-selected genomic DNA library and sequenced (GenBank accession no. AF236075). An ORF that encodes a putative 322-aa protein was identified and termed Salmonella-secreted effector I (sseI). The first 142 amino acid residues of SseI and SspH2 are 89% identical (Fig. 1). The remainder of SseI is not similar to SspH2 or any other known protein. sseI is preceded by 256 bp that are 99% identical to the DNA immediately upstream of sspH2. DNA sequences farther than 256 bp upstream of sseI have similarity to bacteriophage tail fiber and tail fiber assembly genes. blast searches of the S. typhimurium genome revealed that sseI lies within the Gifsy-2 prophage, which is competent for lytic growth and lysogeny of Gifsy-2 minus strains (21). sseI is absent from the currently available S. typhi genome sequence.
Figure 1.
Alignment of STE secretion/translocation signals. Residues in bold are high consensus; at least five of seven STE contain these residues. Residues in gray are low consensus; three or four of seven STE contain these residues. Residues that are not shaded have two or fewer of seven STE containing the residue. Alignments were performed with the blosum62 method (28). Residues that have similar properties are grouped as following: !, any one of IV; $, any one of LM; %, any one of FY; and #, any one of NDQE. The consensus sequence is shown under the STE alignment. The percent identity to SspH2 varies between proteins. SspH2142 is 89% identical to SseI142. SseJ111 is 38% identical to SspH2109. SifB115 is 28% identical to SspH2112. SifA113 is 32% identical to SspH2111. SspH2142 is 45% identical to SspH1139. SspH2128 is 37% identical to SlrP125.
The similarity of the amino termini of SseI, SspH1, SspH2, and SlrP suggested that several TTSS effectors in the salmonellae might use a common amino acid sequence to direct translocation (16). Therefore, blast searches of sequences generated in the S. typhimurium and S. typhi genome-sequencing projects (http://www.sanger.ac.uk/and http://genome.wustl.edu/gsc/) were performed. Three additional ORFs with sequence similarity to the amino termini of SspH1, SspH2, SlrP, and SseI were identified (Fig. 1). Two of these genes are previously unidentified and were termed sseJ and sifB. The third is sifA, a previously identified S. typhimurium virulence factor required for formation of Salmonella-induced filaments (sif) in epithelial cells (22). SifA had not been previously shown to be associated with SPI1 or SPI2 TTSS.
sseJ is predicted to encode a 408-aa protein. Amino acid residues 144–408 of the protein are 30% and 29% identical to glycerophospholipid:cholesterol acyltransferase (GCAT) and lecithinase encoded by Aeromonas and Vibrio species, respectively (23, 24). The catalytic triad and five conserved blocks within GCAT, lecithinase, and other related acyltransferases (25, 26) are present in SseJ. sseJ is not present in the currently available S. typhi genome sequence.
sifB was identified in the S. typhi and S. typhimurium genome sequences; however, at the time of this publication, the S. typhimurium sifB gene was not completely sequenced, and the reported gene in the S. typhi database was predicted to encode a 143-aa protein. Sequences of this chromosomal region were also reported for S. typhimurium in a study describing the pcgL gene that lies 1.5 kb upstream of sifB (27). This sequence predicts that sifB encodes a 10-aa protein. To determine the sequence of sifB, DNA sequences containing sifB were PCR amplified from S. typhimurium 14028s and S. typhi Ty2, and the products from multiple independent reactions were sequenced (GenBank accession nos. AF236076 and AF236077). Frameshift mutations, which predict premature termination of SifB, exist between our data and the S. typhi genome and pcgL region sequences. Our sequence analysis reveals that sifB is predicted to encode 316-aa proteins in S. typhimurium and S. typhi that are 98% identical. SifB is 30% identical to SifA over the length of the two proteins. The effector domains of SifB and SifA are not similar to known proteins.
An alignment of the putative translocation signals of all seven proteins is shown in Fig. 1. Notably, five proteins contain consensus residues in the amino-terminal 15 amino acid residues, which are not shared by SspH1 and SlrP. The most conserved cluster occurs at amino acid residues 34–41 of the alignment, where a WEK(I/M)XXFF motif is present in all seven proteins (with a few conservative changes). Striking clustering of conserved residues also occurs at amino acid residues 48–58 and 104–118 in the alignment.
sseI, sseJ, sifA, and sifB Expression in SPI2-Inducing Media Depends On SsrA/SsrB.
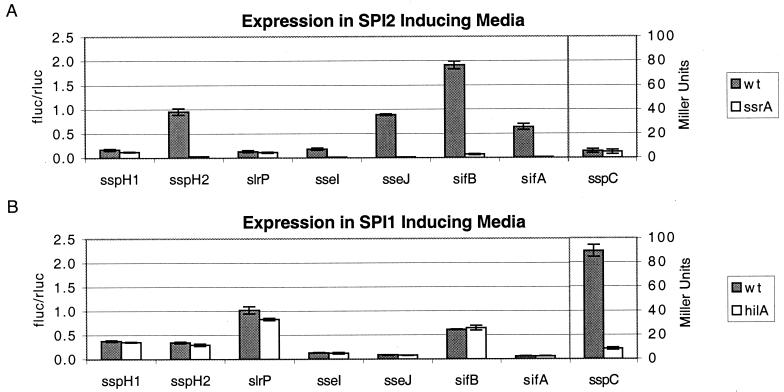
The similarity of the amino termini of SlrP, SseI, SseJ, SifA, and SifB to the SspH1 and SspH2 translocation signals suggested that these proteins are also translocated by TTSS. As coordinate regulation with TTSS would support this hypothesis, transcriptional fusions to the firefly luciferase gene (fluc) were created in the pGPLFR03 suicide vector and integrated into the S. typhimurium chromosome. pGPLFR03 also encodes a constitutively expressed Renilla luciferase (rluc) gene that permits normalization of Fluc activity.
S. typhimurium strains expressing various STE∷fluc transcriptional fusions were grown in media that induce SPI1 or SPI2 gene expression. Expression of the fusions in a wild-type background was compared with expression in strains carrying regulatory mutations in either the SPI1 (hilA) or SPI2 (ssrA) regulon. Expression of five STE (sspH2, sseI, sseJ, sifA, and sifB) in a medium that induces SPI2 gene expression depended on the SPI2-encoded two-component regulatory system SsrA/SsrB (Fig. 2). The fold regulation ranged from 31 to 156, depending on the promoter being examined. Expression of the STE in SPI1-inducing media was not dependent on HilA, whereas expression of a component of the SPI1 translocon, sspC (29), was highly dependent on HilA as has been previously shown. Similar to sspH1, slrP expression was not regulated by either HilA or SsrA/SsrB.
Figure 2.
Regulation of STE expression. S. typhimurium wild-type or regulatory mutants expressing transcriptional fusions of STE genes to firefly luciferase in the pGPLFR03 expression vector were grown overnight in LB and back diluted in SPI2-inducing conditions (A) or SPI1-inducing conditions (B), and firefly luciferase activity was determined and normalized for cell number by dividing by the activity of the constitutively expressed Renilla luciferase. sspC∷Tn5-lacZY expression is presented as Miller units (as a control for SPI1-regulated gene expression).
Determination of a Minimal Translocation Signal for SspH2.
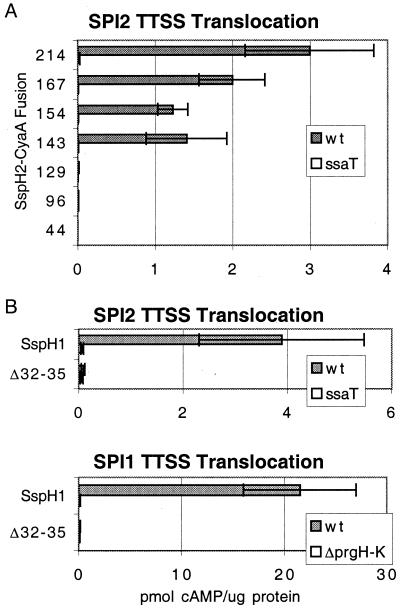
Previous work using fusions to the Bordetella pertussis cyaA gene as a translocation reporter determined that the amino-terminal 214 and 208 amino acids of SspH2 and SspH1 are sufficient for translocation (16). In this assay, putative translocation signals are fused to the catalytic domain of the CyaA adenylate cyclase toxin of B. pertussis and expressed from the lac promoter on a low-copy plasmid. When in the presence of calmodulin in the eukaryotic cell cytosol, CyaA catalyzes the conversion of ATP to cAMP. Thus, increases in cAMP levels in eukaryotic cells correlate with the translocation of CyaA fusion proteins during S. typhimurium infection (30). Translocation by SPI1 and SPI2 TTSS can be independently examined by analyzing bacteria grown in media that induce only one of the two TTSS (16). Because the similarity of the seven proteins is limited to the amino-terminal 142 amino acids or less, the minimal translocation signal for SspH2 was determined.
Various CyaA fusions to SspH2 were created and analyzed for evidence of SPI2 TTSS translocation. Intracellular cAMP levels were determined in RAW264.7 macrophages infected with S. typhimurium expressing SspH2-CyaA fusions under conditions that induce SPI2 but not SPI1 TTSS activity (16). S. typhimurium expressing CyaA fusions to 143 or more residues of SspH2 were able to cause host cell cAMP levels to rise, whereas expression of shorter fusions did not raise intracellular cAMP levels (Fig. 3A), despite comparable production of protein and adenylate cyclase activity (data not shown). These increases were dependent on ssaT, a component of the SPI2 TTSS (9). The data suggest that shorter fusions are insufficient to direct translocation, and that proteins with sequences similar to the amino-terminal 143 residues of SspH2 can also be translocated by the SPI2 TTSS. In agreement with this hypothesis, SspH1140CyaA (residue 140 of SspH1 aligns with residue 143 of SspH2) was competent for translocation by both SPI1 and SPI2 TTSS (Fig. 3 B and C).
Figure 3.
Analysis of residues required for translocation. Wild-type S. typhimurium or strains carrying mutations in SPI2 TTSS (ssaT) or SPI1 TTSS (ΔprgH-K) expressing various CyaA fusions were used to infect RAW264.7 cells at a multiplicity of infection (MOI) of 10. Macrophages were infected for 1 h with bacteria in late logarithmic growth for SPI1 TTSS expression (C) or for 1 h with late stationary phase bacteria plus 5 or 6 h of gentamycin treatment for SPI2 TTSS expression (A and B). Infected macrophages were lysed in 0.1 M HCl; cellular cAMP levels were determined by enzyme immunoassay and were normalized for protein content determined by the Bradford assay and are presented as pmol of cAMP per μg of protein. SspH1140 and SspH11–31,36–140CyaA (Δ32–35) were used in B and C. Note that cAMP production may be variable between assays, and infection of bacteria expressing SspH1140CyaA results in cAMP increases that are similar to those seen with SspH1208CyaA in a direct comparison (data not shown).
Deletions Within the Conserved WEK(I/M)XXFF Motif Eliminate the Ability of SspH1-CyaA Fusions to Be Translocated.
To determine whether residues conserved among the seven STE are required for translocation by SPI1 and/or SPI2 TTSS, a SspH1140CyaA fusion carrying a deletion in the WEKIQVFF motif was created. This construct deletes residues 32–35 of SspH1, EKIQ, removing the conserved glutamate, lysine, and isoleucine residues. Although the Δ32–35 construct and SspH1140CyaA were expressed at similar levels, expression of the deletion mutant had no effect on intracellular cAMP levels in either SPI1 or SPI2 TTSS translocation assays (Fig. 3 B and C), suggesting that the conserved residues are essential for translocation by both SPI1 and SPI2 TTSS.
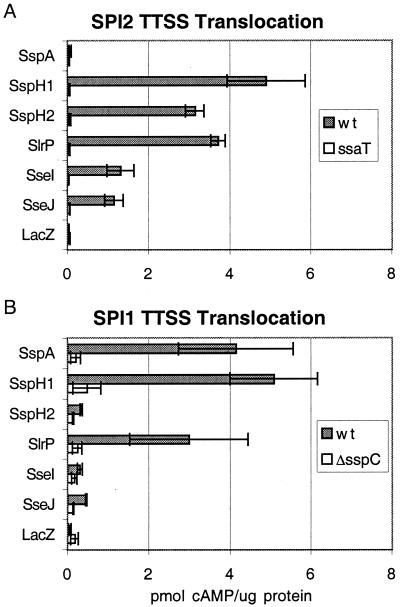
The Conserved Amino Termini of SlrP, SseI, and SseJ Function as TTSS Translocation Signals.
To determine whether SseI, SseJ, and SlrP are translocated by TTSS, CyaA reporter fusions were constructed for the putative translocation signals of each protein. When RAW264.7 macrophages were infected with S. typhimurium expressing SspH1-, SspH2-, SlrP-, SseI-, and SseJ-CyaA under SPI2-inducing conditions, intracellular cAMP levels increased (Fig. 4A). These increases were dependent on ssaT, indicating that these proteins contain SPI2 TTSS translocation signals. In contrast, infection of macrophages with strains expressing SspA-CyaA (an SPI1 TTSS-translocated effector) or LacZ-CyaA did not alter host cell cAMP levels (Fig. 4A). A variety of CyaA fusions to the amino termini of SifA and SifB were constructed, and all were unstable on expression in Salmonella. Infection of macrophages with bacteria expressing these fusions did not result in increases in intracellular cAMP in either SPI1 or SPI2 TTSS assays (data not shown).
Figure 4.
Translocation of STE-CyaA fusion proteins. Wild-type S. typhimurium or strains carrying mutations in SPI2 TTSS (ssaT) or SPI1 TTSS (ΔsspC) expressing various CyaA fusions were used to infect RAW264.7 cells as described in Fig. 3, assaying translocation by SPI2 (A) or SPI1 (B) TTSS. SspH1208, SspH2214, SlrP191, SseI206, SseJ147 and SspA158 and LacZ21CyaA fusions were analyzed.
SspH1 is translocated by both SPI1 and SPI2 TTSS (16). Because the SlrP translocation signal is most similar to SspH1 (Fig. 1), SlrP was hypothesized also to be translocated by SPI1 TTSS. To analyze translocation of various STE by SPI1 TTSS, RAW264.7 macrophages were infected for 1 h with bacteria expressing various STE-CyaA fusions under conditions where SPI1 TTSS is expressed in the absence of SPI2 TTSS activity. When bacteria expressing SspA-, SlrP-, and SspH1-CyaA were used to infect RAW264.7 macrophages, host cell cAMP levels rose (Fig. 4B). These increases were eliminated when the CyaA fusions were expressed in bacteria that are defective for the SPI1 TTSS translocon protein SspC. Bacteria expressing SspH2-, SseI-, SseJ-, and LacZ-CyaA fusions did not stimulate cAMP increases in infected cells under these conditions (Fig. 4B).
Discussion
This work describes a family of TTSS effectors that have similar translocation signal domains. Evidence suggests that the seven proteins that constitute this family are translocated across the phagosome membrane by the SPI2 TTSS after S. typhimurium are phagocytosed by mammalian cells. Two of these family members, SspH1 and SlrP, are also translocated across the plasma membrane by the SPI1 TTSS, which functions on contact with mammalian cells. Therefore, this work extends the complexity and diversity of type III secretion by salmonellae.
In addition to containing translocation signals recognized by both virulence-associated TTSS, SspH1 and SlrP are also distinct from the other family members in that their expression is not regulated by the SPI2 regulators SsrA/SsrB. This, and the fact that they are not regulated by the SPI1 regulator HilA, suggests that these two genes may be constitutively expressed, allowing their protein products to be available for translocation when bacteria reside within either SPI1- or SPI2-inducing environments. The amino acid sequence of SspH1 and SlrP also sets them apart from other family members. Although they do contain the essential WEK(I/M)XXFF motif located in the translocation domain, they lack the consensus residues present in the amino-terminal nine residues of the other family members. SspH1 and SlrP both contain a conserved sequence, MFNIXNXQ, at the amino terminus, whereas the remainder of the family contains a MPX(I/V)GXGX(L/F) sequence. This difference could be crucial to conferring recognition by the SPI1 TTSS in addition to the SPI2 TTSS. This specificity may not be conferred at the amino acid level, as sequences encoding the amino-terminal 15 amino acid residues have been shown to contain frameshift-insensitive signals for secretion in Yersinia, suggesting an mRNA secretion signal (12). The similarity seen at the amino acid level of the amino-terminal eight or nine residues could be a result of similar mRNA secondary structures with the corresponding bases. Indeed, of 27 nucleotides from −3 to +24 in sspH1 and slrP, 22 are identical. Of the six conserved amino acid residues in the amino-terminal eight residues of SspH1 and SlrP, all but one have identical codons. Therefore, the additional ability of members of this family to be translocated by the SPI1 TTSS could require a specific mRNA signal.
Despite the size and diversity of this family, it is clear that the conserved translocation domain is not essential for translocation by the SPI2 TTSS, because at least one translocated effector, SpiC (10), does not share any homology with the family. In addition, other SPI2-encoded proteins that are hypothesized to be translocated (SseA–G) have unique amino termini, as is typical of all TTSS effectors characterized to date (7, 31). In general, TTSS effectors have unique chaperones that bind to the translocation domain and facilitate translocation, seemingly obviating the need for a common signal (15). Therefore, the conservation observed suggests that these seven effectors could share a chaperone.
All of the STE identified are located outside of the pathogenicity islands and therefore may have been acquired after the pathogenicity islands through horizontal transmission. The guanine plus cytosine (G + C) content of the genes encoding the seven family members is variable, ranging from 38% in sifB to 55% in sspH2, indicating that these effectors may have been acquired at different times or from different sources. Those with low G + C content could have been acquired more recently by horizontal transmission. Horizontal gene transmission by temperate bacteriophages is an efficient means of dissemination of genetic information. sseI, sspH1, and sspH2 appear to be located within lysogenic bacteriophages. sspH2 is in the midst of sequences that could include genes for an as-yet-uncharacterized bacteriophage. sseI and sspH1 are within the Gifsy-2 (21) and Gifsy-3 prophages (Linello Bossi, personal communication), respectively. Both of these prophages are competent for lytic growth and lysogeny of naive Salmonella. This suggests that sseI and sspH1 are currently disseminating throughout the salmonellae. Indeed, sspH1 is present in only a minority of Salmonella serotypes, whereas sspH2 is more prevalent (16, 17). Several examples of bacteriophage-mediated horizontal transmission of virulence factors exist (32). In salmonellae, SopEφ carries the SPI1 TTSS-translocated effector sopE and is capable of lytic growth and lysogeny of naive strains (33). The virulence-associated superoxide dismutase gene sodC1 is also within Gifsy-2 (21). Thus, in Salmonella, bacteriophage-mediated transfer of TTSS effectors and other virulence factors appears to be a common mechanism for acquisition of new pathogenic traits.
The G + C content of the translocation and effector domains within each gene is very similar for all family members except sseI. The sseI translocation-encoding domain has a 51% G + C content, whereas the effector-encoding domain has a 33% G + C content. This suggests that a mechanism for the evolution of new TTSS effectors could involve reassortment between DNA sequences encoding a translocation signal and an effector domain. In support of this hypothesis, sseI and sspH2 share 96% identity in their DNA sequences from 256 nucleotides upstream of the start codon through the end of the translocation domain. This suggests that the 5′ region and sequences encoding the translocation signal were brought together with another effector domain by a genetic event. One could also speculate that a similar mechanism could have generated sseJ. The carboxyl-terminal domain of SseJ is highly similar to GCAT and lecithinase, proteins secreted by the general secretory pathway in Aeromonas and Vibrio species. It is possible that a genetic event replaced the sec-dependent signal sequence with the SPI2 translocation signal to create sseJ. Therefore, salmonellae appear to be able to use a variety of genetic mechanisms to disseminate and evolve genes encoding TTSS effectors. This suggests that Gram-negative bacteria could emerge with different or unique virulence characteristics because of the dissemination and amplification of conserved translocation domains.
SPI2 was likely introduced into the salmonellae as a single genetic horizontal transmission event. Strains carrying SPI2 are more virulent and likely grew in the overall population because of their selective advantage. What is the advantage conferred by the acquisition of TTSS effectors? For SpiC, the inhibition of phagosome–lysosome fusion promotes bacterial replication within eukaryotic cells. Other effectors could then have been introduced to further alter phagosome trafficking and killing mechanisms or to effect other mammalian cell processes. SifA likely also functions to alter the Salmonella-containing vacuole as it promotes the formation of filamentous endosomal structures in epithelial cells. SifB shares sequence similarity with SifA, suggesting that it also may have similar ancestry and/or molecular function. Dissecting the roles of this family of proteins in conferring a selective advantage to salmonellae within its mammalian environments should lead to greater understanding of how bacteria exploit host functions and how bacterial pathogens can emerge with different properties.
Acknowledgments
We thank Andreas Bäumler and Reneé Tsolis for critical discussion regarding SlrP, Linello Bossi for sharing unpublished data regarding Gifsy-3, Christina Scherer and Philip Bronstein for critical review of this manuscript, and Philip Bronstein for creation of SspA-CyaA. This work was supported by the Poncin Scholarship Fund (to E.A.M.) and by Grant RO1 AI30479 (to S.I.M.) from the National Institutes of Health.
Abbreviations
- TTSS
type III secretion system
- SPI
Salmonella pathogenicity island
- STE
Salmonella-translocated effector
- Sse
Salmonella-secreted effector
- Sif
Salmonella-induced filament
- G + C
guanine plus cytosine
Footnotes
References
- 1.Hueck C J. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsolis R M, Adams L G, Ficht T A, Bäumler A J. Infect Immun. 1999;67:4879–4885. doi: 10.1128/iai.67.9.4879-4885.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones M A, Wood M W, Mullan P B, Watson P R, Wallis T S, Galyov E E. Infect Immun. 1998;66:5799–5804. doi: 10.1128/iai.66.12.5799-5804.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rakeman J L, Miller S I. Trends Microbiol. 1999;7:221–223. doi: 10.1016/s0966-842x(99)01514-0. [DOI] [PubMed] [Google Scholar]
- 5.Deiwick J, Nikolaus T, Erdogan S, Hensel M. Mol Microbiol. 1999;31:1759–1773. doi: 10.1046/j.1365-2958.1999.01312.x. [DOI] [PubMed] [Google Scholar]
- 6.Lee A K, Detweiler C S, Falkow S. J Bacteriol. 2000;182:771–781. doi: 10.1128/jb.182.3.771-781.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hensel M, Shea J E, Waterman S R, Mundy R, Nikolaus T, Banks G, Vazquez-Torres A, Gleeson C, Fang F C, Holden D W. Mol Microbiol. 1998;30:163–174. doi: 10.1046/j.1365-2958.1998.01047.x. [DOI] [PubMed] [Google Scholar]
- 8.Cirillo D M, Valdivia R H, Monack D M, Falkow S. Mol Microbiol. 1998;30:175–188. doi: 10.1046/j.1365-2958.1998.01048.x. [DOI] [PubMed] [Google Scholar]
- 9.Hensel M, Shea J E, Gleeson C, Jones M D, Dalton E, Holden D W. Science. 1995;269:400–403. doi: 10.1126/science.7618105. [DOI] [PubMed] [Google Scholar]
- 10.Uchiya K, Barbieri M A, Funato K, Shah A H, Stahl P D, Groisman E A. EMBO J. 1999;18:3924–3933. doi: 10.1093/emboj/18.14.3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sory M P, Boland A, Lambermont I, Cornelis G R. Proc Natl Acad Sci USA. 1995;92:11998–12002. doi: 10.1073/pnas.92.26.11998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anderson D M, Schneewind O. Science. 1997;278:1140–1143. doi: 10.1126/science.278.5340.1140. [DOI] [PubMed] [Google Scholar]
- 13.Schesser K, Frithz-Lindsten E, Wolf-Watz H. J Bacteriol. 1996;178:7227–7233. doi: 10.1128/jb.178.24.7227-7233.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boland A, Sory M P, Iriarte M, Kerbourch C, Wattiau P, Cornelis G R. EMBO J. 1996;15:5191–5201. [PMC free article] [PubMed] [Google Scholar]
- 15.Wattiau P, Woestyn S, Cornelis G R. Mol Microbiol. 1996;20:255–262. doi: 10.1111/j.1365-2958.1996.tb02614.x. [DOI] [PubMed] [Google Scholar]
- 16.Miao E A, Scherer C A, Tsolis R M, Kingsley R A, Adams L G, Bäumler A J, Miller S I. Mol Microbiol. 1999;34:850–864. doi: 10.1046/j.1365-2958.1999.01651.x. [DOI] [PubMed] [Google Scholar]
- 17.Tsolis R M, Townsend S M, Miao E A, Miller S I, Ficht T A, Adams L G, Bäumler A J. Infect Immun. 1999;67:6385–6393. doi: 10.1128/iai.67.12.6385-6393.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 19.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 20.Rakeman J L, Bonifield H R, Miller S I. J Bacteriol. 1999;181:3096–3104. doi: 10.1128/jb.181.10.3096-3104.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Figueroa-Bossi N, Bossi L. Mol Microbiol. 1999;33:167–176. doi: 10.1046/j.1365-2958.1999.01461.x. [DOI] [PubMed] [Google Scholar]
- 22.Stein M A, Leung K Y, Zwick M, Garcia-del Portillo F, Finlay B B. Mol Microbiol. 1996;20:151–164. doi: 10.1111/j.1365-2958.1996.tb02497.x. [DOI] [PubMed] [Google Scholar]
- 23.MacIntyre S, Buckley J T. J Bacteriol. 1978;135:402–407. doi: 10.1128/jb.135.2.402-407.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shinoda S, Matsuoka H, Tsuchie T, Miyoshi S, Yamamoto S, Taniguchi H, Mizuguchi Y. J Gen Microbiol. 1991;137:2705–2711. doi: 10.1099/00221287-137-12-2705. [DOI] [PubMed] [Google Scholar]
- 25.Upton C, Buckley J T. Trends Biochem Sci. 1995;20:178–179. doi: 10.1016/s0968-0004(00)89002-7. [DOI] [PubMed] [Google Scholar]
- 26.Brumlik M J, Buckley J T. J Bacteriol. 1996;178:2060–2064. doi: 10.1128/jb.178.7.2060-2064.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hilbert F, del Portillo F G, Groisman E A. J Bacteriol. 1999;181:2158–2165. doi: 10.1128/jb.181.7.2158-2165.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Corpet F. Nucleic Acids Res. 1988;16:10881–90. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bajaj V, Lucas R L, Hwang C, Lee C A. Mol Microbiol. 1996;22:703–714. doi: 10.1046/j.1365-2958.1996.d01-1718.x. [DOI] [PubMed] [Google Scholar]
- 30.Sory M P, Cornelis G R. Mol Microbiol. 1994;14:583–594. doi: 10.1111/j.1365-2958.1994.tb02191.x. [DOI] [PubMed] [Google Scholar]
- 31.Beuzon C R, Banks G, Deiwick J, Hensel M, Holden D W. Mol Microbiol. 1999;33:806–816. doi: 10.1046/j.1365-2958.1999.01527.x. [DOI] [PubMed] [Google Scholar]
- 32.Miao E A, Miller S I. Proc Natl Acad Sci USA. 1999;96:9452–9454. doi: 10.1073/pnas.96.17.9452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mirold S, Rabsch W, Rohde M, Stender S, Tschape H, Russmann H, Igwe E, Hardt W D. Proc Natl Acad Sci USA. 1999;96:9845–9850. doi: 10.1073/pnas.96.17.9845. [DOI] [PMC free article] [PubMed] [Google Scholar]