Abstract
The highly organized structure of M13 bacteriophage was used as an evolved biological template for the nucleation and orientation of semiconductor nanowires. To create this organized template, peptides were selected by using a pIII phage display library for their ability to nucleate ZnS or CdS nanocrystals. The successful peptides were expressed as pVIII fusion proteins into the crystalline capsid of the virus. The engineered viruses were exposed to semiconductor precursor solutions, and the resultant nanocrystals that were templated along the viruses to form nanowires were extensively characterized by using high-resolution analytical electron microscopy and photoluminescence. ZnS nanocrystals were well crystallized on the viral capsid in a hexagonal wurtzite or a cubic zinc blende structure, depending on the peptide expressed on the viral capsid. Electron diffraction patterns showed single-crystal type behavior from a polynanocrystalline area of the nanowire formed, suggesting that the nanocrystals on the virus were preferentially oriented with their [001] perpendicular to the viral surface. Peptides that specifically directed CdS nanocrystal growth were also engineered into the viral capsid to create wurtzite CdS virus-based nanowires. Lastly, heterostructured nucleation was achieved with a dual-peptide virus engineered to express two distinct peptides within the same viral capsid. This work represents a genetically controlled biological synthesis route to a semiconductor nanoscale heterostructure.
Exploiting the biologically evolved, self-assembling structures of viral capsids is a possible mechanism in the ordering of technologically important materials. A key characteristic of materials synthesized by natural biological systems is the hierarchical organization of structures on many length scales with controlled size, shape, alignment, and orientation (1–8). There has been much interest in using biological templates in vitro to modulate the growth of inorganic, semiconductor, magnetic, and other technologically important materials (9–14). Our previous work, along with that of others, has shown that biological combinatorial approaches can identify amino acid sequences capable of interacting with a variety of materials (11, 15). Here, we present a system that exploits the self-assembling, crystalline structure of the viral coat to nucleate and template II-VI semiconductor nanocrystals into highly oriented quantum dot nanowires. To accomplish this, the pVIII major coat protein was engineered to display evolutionary-selected peptides, enabling directed nanocrystal growth into viral–semiconductor hybrid nanowires that were crystallographically oriented over the micrometer size range.
The helical major coat protein, pVIII, of viruses can be engineered to express fusion proteins that readily self-assemble into a highly oriented viral coat structure. A 5-fold axis of rotation relates symmetric points on the coat (16, 17). Peptide sequences engineered into the pVIII protein for use as nanocrystal templates were selected based on affinity toward either ZnS or CdS, specificity for ZnS over CdS (or vice versa), and the ability to affect nucleation and growth of nanocrystals during synthesis (Fig. 6 and Supporting Text, which are published as supporting information on the PNAS web site, www.pnas.org). We have previously identified a conformationally constrained peptide, termed A7 (Cys-Asn-Asn-Pro-Met-His-Gln-Asn-Cys) and an unconstrained peptide, Z8 (Val-Ile-Ser-Asn-His-Ala-Glu-Ser-Ser-Arg-Arg-Leu), both of which specifically recognize and control growth of ZnS. For example, MS data indicate that the A7 peptide, with the disulfide bond formed, bound four Zn(II) ions with at least 3 orders of magnitude more strongly than the S2– (Fig. 7, which is published as supporting information on the PNAS web site; see Supporting Text). Further, we have demonstrated that A7–ZnS viral semiconductor complexes can be assembled into self-supporting thin films (12). Similarly, the peptide J140, with a sequence of Ser-Leu-Thr-Pro-Leu-Thr-Thr-Ser-His-Leu-Arg-Ser, was selected for recognition of single-crystal CdS and nucleation control over CdS. Using high-resolution analytical electron microscopy and photoluminescence (PL), we characterized collections of single M13 phage particles engineered to express semiconductor nucleating peptides upon the viral capsid that direct highly oriented, phase-specific nanocrystal growth. This approach combines the use of perfectly ordered and assembled natural viral templates with the flexibility of genetic control over material synthesis to assemble semiconductor wires in supramolecular hybrid structures.
Materials and Methods
Genetic Engineering for pVIII Peptide Display. Primers encoding the A7 and Z8 peptides were used to PCR-amplify the major coat pVIII gene from a derivative of pfdisplay8 (18) (a gift of Richard Perham, University of Cambridge, Cambridge, U.K.) such that a Gly-Gly-Gly-Ser flexible linker separated the peptides from Asp+4 of the mature pVIII sequence. The extended pVIII genes were separately cloned into pAK400 (19) (a gift of Andreas Plückthun, University of Zurich, Zurich) such that the fusion proteins were under lac promoter control and targeted to the inner membrane by using the pelB leader. The ORF was verified by using Applied Biosystems cycle sequencing. The vectors pAKdisplay.A7 and pAKdisplay.Z8 were separately rescued by superinfection of Escherichia coli TG-1 with M13KO7, and peptide display was induced with 1 mM isopropyl β-D-thiogalactoside in glucose-free terrific broth at 25°C overnight with chloramphenicol and kanamycin selection. Cultures were clarified by centrifugation and filtration through 0.22-μm filters, and the resulting phage was precipitated with polyethylene glycol twice. A control vector displaying the minimal FLAG peptide Asp-Tyr-Lys-Asp was used to confirm peptide display with anti-FLAG M1/anti-M13 HRP capture ELISA. Coomassie staining of a 17% Laemmli PAGE gel of 1010 transforming units indicated both A7 and Z8 peptides were displayed by virtue of retardation of pVIII migration when compared with WT phage and increased propensity to stain. CdS peptides were displayed in a similar manner. The pVIII-J140 fusion peptide, with the linker Gly-Ala-Ser-Gly-Ala, was cloned into pMoPac32, an ampicillin-resistant derivative of pAK400, to generate pMoPac32.J140.
To generate virus constructs simultaneously displaying A7 and J140 peptides the above superinfection scheme was applied to TG-1 cells bearing both pAKdisplay.A7 and pMoPac32.J140. Although the vectors both have a pUC origin, the presence of different antibiotic selection markers ensured both were maintained in the short term for double display under chloramphenicol and ampicillin selection. This approach avoided virus genome engineering and is presumably limited only by the number of different resistance markers available for multiplex display on the same virus construct (18, 20, 21).
Inorganic Synthesis. Twenty microliters of engineered viruses (8 × 1013 plaque-forming units/ml) was placed in a 1-ml aqueous ZnCl2 solution (1 mM) and allowed to interact for 12 h before introduction of 1 ml aqueous Na2S (1 mM). The mixed solution temperature was lowered to either 0°C or —25°C for 24 h for the virus–ZnS interaction, followed by aging at room temperature for 12 h. CdCl2 was used as Cd(II) source in the synthesis of CdS nanowires. The viral nanowires were transferred to a carbon film-supported copper transmission electron microscopy (TEM) grid, rinsed in the deionized water, dried in a dessicator, and visualized by a JEOL 2010F field emission transmission electron microscope with scanning capabilities and an Oxford (High Wycombe, U.K.) energy dispersive x-ray spectroscopy (EDS) detector system. The viral nanowires were also transferred to silicon wafer substrates and characterized by PL.
Electron Microscopy Methods. The JEOL 2010F electron microscope was operated at an accelerating voltage of 200 kV unless otherwise stated, in different modes including conventional TEM, electron diffraction (ED), high-resolution TEM (HRTEM), scanning TEM (STEM), and high-resolution EDS. Whereas HRTEM images were formed by phase contrast, annular dark field (ADF) STEM images were atomic number (Z) contrast images. Under ADF STEM mode, high-angle scattered electrons were a result of scattering by nuclei, and the probability of an electron being scattered to such high angles by an atom was approximately proportional to (Z1.7) (22). EDS elemental mapping was performed under ADF STEM mode with a larger scanning electron probe size (3 nm) to ensure a high EDS signal-to-noise ratio. High-angle ADF (HAADF) STEM mode with a smaller scanning electron probe size (0.5 nm) was applied when imaging nanocrystals bound to the viruses. To ensure the correspondence between ED pattern and nanowire STEM image, the image rotation between TEM mode and STEM mode was calibrated by using a Gatan (Pleasanton, CA) digital micrograph while STEM images were acquired.
PL Methods. The viral CdS nanowires were transferred to a Si(100) wafer, rinsed, dried, and characterized by PL. The wafer was then mounted in a liquid-He flow Cryostat. The second harmonic wave of a mode-locked Ti:sapphire laser was used as an excitation source (350 nm). The pulse width was several picoseconds. A conventional far-field micro-PL system was used to collect the PL signal. The PL polarization of the nanowires was also characterized. Because the nanowires on the Si substrate were randomly distributed, a nanowire could be readily found whose long axis was either parallel or perpendicular to the polarization direction of the linearly polarized excitation light. Otherwise, the polarization direction of the excitation light could be rotated by a right angle by using a waveplate to perform the polarization experiment. The PL was collected by a microobjective, passed through a polarization analyzer, and detected by an Acton (Acton, MA) 0.5-m monochromator equipped with a liquid nitrogen-cooled charge-coupled-device array detector mounted on the exit port of the imaging spectrograph. A depolarizer was placed before the entrance slit of the monochromator to remove the polarization-dependent response of the monochromator and detector. All of the PL spectra were corrected for the spectral response of the apparatus. The PL experiment was performed at T = 6 K, with an average excitation intensity of ≈2 mW/cm2. The resolution of the PL emission spectrum was ≈0.05 nm. The spatial resolution of PL image mode was ≈200 nm.
Results and Discussion
Phagemid constructs, which contained bacterial phagemid DNA encased in a viral protein coat, were used as the basic building block of the viral wire semiconductor template (Fig. 1a). The A7, Z8, and J140 peptide nucleotide sequences were fused to the fd gene VIII and expressed from a phagemid vector, resulting in a maximum of one peptide displayed per pVIII protein. As expected, the length of the virus was proportional to the genome packaged in the vector (Fig. 8, which is published as supporting information on the PNAS web site; Supporting Text). Because of the viral assembly process, the peptide–pVIII fusion proteins were spatially interspersed with WT pVIII (nonengineered pVIII) during expression of the pVIII proteins that assemble to form the capsid coat (18), affording a template for the nucleation of materials into a wire-like structure.
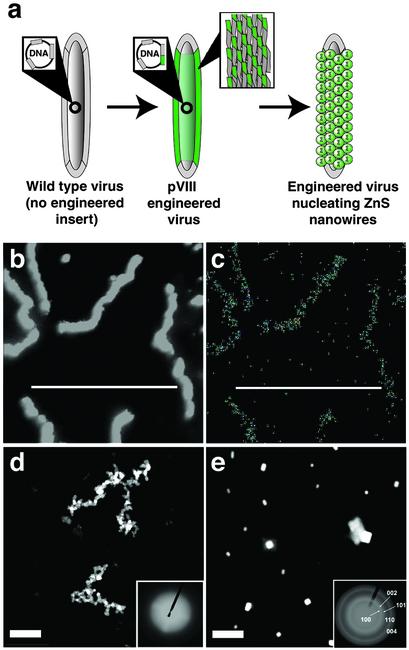
Fig. 1.
Images of A7–pVIII-engineered viruses directing ZnS nanocrystal synthesis at 0°C. (a) Illustration depicting A7 peptide expression on the pVIII protein upon phage amplification and assembly, then subsequent nucleation of ZnS nanocrystals. Call-outs depict insertion of the A7 nucleotide sequences, resulting in A7 fusion protein shown as green-shaded areas. Additionally, the call-out of the engineered virus shows detail of the WT pVIII protein (gray) and the A7-engineered pVIII protein (green) composing the viral coat. (b) ADF STEM images showing the morphology of the ZnS-virus nanowires. The ZnS-virus nanowires were 560 nm long and 20 nm wide. (c) EDS mapping of S (an identical EDS mapping of Zn can be found in Supporting Text). (d) Typical HAADF STEM images of interconnected and branched viral nanowires formed through the specific binding of preformed ZnS noncrystalline nanoparticles on the viruses expressing the A7 peptide. (Inset) ED pattern of the nanowires. (e) HAADF STEM image of the control experiment using the A7 peptide-engineered viruses in CdS nanocrystal growth conditions. No viral wires were visible, only randomly precipitated large CdS particles. (Inset) ED pattern of control experiment. (Scale bars: 800 nm.)
For the A7-displaying constructs incubated at 0°C, ZnS nanocrystals were nucleated along the length of the viruses, forming nanowires. These constructs were imaged by ADF STEM, also referred to as Z-contrast imaging (Fig. 1b) (22). The contrast arose from average atomic number differences between semiconductor–viral complexes and the carbon support film. EDS elemental mapping (Fig. 1c) confirmed ZnS formation along the length of the virus. The EDS spectra collected from the whole area of Fig. 1b verified ZnS formation with little excess NaCl precipitation on the wires (Fig. 9, which is published as supporting information on the PNAS web site; Supporting Text).
Important control experiments were performed. To differentiate between peptide-induced nanocrystal ZnS nucleation and postgrowth ZnS nanocrystal binding, noncrystalline ZnS particles were preformed at room temperature then incubated with the engineered viruses at 0°C. As expected, the viruses bound the preformed particles, but did not form oriented crystalline structures (Fig. 1d). These results are consistent with the notion that the oriented crystalline nanocrystals observed with the A7 fusion in Fig. 1b were a result of peptide-nucleated crystallization of ZnS on the engineered viruses, not binding of preformed particles. In another control experiment, A7–pVIII-engineered viruses were interacted with CdS precursor solutions. No nanowires were observed under TEM/STEM (Fig. 1e). Large CdS precipitates were visible, similar to what was observed in the absence of engineered peptide. Additionally, nanowires were not observed in control experiments using WT constructs (no engineered peptide fusion) incubated in either salt precursor solution.
For the 0°CA7–pVIII ZnS synthesis, HAADF STEM imaging with a scanning electron probe of 0.5 nm revealed that the ZnS nanocrystals (3–5 nm) were formed along the length of the viral constructs (Fig. 2 a and b). From the HAADF STEM image (Fig. 2b), the average density of nanocrystals on the virus was estimated to be ≈8–16 quantum dots per 10 nm of virus. The selected area ED pattern (Fig. 2b Inset), obtained with the electron beam perpendicular to the long axis of the virus, indicated that ZnS nanocrystals were well crystallized in a hexagonal wurtzite structure. The ED pattern showed single crystal-type behavior, even though the sample area was composed of many nanocrystals. This behavior suggested that the nanocrystals on the virus were preferentially oriented with their c-axes perpendicular to the viral surface (Figs. 10 and 11, which are published as supporting information on the PNAS web site; see Supporting Text). Because of the 3D structure of the virus and its flattening under TEM conditions (23, 24), the binding ability of A7 peptide to ZnS resulted in nanocrystals that were in close contact, were highly oriented, and appeared to overlap in two layers of close-packed nanocrystals along the electron beam advancing direction when imaged by HRTEM/STEM. Fast Fourier transformation (FFT) of the lattice image taken from such overlapped layers, representing a [001] zone-axis single-crystal-type diffractogram, showed the 3D preferred orientation of the polycrystalline area.
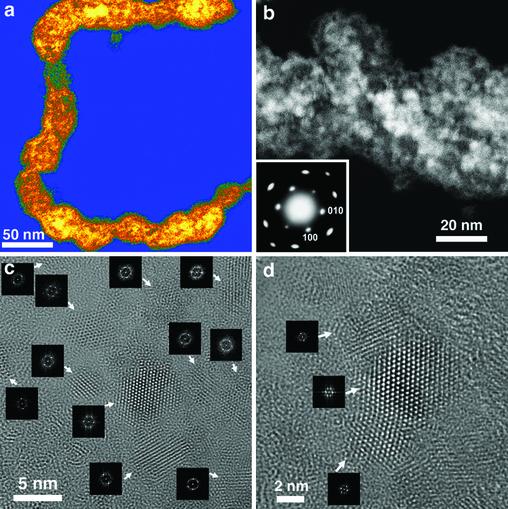
Fig. 2.
Higher-magnification images of A7–pVIII-engineered viruses directing ZnS nanocrystal synthesis at 0°C. (a) HAADF STEM image of an individual viral ZnS-virus nanowire. (b) HAADF STEM image of a straight region of a viral nanowire at higher magnification showing the close-packed ZnS nanocrystal morphology. In this image, some areas were brighter as a result of overlapped ZnS nanocrystals (Fig. 12, which is published as supporting information on the PNAS web site). (Inset) ED pattern, taken from the area shown in b, shows the hexagonal wurtzite ZnS structure. (c) HRTEM image of a less close-packed area on the virus shown in a. (Insets) FFT of the visible individual nanocrystals as denoted by arrows. (d) HRTEM image of a less close-packed area on the virus shown in a. (Insets) FFT of the visible individual nanocrystals as denoted by arrows.
In a few regions where nanocrystals were not overlapped, individual nanocrystals (3–5 nm) were well resolved by HRTEM lattice imaging (Fig. 2 c and d and Supporting Text). Similar to the close packed layers, these nanocrystals were preferentially oriented with their c-axes perpendicular to the viral surface. From the FFT diffractograms of the visible nanocrystals (Fig. 2 c and d Insets), the standard deviation of the ab-plane orientation was estimated to be 9° (Figs. 13 and 14, which are published as supporting information on the PNAS web site; Supporting Text). The distribution in nanocrystal size was likely a result of the spacing and distribution between the WT pVIII protein, necessary for viral assembly, and the peptide–pVIII fusion protein along the viral coat (18). Additionally, the same experimental set-up was scaled up for analysis by x-ray diffraction. The nanocrystal size calculated from the x-ray diffraction pattern was 3.7 nm (Fig. 15, which is published as supporting information on the PNAS web site; Supporting Text) and was consistent with the nanocrystal size (3–5 nm) resolved by HRTEM and STEM direct imaging. Moreover, only (00l) type diffraction peaks were observed in the diffraction patterns, suggesting that the ZnS nanocrystals were preferentially oriented with c-axis perpendicular to the sample surface, results that are consistent with the ED analysis.
Nanowire synthesis at —25°C of A7–pVIII constructs with ZnS resulted in engineered viruses that formed a layered ZnS structure caused by the viral–ZnS interactions during a phase separation of the water and viral solutions. The ZnS nanocrystals were templated along the viral structure in high density, but did not show the single-crystal-like ED behavior seen with synthesis at 0°C (Fig. 3). The low-magnification TEM image (Fig. 3a) and selected area ED pattern (Fig. 3a Inset) taken at a lower electron accelerating voltage (80 kV at JEOL 200 CX), HAADF STEM images (Fig. 3 b and d), and HRTEM images (Fig. 3c) together showed that viruses were coated by ZnS wurtzite-structured nanocrystals (≈5 nm) and assembled into a polycrystallinelayered hybrid material (Fig. 3d Inset). The ED pattern (Fig. 3a Inset), HAADF STEM, and HRTEM images (Fig. 3 b–d) revealed that the layered hybrid material contained close-packed ZnS nanocrystals.
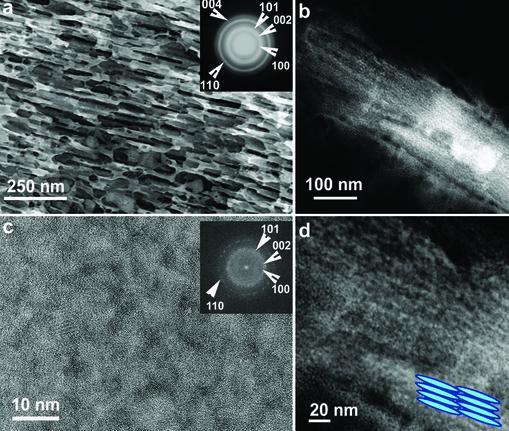
Fig. 3.
Images and characterization of A7–pVIII-engineered viruses directing ZnS nanocrystal synthesis at —25°C. (a) TEM image of the layered structure taken at 80 kV accelerating voltage. (Inset) ED of the layered structures showing the wurtzite ZnS structure and the polycrystalline status of the sample. (b) HAADF STEM image of a layered structure. (c) HRTEM of the layered structure showing close-packed and less oriented ZnS nanocrystals. (Inset) FFT of the whole image, which is consistent with the ED pattern in a.(d) HAADF STEM image of the layered structure shown in b at a higher magnification. (Inset) Schematic illustration.
When the other ZnS-specific peptide, the linear Z8, was displayed on the pVIII protein of the virus, ZnS nanocrystals were nucleated at 0°C with a preferred orientation of [001] perpendicular to the virus surface, but with cubic zinc blende structure (Figs. 16 and 17, which are published as supporting information on the PNAS web site). This finding was consistent with Z8-directed growth when displayed on the pIII minor coat protein of a virus (C.E.F., C.M., J. Williams, E. Ryan, and A.M.B., unpublished data). The Z8 peptide-engineered viruses showed lower density of nucleation (1.6–3.2 quantum dots per 10 nm of the virus) compared with the A7-engineered virus. Presumably because of increased length of the peptide, the Z8 fusions were displayed in lower copy number on the virus relative to the WT pVIII protein as verified by MS, resulting in reduced nanocrystal formation along the virus (17). Thus the inorganic nucleation also provided a probe into the extent of peptide expression relative to the previous A7 results.
When viruses engineered to express the CdS-specific J140 peptide were incubated with CdS precursor solutions, wurtzite-structured CdS nanocrystals (3–5 nm) were assembled into nanowires, as confirmed by STEM (Fig. 4a), EDS mapping (Fig. 4 b and c), and HRTEM (Fig. 18, which is published as supporting information on the PNAS web site; Supporting Text). The density of CdS nanocrystals on the virus was estimated to be three to six quantum dots per 10 nm of the virus (Fig. 4a Inset). These results confirmed that the viral nanowire template approach was extendable to other semiconductor systems. In a control experiment (Fig. 4d), when ZnS salt precursor solutions were exposed to the CdS-specific J140 viral template, no ZnS nanowires were observed, rather just μm-sized precipitates. The PL image of the J140-directed CdS nanowires (Fig. 4e) was consistent with the length scale of viruses observed by TEM and atomic force microscopy. The polarized PL spectra (Fig. 4f; Fig. 19, which is published as supporting information on the PNAS web site) of an individual nanowire, when excited by linear polarized light, displayed emission intensity strongest in the polarization direction parallel to the nanowires, independent of the excitation direction. Such behavior is likely derived from the anisotropic dielectric contrast between the nanowire and its surroundings (air or vacuum) when the nanowire was modeled as a quasi-continuous cylinder with an effective dielectric constant greater than its surroundings (25, 26).
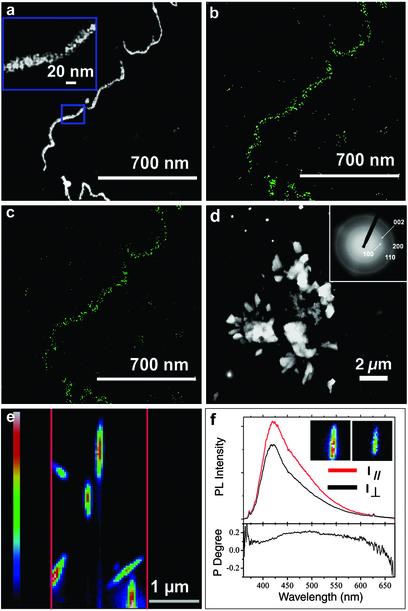
Fig. 4.
Images and characterization of the J140–pVIII-engineered viruses directing CdS nanocrystal synthesis at 0°C. (a–c) ADF STEM image (a) of viral CdS–virus nanowires and corresponding EDS mapping of elements S (b) and Cd (c). (a Inset) HAADF image of the framed portion in a.(d) HAADF STEM image of the control experiment, where J140-engineered viruses were exposed to ZnS growth conditions. No viral wires were seen, only large precipitates were visible. (Inset) ED of d. (e) A typical PL image of CdS virus nanowires on the silicon substrate. The parallel red lines indicate the entrance slit to the monochromator and thus the sample area detectable by a charge-coupled-device camera. Excitation source was at 350 nm. The color bar (Left) indicates the linear PL intensity scale. The width of the nanowires is larger than that resolved by STEM because the nanowires (20 nm wide) are narrower than the spatial resolution (200 nm) of PL imaging. (f) PL spectra (Upper) and polarization degree (Lower) of a single nanowire. The polarization degree (P) is calculated from P = (I ∥ — I⊥)/(I ∥ + I⊥), where I∥ and I⊥ are PL intensity in the polarization direction that is parallel (red line Upper) and perpendicular (black line Upper) to the length of an individual nanowire, respectively. (Insets) PL images detected along the direction parallel (Left) and perpendicular (Right) to the length of the nanowire. An integrated PL polarization anisotropy ratio of up to 1.5 was observed.
Heterostructured nucleation was achieved with a dual-peptide virus engineered to express both the J140 and A7 peptides within the same viral capsid. When viruses were incubated with both Zn(II), Cd(II), and S2— anions (molar ratio: Zn/Cd/S = 1:1:2) at —25°C, the viruses nucleated both CdS and ZnS nanocrystals with a stochastic distribution. Both CdS and ZnS nanocrystals appeared on the same phage constructs (Fig. 5). The HAADF STEM images (Fig. 5 b and c) indicated that the nanocrystals were uniform in size (5 nm). Following the HAADF STEM Z-contrast imaging principle (22, 27), the brighter nanocrystals were determined to be CdS and the darker nanocrystals were determined to be ZnS (Fig. 5 b and c). In a complementary approach, differently sized preformed ZnS (3–5 nm) and CdS (≈20 nm) nanocrystals were interacted with the A7–J140 phage template, resulting in formation of a heterostructured nanowire containing both semiconducting nanocrystals. The Z-contrast, the size difference between ZnS and CdS nanocrystals, PL, EDS, and ED, all indicated the stochastic binding of the two materials on one viral wire (Fig. 20, which is published as supporting information on the PNAS web site; Supporting Text).
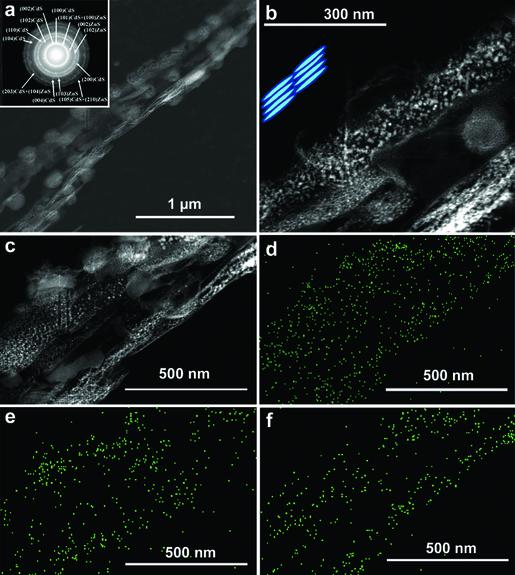
Fig. 5.
Images and characterization of ZnS–CdS hybrid nanowires prepared from viruses expressing a stochastic mixture of both the A7–pVIII and J140–pVIII fusion proteins by using CdS/ZnS nanocrystal synthesis at —25°C. (a) HAADF STEM image of a viral CdS and ZnS hybrid layered structure. (Inset)ED pattern of the layered structure showing the coexistence of wurtzite CdS and ZnS phases. (b) HAADF STEM image of the layered structure at higher magnification. (Inset) Cartoon illustrating the layered structure composed of viruses and nanocrystals. (c–f) HAADF STEM image (c) of the layer structure and its corresponding EDS mapping of elements S (d), Zn (e), and Cd (f).
Based on results showing that nanocrystal growth occurred only on viruses expressing the material-specific peptide and that preformed nanocrystals bound but were not organized on the virus (Fig. 21, which is published as supporting information on the PNAS web site), and the MS data of ion binding, we believe that the crystal growth on the virus followed a mechanism of peptide-templated growth similar to naturally evolved biomineralization systems. In our case, the nanocrystal growth results from the uniform conformation of expressed peptide on the engineered virus, from specific molecular interaction between a specific peptide and inorganic ions (e.g., Zn2+, S2—), and from the slow process of nucleation and growth at low temperature (although this temperature is lower than natural biomineralization systems). Competing mechanisms to peptide-directed nucleation include the binding of small particulates, based on the solubility product of ZnS. However, based on the preformed particle experiment and preliminary modeling of the A7 peptide on the surface of ZnS showing a peptide interaction with three amides and a histidine regularly spaced and pointed toward the ZnS surface (data not shown), we believe the dominant mechanism for nucleation arises from the nucleation role of the peptide, not solely recognition of a preformed particle. Moreover, although the crystal growth mechanisms for our systems are still being investigated, we believe that the viral coat organization combined with the ease of genetic manipulation to express a variety of selected peptides provides a platform for studying peptide-mediated nucleation of materials.
Nanowires with oriented nanocrystals of a single material or interveined with different types of materials have technological potential in device fabrication (28, 29). Our results demonstrate the potential application of engineered viruses to serve as the template to assemble nanocrystals along a nanowire, including heterostructures and superlattices beginning to be studied in our lab. In the future, specific receptors could be placed at each end of the viral nanocrystalline wires as fusions to terminal proteins. Self-assembly of these constructs in a programmable fashion might then be used to create complex nanowire arrays of varied one-, two-, and three-dimensional geometries.
Supplementary Material
Acknowledgments
We thank Dr. Moungi Bawendi and Dr. Evelyn Hu for helpful discussions on PL data. We acknowledge the use of the core microscopy facilities in the Texas Materials Institute, Center for Nano- and Molecular Science and Technology, and the Institute for Cellular and Molecular Biology at the University of Texas. This work was supported by an Army Research Office and National Science Foundation Nanoscale Interdisciplinary Research Team, the Welch Foundation, and the Beckman Foundation.
Abbreviations: TEM, transmission electron microscopy; HRTEM, high-resolution TEM; STEM, scanning TEM; EDS, energy dispersive x-ray spectroscopy; ED, electron diffraction; ADF, annular dark field; HAADF, high-angle ADF; PL, photoluminescence; FFT, fast Fourier transformation.
References
- 1.Li, M., Schnablegger, H. & Mann, S. (1999) Nature 402, 393–395. [Google Scholar]
- 2.Zhang, B., Davis, S. A., Mann, S. & Mendelson, N. H. (2000) Chem. Commun. 9, 781–782. [Google Scholar]
- 3.Weiner, S. & Wagner, H. D. (1998) Annu. Rev. Mater. Sci. 28, 271–298. [Google Scholar]
- 4.Weiner, S. & Addadi, L. (1997) J. Mater. Chem. 7, 689–702. [Google Scholar]
- 5.Weiner, S., Addadi, L. & Wagner, H. D. (2000) Mater. Sci. Eng. C 11, 1–8. [DOI] [PubMed] [Google Scholar]
- 6.Belcher, A. M., Wu, X. H., Christensen, R. J., Hansma, P. K., Stucky, G. D. & Morse, D. E. (1996) Nature 381, 56–58. [Google Scholar]
- 7.Cha, J. N., Shimizu, K., Zhou, Y., Christiansen, S. C., Chmelka, B. F., Stucky, G. D. & Morse, D. E. (1999) Proc. Natl. Acad. Sci. USA 96, 361–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aizenberg, J., Tkachenko, A., Weiner, S., Addadi, L. & Hendler, G. (2001) Nature 412, 819–822. [DOI] [PubMed] [Google Scholar]
- 9.Field, M., Smith, C. J., Awschalom, D. D., Mendelson, N. H., Mayes, E. L., Davis, S. A. & Mann, S. (1998) Appl. Phys. Lett. 73, 1739–1741. [Google Scholar]
- 10.Shenton, W., Douglas, T., Young, M., Stubbs, G. & Mann, S. (1999) Adv. Mater. 11, 253–256. [Google Scholar]
- 11.Whaley, S. R., English, D. S., Hu, E. L., Barbara, P. F. & Belcher, A. M. (2000) Nature 405, 665–668. [DOI] [PubMed] [Google Scholar]
- 12.Lee, S.-W., Mao, C., Flynn, C. E. & Belcher, A. M. (2002) Science 296, 892–895. [DOI] [PubMed] [Google Scholar]
- 13.Hartgerink, J. D., Beniash, E. & Stupp, S. I. (2001) Science 294, 1684–1688. [DOI] [PubMed] [Google Scholar]
- 14.Brown, S. (2001) Nano Lett. 1, 391–394. [Google Scholar]
- 15.Brown, S. (1997) Nat. Biotechnol. 15, 269–272. [DOI] [PubMed] [Google Scholar]
- 16.Zwick, M. B., Shen, J. & Scott, J. K. (2000) J. Mol. Biol. 300, 307–320. [DOI] [PubMed] [Google Scholar]
- 17.Iannolo, G., Minenkova, O., Petruzzelli, R. & Cesareni, G. (1995) J. Mol. Biol. 248, 835–844. [DOI] [PubMed] [Google Scholar]
- 18.Malik, P. & Perham, R. M. (1996) Gene 171, 49–51. [DOI] [PubMed] [Google Scholar]
- 19.Krebber, A., Bornhauser, S., Burmester, J., Honegger, A., Willuda, J., Bosshard, H. R. & Pluckthun, A. (1997) J. Immunol. Methods 201, 35–55. [DOI] [PubMed] [Google Scholar]
- 20.Malik, P. & Perham, R. N. (1997) Nucleic Acids Res. 25, 915–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Enshell-Seijffers, D., Smelyanski, L. & Gershoni, J. M. (2001) Nucleic Acids Res. 29, e50/1–e50/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Voyles, P. M., Muller, D. A., Grazul, J. L., Citrin, P. H. & Gossmann, H. J. L. (2002) Nature 416, 826–829. [DOI] [PubMed] [Google Scholar]
- 23.Kellenberger, E., Haner, M. & Wurtz, M. (1982) Ultramicroscopy 9, 139–150. [DOI] [PubMed] [Google Scholar]
- 24.Sarkar, N. H., Manthey, W. J. & Sheffield, J. B. (1975) Cancer Res. 35, 740–749. [PubMed] [Google Scholar]
- 25.Muljarov, E. A., Zhukov, E. A., Dneprovskii, V. S., Lomonosov, M. V. & Masumoto, Y. (2000) Phys. Rev. B Condens. Matter 62, 7420–7432. [Google Scholar]
- 26.Wang, J., Gudiksen, M. S., Duan, X., Cui, Y. & Lieber, C. M. (2001) Science 293, 1455–1457. [DOI] [PubMed] [Google Scholar]
- 27.Keyse, R. J., Garratt-Reed, A. J., Goodhew, P. J. & Lorimer, G. W. (1998) Introduction to Scanning Transmission Electron Microscopy (Springer, New York).
- 28.Wu, Y., Fan, R. & Yang, P. (2002) Nano Lett. 2, 83–86. [Google Scholar]
- 29.Gudiksen, M. S., Lauhon, U. J., Wang, J., Smith, D. C. & Lieber, C. M. (2002) Nature 415, 617–620. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.