Abstract
To identify potential molecular determinants of tumor biology and possible clinical outcomes, global gene-expression patterns were analyzed in the primary tumors of patients with metastatic renal cell cancer by using cDNA microarrays. We used grossly dissected tumor masses that included tumor, blood vessels, connective tissue, and infiltrating immune cells to obtain a gene-expression “profile” from each primary tumor. Two patterns of gene expression were found within this uniformly staged patient population, which correlated with a significant difference in overall survival between the two patient groups. Subsets of genes most significantly associated with survival were defined, and vascular cell adhesion molecule-1 (VCAM-1) was the gene most predictive for survival. Therefore, despite the complex biological nature of metastatic cancer, basic clinical behavior as defined by survival may be determined by the gene-expression patterns expressed within the compilation of primary gross tumor cells. We conclude that survival in patients with metastatic renal cell cancer can be correlated with the expression of various genes based solely on the expression profile in the primary kidney tumor.
Although metastatic renal cell cancer (RCC) carries a uniformly dismal prognosis, the clinical course can be highly variable (1, 2). Typically, patients experience progressive disease with further spread and growth of metastasis resulting in death, frequently within 1 year of diagnosis. However, some patients have a protracted clinical course, living for years with slowly progressive disease. Options for systemic treatment include agents such as IFN or IL-2 (3, 4). However, the majority of patients with advanced disease do not respond to therapy (5). Despite the wide range of clinical behavior, we have yet to identify reliable prognostic indicators for stage IV RCC. To identify potential molecular determinants of tumor biology, we analyzed global gene-expression patterns in the primary tumors of patients with metastatic RCC using cDNA microarrays. We demonstrate at least two patterns existing within similarly staged RCC that correlate with overall survival. In addition, we define a subset of genes most predictive for survival outcome.
Experimental Procedures
Clinical Material. We obtained 58 archived frozen tissue fragments from patients with sporadic, stage IV kidney tumors and eight matched, grossly normal-appearing kidney fragments from these patients. Multiple specimens were excised from all the tumors used in this study, and care was taken to ensure that only viable, nonnecrotic tumor was removed. There were 42 men and 16 women ranging in age from 21 to 66 years old (mean 50.2); 51 tumors had clear cell histology, 6 papillary and 1 undifferentiated histology (see Table 2, which is published as supporting information on the PNAS web site, www.pnas.org). All patients had a good prenephrectomy performance status (Eastern Cooperative Oncology Group 1 or 2). Nephrectomies were performed at the National Cancer Institute between April, 1994 and November, 2000. Patient survival was calculated from the date of nephrectomy to the date the patient died or was last known alive; survival ranged from 28 to 1,711 days (mean 499). The majority of patients were deemed ineligible for postoperative IL-2 treatment here at the National Institutes of Health (6). Thus most patients received adjuvant therapy for their metastatic disease outside the National Institutes of Health. No detailed records of treatment were kept for these patients, although periodic contact was maintained to check for survival. All deaths were secondary to the RCC metastatic disease. All the pathology was reviewed by M.M.
Microarray Procedures. Total RNA was isolated from the tumor samples by grinding the tissue fragments into fine powder by mortar and pestle on dry ice and homogenizing the powder in 5 ml of Trizol (GIBCO/BRL). This procedure yielded ample amounts (0.5–1 mg) of a full range of RNA species (as seen by a full smear on gel electrophoresis) for array analysis. The total RNA, however, yielded only moderate signal intensities on the arrays with some nonspecific background, which most likely was due to a relatively low percentage of mRNA compared with ribosomal RNA. Messenger RNA isolated from total RNA with a MicroFastrak 2.0 kit (Invitrogen) was used to improve the signal intensity, but only very small amounts of mRNA could be obtained from the total RNA. We thus decided to amplify the mRNA to maximize the signal intensity and reduce the nonspecific background. The mRNA was reverse-transcribed by priming with oligo(dT) into single-stranded DNA and double-stranded DNA transcribed from a primer added during the reverse-transcribed reaction (7). One round of in vitro transcription amplified ≈2 μg of mRNA into 30–60 μg of amplified mRNA. This method of “linear” mRNA amplification maintains the fidelity of the original mRNA species isolated from the tumor samples (8). The amplified tumor and normal mRNA were reverse-transcribed, incorporating a Cy3 (green)-labeled thymidine into the newly formed DNA molecule. All tumor and normal DNA was hybridized against a standard reference consisting of amplified mRNA from six pooled tumor cell lines labeled with Cy5 (red) onto microarray chips spotted with 6,400 cDNA PCR products (see Table 3, which is published as supporting information on the PNAS web site, for a list of cells). The gene set spotted on the arrays represents all the available, sequence-confirmed cDNA species available from Research Genetics (Invitrogen) at the time the arrays were printed. Thus there was no attempt to produce a cancer- or kidney-specific gene set.
Immunohistochemistry. We performed immunohistochemical studies on nine tumors selected for high or low expression of vascular cell adhesion molecule (VCAM) as demonstrated on arrays. We first stained each tissue section with CD31 to identify blood vessels and then counterstained with VCAM (DAKO). VCAM is highly expressed in normal Bowman's capsule and was used as a positive control in the histologically normal-appearing areas from each tissue section. The staining was quantitated by using a 0–3 scale with 3 being the strongest level of staining as compared with the other samples in the study.
Data Analysis. The microarrays were scanned by using a GenePix 4000 microarray scanner (Axon Instruments, Foster City, CA) and analyzed with GENEPIX PRO 3.0 software. Spots were excluded if there was an obvious problem with the hybridization or the spot was defective. Log base-2 ratios of local background-subtracted intensity levels were analyzed. Log ratios were considered missing for spots with either defects or intensity <50 in both channels and were truncated at ±6. A spot was filtered out from analysis if the log ratio was missing in >30% of the arrays. Log ratios for each microarray were median-centered to adjust for dye bias and photomultiplier tube voltage settings. The “raw” array data are available in Supporting Data Set 1, which is published as supporting information on the PNAS web site. The expression profiles of the samples were hierarchically clustered by using average-linkage clustering with Pearson correlation as the similarity metric based on median-centered genes, and a dendrogram was constructed (9).
We evaluated the reproducibility of the two major clusters determined by the dendrogram using previously described methods (10). Briefly, we perturbed the log ratios by adding Gaussian noise and reclustering the patients 1,000 times. In >94% of cases, two samples that clustered together in the original data remained clustered together after perturbation. Some analyses, such as survival analyses, required a single expression profile per patient. In 21 of the 25 patients with multiple arrays, the Pearson correlation among log-ratio expression profiles was considered to be satisfactory, and we averaged the expression profiles. For the remaining four patients, we averaged the expression profiles that were technically best and well correlated.
We identified genes associated with survival using the Cox proportional-hazards model. We required that the log ratio be statistically significant at the 0.001 level. We used a “leave-oneout” cross-validation procedure to place patients in one of two prognosis groups to estimate the extent to which we could predict survival using a multivariate gene-expression profile classifier. The procedure cross-validated both the selection of significant genes and the prognostic classification of each patient based on a training set not including that patient. We performed a permutation test to evaluate the significance of the difference between the survival curves. The cross-validation procedure used for the survival analysis and permutation test is described in Explanation of Statistical Methods, which is published as supporting information on the PNAS web site. We assessed the association between covariates and survival using the Cox proportional-hazards model.
Results
Total Gene-Expression Patterns. We initially set out to determine whether global gene-expression patterns in the primary tumors of patients with metastatic RCC demonstrated subgroups of tumors based on overall patterns of gene expression. We began by arraying amplified RNA isolated from frozen archived tissue samples and using a hierarchical average-linkage clustering program that grouped patients by the similarity or differences between overall gene expression (11). A dendrogram was generated in which patients with the most similar overall gene-expression patterns were grouped together. To evaluate the reproducibility of the array methodology, replicate arrays were performed by using the same RNA samples for 22 of the patients. We determined the Pearson correlation among log-ratio expression profiles between the replicates using 0.5 as a stringent cutoff for the samples to be considered as having a significant degree of correlation. With this criterion, 20 of 22 patients demonstrated satisfactory reproducibility. In 6 of the 58 patients studied, we performed array analysis using RNA isolated from three separate areas of the same tumor. When we assessed the Pearson correlation among log-ratio expression profiles between the three areas of tumor, only two of the six patients had correlations >0.5 for all three sampled areas of tumor areas, indicating the possibility of some degree of intratumor genetic expression heterogeneity.
A total of 103 arrays were performed including the repeated samples and 8 matched normal samples. cDNAs (4,922) were evaluable for all samples and used for unsupervised clustering. Two major groups of patients identified by the dendrogram were analyzed further. The eight normal samples clustered together in this analysis as a subcluster of one of the two major tumor clusters.
We next asked whether the two clusters reflected clinical behavior by asking whether the two groups demonstrated any difference in overall survival. Using the methods described, we averaged data from tumors with multiple arrays from the same tumor and excluded the matched normal expression profiles such that each patient had only one profile to provide a more clearly defined cluster analysis (Fig. 1a). A Kaplan–Meier curve of the resulting unsupervised cluster groups demonstrated a nearly significant survival difference (Fig. 1b). When we excluded the four patients whose replicate arrays did not meet the criteria for averaging, results were similar, although the P value did become significant (Fig. 1c).
Fig. 1.
Unsupervised dendrogram and resulting survival curves. (a) Dendrogram created by unsupervised clustering demonstrating two major groups of patients. (b) Kaplan–Meier survival curve based on the two groups of patients from a. (c) Survival curve from b depicting patients with <0.5 Pearson correlation between arrays removed.
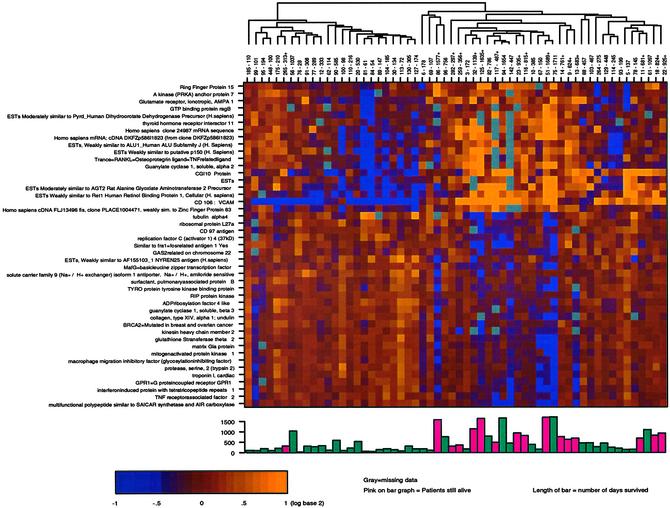
Gene Expression Associated with Survival. Because we were seeing a survival difference based on unsupervised gene-expression patterns derived from the cluster analysis in the primary tumor in patients with stage IV RCC, our next goal was to define more clearly the genes most significantly associated with survival. To do this, we used a different method to analyze the data. Using the Cox proportional-hazards model, we identified 45 genes most significantly associated with longer or shorter survival at the 0.001 significance level (Table 1), a number in excess of the number expected by chance (P = 0.017). We reclustered our 58 patients using only these genes. The two major clusters had 24 patients in one group and 34 patients in the other. The data are shown in matrix format in Fig. 2, with each column representing all the measured expression levels for a single patient and each row representing all the hybridization results for a single gene (12). The expression level of each gene, relative to its median expression levels across all samples, was represented by color intensity, with yellow indicating expression greater than the median and blue indicating expression less than the median. The highest intensity indicates a ≥2-fold difference in gene expression. A bar graph below the cluster diagram depicts the length of survival for each of the patients. The bars in red represent patients alive at last evaluation.
Table 1. The 45 genes most significantly associated with survival based on Cox proportional-hazards regression analysis.
| Proportion* | P value survival† | clust2/clust1‡ | P value clust2/clust1§ | Expression for longer survival¶ | Gene description | |
|---|---|---|---|---|---|---|
| 1 | 0.71 | 0.0001 | 1.86 | 0.0092 | Up | VCAM-1 |
| 2 | 0.41 | 0.0007 | 1.45 | 0.0022 | Up | CGI-10 protein |
| 3 | 0.41 | 0.0002 | 1.41 | 0.0061 | Up | EST |
| 4 | 0.35 | 0.0001 | 1.46 | 0.0027 | Up | EST |
| 5 | 0.31 | 0.0001 | 1.18 | 0.1175 | Up | EST |
| 6 | 0.29 | 0.0007 | 1.10 | 0.3698 | Up | cDNA DKFZp586l1823 |
| 7 | 0.25 | 0.0006 | 1.06 | 0.5411 | Up | EST |
| 8 | 0.24 | 0.0007 | 1.11 | 0.3292 | Up | Guanylate cyclase 1, soluble α2 |
| 9 | 0.23 | 0.0009 | 1.17 | 0.1185 | Up | Homo sapiens clone 24987 |
| 10 | 0.23 | 0.0008 | 1.12 | 0.1576 | Up | Ring finger protein 15 |
| 11 | 0.22 | 0.0005 | 0.94 | 0.4678 | Up | Glutamate receptor, AMPA 1 |
| 12 | 0.19 | 0.0006 | 0.99 | 0.8342 | Up | EST |
| 13 | 0.18 | 0.0006 | 1.16 | 0.1125 | Up | TRANCE, TNF-related ligand |
| 14 | 0.17 | 0.0004 | 1.18 | 0.0190 | Up | Thyroid hormone receptor interactor 11 |
| 15 | 0.16 | 0.0005 | 1.15 | 0.0555 | Up | EST |
| 16 | 0.14 | 0.0002 | 1.02 | 0.8083 | Up | A kinase (PRKA) anchor protein 7 |
| 17 | 0.13 | 0.0007 | 1.02 | 0.7726 | Up | cDNA FLJ13496 fis |
| 18 | 0.11 | 0.0000 | 1.11 | 0.1456 | Up | GTP-binding protein ragB |
| 19 | 0.16 | 0.0002 | 0.70 | 0.0000 | Down | Tubulin-α4 |
| 20 | 0.12 | 0.0002 | 0.83 | 0.0010 | Down | fos-related antigen 1 |
| 21 | 0.12 | 0.0004 | 1.06 | 0.3067 | Down | EST |
| 22 | 0.12 | 0.0003 | 0.93 | 0.1250 | Down | Multifunctional polypeptide similar to SAICAR synthetase |
| 23 | 0.12 | 0.0006 | 1.00 | 0.9833 | Down | Interferon-induced protein with tetratricopeptide repeats 1 |
| 24 | 0.10 | 0.0003 | 1.03 | 0.5846 | Down | Protease, serine, 2 (trypsin 2) |
| 25 | 0.09 | 0.0000 | 0.82 | 0.0001 | Down | Ribosomal protein L27a |
| 26 | 0.09 | 0.0005 | 1.03 | 0.5952 | Down | ADP-ribosylation factor 4-like |
| 27 | 0.09 | 0.0001 | 1.01 | 0.8006 | Down | Solute carrier family 9 (Na+/H+ exchanger) isoform 1 |
| 28 | 0.07 | 0.0006 | 0.91 | 0.0475 | Down | GAS2-related on chromosome 22 |
| 29 | 0.07 | 0.0003 | 1.06 | 0.2109 | Down | Guanylate cyclase 1, soluble, β3 |
| 30 | 0.07 | 0.0001 | 1.01 | 0.7728 | Down | Matrix Gla protein |
| 31 | 0.07 | 0.0007 | 0.98 | 0.6387 | Down | GPR1, G protein-coupled receptor |
| 32 | 0.07 | 0.0001 | 1.02 | 0.6866 | Down | Troponin I, cardiac |
| 33 | 0.07 | 0.0002 | 0.95 | 0.2839 | Down | TNF receptor-associated factor 2 |
| 34 | 0.07 | 0.0000 | 0.97 | 0.4729 | Down | Glutathione S-transferase θ2 |
| 35 | 0.07 | 0.0002 | 1.05 | 0.2258 | Down | BRCA2 |
| 36 | 0.05 | 0.0008 | 0.80 | 0.0000 | Down | Replication factor C (activator 1) 4 (37 kDa) |
| 37 | 0.05 | 0.0002 | 1.01 | 0.8859 | Down | TYRO protein tyrosine kinase-binding protein |
| 38 | 0.05 | 0.0001 | 0.93 | 0.1051 | Down | MafG |
| 39 | 0.04 | 0.0006 | 0.95 | 0.2372 | Down | Collagen XIV, α1; undulin |
| 40 | 0.04 | 0.0006 | 1.01 | 0.8032 | Down | Kinesin heavy chain member 2 |
| 41 | 0.03 | 0.0002 | 1.05 | 0.2256 | Down | Macrophage migration inhibitory factor |
| 42 | 0.03 | 0.0001 | 1.02 | 0.6427 | Down | Mitogen-activated protein kinase 1 |
| 43 | 0.02 | 0.0000 | 0.91 | 0.0164 | Down | CD 97 antigen |
| 44 | 0.02 | 0.0006 | 1.00 | 0.9715 | Down | Surfactant, pulmonary-associated protein B |
| 45 | 0.02 | 0.0005 | 1.03 | 0.4154 | Down | RIP protein kinase |
Proportion of patients with a log-ratio value of the gene differing from the median >0.5 for each gene. The median expression of a gene is defined as the median level of expression of that gene across all 58 patients. The higher the proportion means a greater variability in expression of that gene across the patient population.
The level of significance of the gene's association with survival.
Average fold difference of the expression of the gene between the two major patient groups identified by unsupervised clustering (Fig. 1a). Cluster 1 patients had the shorter mean survival (351 vs. 611 days). The average fold difference is 2 to the power of the difference in mean log ratios for the two clusters.
Level of significance comparing expression of the genes between the two clusters based on the unsupervised analysis.
Relative expression level of each gene related to longer patient survival. Calculations were based on the sign of the regression coefficient associated with each gene in the Cox proportional-hazards model. A negative coefficient yields improved survival with increased (up) expression, and a positive coefficient yields improved survival with decreased (down) expression.
Fig. 2.
Cluster matrix of 45 genes most significantly associated with survival. The colors in the matrix represent a maximum of 2-fold up-regulation (brightest yellow) or 2-fold down-regulation (brightest blue) relative to the median for each gene. The patients (columns) are labeled by patient number followed by the number of days alive after nephrectomy. The bar graph below the matrix graphically depicts the length of patient survival; red bars represent patients who are still alive. The dendrogram above the patient list graphically depicts the two major patient groups and minor subgroups within the two larger groups.
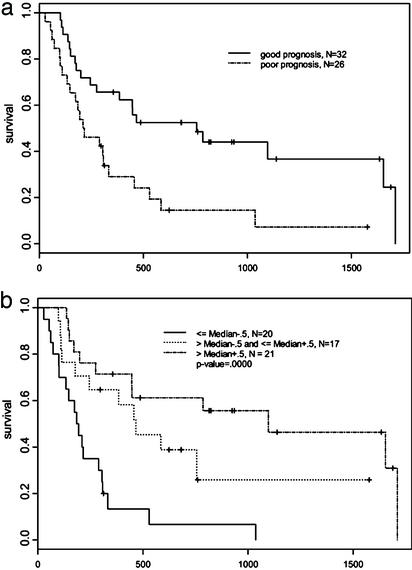
To determine the extent that survival could be predicted based on a multivariate prognostic index, we used the leave-one-out cross-validation model described in Explanation of Statistical Methods to classify patients into either a longer or shorter survival group (Fig. 3a). The difference in survival was statistically significant as determined by the permutation distribution of the cross-validated log-rank χ2 statistic (P < 0.05). The results demonstrate that the 34 patients in the longer-surviving group had a median time to death of 556 versus 180 days for the shorter-surviving group. Note that 15 of the 16 surviving patients depicted on the bar graph (Fig. 2) fall within the longer-surviving group, suggesting that the survival difference between the groups may continue to increase. No other clinical variables including age, gender, tumor grade, and adjuvant therapy had a significant impact on survival outcome.
Fig. 3.
Survival curves based on supervised analysis. (a) Kaplan–Meier plots of overall survival of 58 patients grouped into good prognosis and poor prognosis by their gene-expression outcome predictor generated by the leave-one-out cross-validated procedure. (b) Survival curves of patients grouped by the median of log ratio +0.5 and –0.5 for VCAM-1.
VCAM-1 as a Prognostic Indicator. Two major patterns of gene expression are apparent (Fig. 2) that divide patients into longerand shorter-surviving groups. The shorter-surviving group had down-regulation of genes located near the top of the diagram (blue) and up-regulation near the bottom (yellow). This pattern is reversed for the longer-surviving group. Although these global patterns are clearly evident, actual patient-to-patient variability can be quite large. To identify which of the 45 survival genes were most consistently up- or down-regulated across the patient population, we determined for each gene the percentage of patients with the expression level different from its median value by >0.5. From this analysis it is clear that regulation of one gene in particular, VCAM-1, was up- or down-regulated (defined as log-ratio value different than the median by at least 0.5, corresponding to a 1.4-fold effect) in 71% of the patients. A Kaplan–Meier survival curve was constructed for low, median, and high levels of VCAM-1 expression, and we found a significant difference in survival between the patients expressing high versus low levels of VCAM-1, with the higher expressers having the longer survival time (Fig. 3b). Other genes significantly associated with survival were altered 1.4-fold in a smaller percentage of patients. Immmunohistochemical VCAM-1 analysis of nine of the tumor samples revealed heterogeneous staining. Strikingly, almost all of the VCAM-1 staining occurred on the tumor cells, and very little staining occurred on either the vasculature or infiltrating cells in all nine samples (Fig. 4, which is published as supporting information on the PNAS web site). Also, there was strong VCAM-1 staining seen on the cells in Bowman's capsule in the normal kidney adjacent to the tumor.
Discussion
We performed microarray analysis on the primary tumors of 58 patients with stage IV RCC to determine whether the natural history of the disease could be determined based on gene expression alone. This represents a first step into the understanding of the molecular basis for the clinical behavior of metastatic RCC. Importantly, all patients underwent resection of their primary tumor; therefore the primary tumor itself had no further direct impact on survival.
We began the analysis with an unsupervised hierarchical clustering of the gene set, and as expected with this type of analysis, two major groups of patients were identified. A Kaplan–Meier survival analysis demonstrated a small but statistically significant difference in length of survival between the two groups. Finding patients with longer survival based only on unsupervised clustering suggested that gene-expression patterns relevant to the biological behavior of metastatic RCC may exist. However, it is difficult to define associations between genes and clinical parameters such as survival based only on unsupervised global cluster analysis. The Cox proportional-hazards model was used to identify the genes that were significantly associated with survival at a P < 0.001 level, and 45 genes were identified. The 58-patient cohort then was reclustered based only on these 45 genes, and mean survival was ≈1 year longer in the better-prognosis group compared with the worse-prognosis group. Predictive models were constructed based on the leave-one-out cross-validation technique to place patients into a long- or short-survival group with the predictive genes redetermined for each leave-one-out training set. The Kaplan–Meier survival curve based on these two patient groups demonstrated a statistically significant survival difference as determined by permutation analysis of the entire cross-validation procedure. Therefore, there is a structure within gene-expression patterns, defined by the array data, on which a statistical model can be constructed to predict survival outcome in stage IV RCC. This is a significant finding, because no prognostic indicators for stage IV RCC currently exist.
The strongest existing prognostic indicators for RCC are stage, grade, and Eastern Cooperative Oncology Group status (13, 14). Among stage IV tumors, no clinical parameters have been identified that predict improved survival (15). Other groups have reported microarray profiles for RCC. Consistent with our findings, Takahashi et al. (16) examined 29 RCCs and identified two cluster groups predictive of survival. However, they included all stages of RCC and report a marginal improvement in survival prediction over stage alone. Because all patients in our study had advanced metastatic disease, our cluster groups may reflect inherent biologic differences relevant to survival more accurately. Another group reports paired analysis of 37 tumor and normal specimens in an effort to find genes differentially regulated in RCC but make no reference to profiles within RCC (17). Our results represent a large cohort and eliminate prognostic contributions of stage and grade.
Examination of a subset of 14 patients for Von Hippel Lindau (VHL) mutation yielded no correlation between VHL status and the survival groups defined by the Cox regression-analysis model (data not shown), suggesting that our cluster is not driven by the presence or absence of the VHL tumorigenesis pathway. Interpreting gene expression from solid tumors adds another level of complexity to the analysis because of their heterogeneous nature. The population of cells within a solid-tumor sample includes vascular, lymphoid, stromal, and interposed normal renal tissue. The possibility of lymphoid infiltration having a major affect on the clustering of patients is unlikely, because the amount of lymphocytic infiltrate within the tumor has not correlated with survival in RCC (18, 19). Also, we did not observe any major difference for immunologic gene expression (i.e., T cell receptor, MHC, transforming growth factor, and IFN-related genes) between the two groups defined by clustering.
VCAM-1 was identified as the single-most predictive gene for survival in our patients, with 71% of the patients demonstrating differential expression and uniform up-regulation in patients with longer survival and down-regulation in patients with shorter survival. VCAM-1 is a cell-surface glycoprotein that interacts with the integrin, very-late antigen-4 (VLA-4), and despite the “vascular” title is expressed on a variety of endothelial cell types (20, 21). Our results with a cohort of metastatic RCC patients demonstrate that increased expression of VCAM-1 correlates with improved survival. Previous studies have demonstrated that the VCAM-1 and VLA-4 interaction was involved in the adhesion between RCC cells and epithelial cells, and it was hypothesized that the VCAM–VLA-4 interaction might play a crucial role in hematogenous metastasis (22, 23). However, these were not clinically based studies. Our study compares VCAM-1 and survival in patients with RCC. All patients presented to us with existing metastatic disease; therefore we cannot directly evaluate how VCAM relates to metastasis in our cohort of patients. However, it is difficult to conceive of how increased VCAM-1 expression could potentiate metastasis and be associated with good prognosis once metastases are established. Thus even though the VCAM-1–VLA-4 association may enable RCC cells to become adherent to epithelium, the RCC–epithelial association may not be linked to the development of metastasis and seems to improve survival. Furthermore, finding relatively high levels of VCAM-1 on normal renal cells in Bowman's capsule implies a normal physiologic role for VCAM-1 in the kidney.
The results of this study infer that there is an underlying organization within the gene-expression profile of the primary tumor of patients with stage IV RCC that is related to patient survival. These patterns can be detected by microarray analysis and applied by using a statistical model to predict survival. We have identified a set of genes expressed in the primary tumor of patients with metastatic RCC that are the most significantly related to survival. The next step will be to determine whether our predictive gene set can define longer- and shorter-surviving groups, prospectively, in patients with stage IV RCC. It would be instructive to determine whether the gene set would also be predictive for survival in patients with earlier-staged RCC. It is likely that these genes would be predictive for survival in all stages of RCC. It is conceivable, however, that genes predictive for longer survival in stage IV would not be predictive or portend a shorter survival at an earlier stage of disease. This is an unlikely scenario, but it needs to be explored. Evaluating our gene set against gene expression found in other types of late-stage cancer would also be useful. Genes predictive for survival in several tissue types might identify those genes most influential in determining the biological behavior and thus clinical outcome of late-stage cancer.
Supplementary Material
Abbreviations: RCC, renal cell cancer; VCAM, vascular cell adhesion molecule.
References
- 1.American Cancer Society (2002) Cancer Facts and Figures (Am. Cancer Soc., Atlanta).
- 2.Vasselli, J. R., Yang, J. C., Linehan, W. M., White, D. E., Rosenberg, S. A. & Walther, M. M. (2001) J. Urol. 166, 68–72. [PubMed] [Google Scholar]
- 3.Wagner, J. R., Walther, M. M., Linehan, W. M., White, D. E., Rosenberg, S. A. & Yang, J. C. (1999) J. Urol. 162, 43–45. [DOI] [PubMed] [Google Scholar]
- 4.Yang, J. C. & Rosenberg, S. A. (1997) Cancer J. Sci. Am. 3, Suppl. 1, S79–S84. [PubMed] [Google Scholar]
- 5.Lee, D. S., White, D. E., Hurst, R., Rosenberg, S. A. & Yang, J. C. (1998) Cancer J. Sci. Am. 4, 86–93. [PubMed] [Google Scholar]
- 6.Royal, R. E., Steinberg, S. M., Krouse, R. S., Heywood, G., White, D. E., Hwu, P., Marincola, F. M., Parkinson, D. R., Schwartzentruber, D. J., Topalian, S. L., et al. (1996) Cancer J. Sci. Am. 2, 91–98. [PubMed] [Google Scholar]
- 7.Wang, E., Miller, L. D., Ohnmacht, G. A., Liu, E. T. & Marincola, F. M. (2000) Nat. Biotechnol. 18, 457–459. [DOI] [PubMed] [Google Scholar]
- 8.Feldman, A. L., Costouros, N. G., Wang, E., Qian, M., Marincola, F. M., Alexander, H. R. & Libutti, S. K. (2002) Biotechniques 33, 906–914. [DOI] [PubMed] [Google Scholar]
- 9.Brown, P. O. & Botstein, D. (1999) Nat. Genet. 21, 33–37. [DOI] [PubMed] [Google Scholar]
- 10.McShane, L. M., Radmacher, M. D., Freidlin, B., Yu, R., Li, M. C. & Simon, R. (2002) Bioinformatics 18, 1462–1469. [DOI] [PubMed] [Google Scholar]
- 11.Eisen, M. B., Spellman, P. T., Brown, P. O. & Botstein, D. (1998) Proc. Natl. Acad. Sci. USA 95, 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alizadeh, A. A., Eisen, M. B., Davis, R. E., Ma, C., Lossos, I. S., Rosenwald, A., Boldrick, J. C., Sabet, H., Tran, T., Yu, X., et al. (2000) Nature 403, 503–511. [DOI] [PubMed] [Google Scholar]
- 13.Abou-Rebyeh, H., Borgmann, V., Nagel, R. & Al Abadi, H. (2001) Cancer 92, 2280–2285. [PubMed] [Google Scholar]
- 14.Tsui, K. H., Shvarts, O., Smith, R. B., Figlin, R. A., deKernion, J. B. & Belldegrun, A. (2000) J. Urol. 163, 1090–1095. [DOI] [PubMed] [Google Scholar]
- 15.Frank, W., Stuhldreher, D., Saffrin, R., Shott, S. & Guinan, P. (1994) J. Urol. 152, 1998–1999. [DOI] [PubMed] [Google Scholar]
- 16.Takahashi, M., Rhodes, D. R., Furge, K. A., Kanayama, H., Kagawa, S., Haab, B. B. & Teh, B. T. (2001) Proc. Natl. Acad. Sci. USA 98, 9754–9759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boer, J. M., Huber, W. K., Sultmann, H., Wilmer, F., von Heydebreck, A., Haas, S., Korn, B., Gunawan, B., Vente, A., Fuzesi, L., et al. (2001) Genome Res. 11, 1861–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kolbeck, P. C., Kaveggia, F. F., Johansson, S. L., Grune, M. T. & Taylor, R. J. (1992) Mod. Pathol. 5, 420–425. [PubMed] [Google Scholar]
- 19.Nakano, O., Sato, M., Naito, Y., Suzuki, K., Orikasa, S., Aizawa, M., Suzuki, Y., Shintaku, I., Nagura, H. & Ohtani, H. (2001) Cancer Res. 61, 5132–5136. [PubMed] [Google Scholar]
- 20.Cybulsky, M. I., Fries, J. W., Williams, A. J., Sultan, P., Eddy, R., Byers, M., Shows, T., Gimbrone, M. A., Jr., & Collins, T. (1991) Proc. Natl. Acad. Sci. USA 88, 7859–7863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pepinsky, B., Hession, C., Chen, L. L., Moy, P., Burkly, L., Jakubowski, A., Chow, E. P., Benjamin, C., Chi-Rosso, G., Luhowskyj, S., et al. (1992) J. Biol. Chem. 267, 17820–17826. [PubMed] [Google Scholar]
- 22.Steinbach, F., Tanabe, K., Alexander, J., Edinger, M., Tubbs, R., Brenner, W., Stockle, M., Novick, A. C. & Klein, E. A. (1996) J. Urol. 155, 743–748. [PubMed] [Google Scholar]
- 23.Tomita, Y., Saito, T., Saito, K., Oite, T., Shimizu, F. & Sato, S. (1995) Int. J. Cancer 60, 753–758. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.