Abstract
The x-ray structure of complete RNA polymerase II from Saccharomyces cerevisiae has been determined, including a heterodimer of subunits Rpb4 and Rpb7 not present in previous “core” polymerase II structures. The heterodimer maintains the polymerase in the conformation of a transcribing complex, may bind RNA as it emerges from the enzyme, and is in a position to interact with general transcription factors and the Mediator of transcriptional regulation.
RNA polymerase II (pol II), the enzyme responsible for all mRNA synthesis in eukaryotes, has been isolated in two forms, a 12-subunit “complete” enzyme and a 10-subunit “core.” The two additional subunits of the complete enzyme, Rpb4 and Rpb7, form a heterodimer and associate reversibly with core. The two enzymes are equivalent in RNA chain elongation, but core pol II is defective in the initiation of transcription. Addition of Rpb4/Rpb7 to core pol II restores initiation activity (1, 2). Rpb4/Rpb7 therefore may be regarded as a general transcription factor, akin to the previously described TFIIB, -D, -E, -F, and -H.
Deletion of the RPB4 gene in yeast results in a temperature-sensitive phenotype, with cessation of growth above 32°C, while deletion of RPB7 is lethal (3–5). Microarray analysis reveals the rapid shutdown of 98% of all yeast mRNA synthesis upon shift of a Δrpb4 strain to a restrictive temperature, consistent with Rpb4/Rpb7 serving as a general transcription factor. Even at a permissive temperature, where constitutive gene transcription is not much affected by RPB4 deletion, transcription of inducible promoters is largely abolished (6). Overexpression of RPB7 suppresses many of the phenotypes of a Δrpb4 strain, but it fails to suppress the activation defect at most promoters tested (7–9). These results confirm the interaction of Rpb4 and Rpb7 in vivo and show that the heterodimer also fits the definition of a transcriptional “coactivator.”
We have investigated the structure of RNA pol II by x-ray crystallography, giving insight into the transcription mechanism. Structures of core pol II were determined at 3.1-Å resolution (form 1), 2.8-Å resolution (form 2), and 3.3-Å resolution (transcribing complex) (10–12). Comparison of these structures revealed a number of changes related to transcription, including the movement of a massive “clamp” by >30 Å between form 2 (“open” state) and the transcribing complex (“closed” state), in which it largely surrounds the DNA template and RNA transcript (11, 12).
For structural studies of complete, 12-subunit pol II, the enzyme was initially isolated from yeast cells grown to stationary phase, where almost all pol II is in the complete form (13). The resulting crystals were poorly ordered, likely because of the persistence of some core pol II. To overcome the difficulty, we prepared a yeast strain bearing an affinity tag on Rpb4 and isolated the complete enzyme, devoid of core pol II, by affinity chromatography. This homogeneous, complete enzyme preparation formed crystals diffracting to ≈4-Å resolution.
Materials and Methods
Yeast strain CB010 with a tandem affinity purification tag integrated at the C terminus of Rpb4 was grown on yeast extract/peptone/dextrose medium to late log phase (14). Yeast cells were resuspended to a density of 0.5 g/ml in 10% glycerol, 50 mM Tris-Cl (pH 8.0), 150 mM potassium chloride, 10 mM DTT, and 1 mM EDTA. Cells were lysed by using a bead beater, and clarified lysate was bound to IgG fast flow beads (Amersham Biosciences). The beads were washed with 10 column volumes of 50 mM Hepes (pH 7.6), 500 mM ammonium sulfate, 1 mM DTT, and 1 mM EDTA, and then with 5 column volumes of 50 mM Hepes (pH 7.6), 100 mM potassium chloride, 1 mM DTT, and 1 mM EDTA before elution by cleavage with tobacco etch virus. The eluate was purified on an 8WG16 antibody column and a DEAE HPLC column as described (15, 16).
pol II was concentrated to 10 mg/ml in a microcon with a 100-kDa molecular mass cutoff in 5 mM Tris·Cl (pH 7.5), 60 mM ammonium sulfate, and 10 mM DTT. Crystals were grown by using the hanging-drop method against 100 mM ammonium phosphate buffer (pH 6.3), 100 mM NaCl, 5 mM dioxane, 1 mM zinc chloride, 5% PEG 6000, and 20–25% PEG 400. Crystals were frozen directly from the mother liquor. Diffraction data were collected at the Advanced Light Source beamline 5.0.2 at 0.98 Å. Diffraction data were reduced by using the HKL package (17).
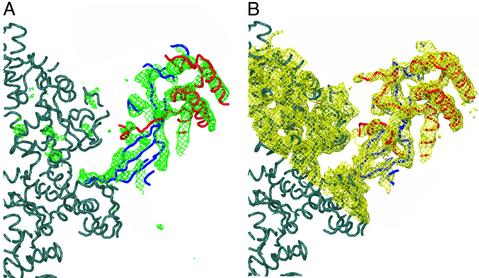
Molecular replacement was carried out with CNS by using the fast direct method (18). The three current pol II models were used as search models. The transcribing complex model (PDB ID code 1I6H) was found to give the best results, and all subsequent steps were performed with this model. Rigid-body refinement and group B refinement were performed with CNS (final Rcryst = 32.5, Rfree = 35.7–4.1 Å). A difference map calculated by using SigmaA-weighted phases revealed a large difference density on the side of the clamp near the back of pol II (Fig. 1). To improve the phases and remove model bias, the SigmaA-weighted phases were used as a starting point for density modification. With only one molecule per asymmetric unit, the calculated solvent content for the complete pol II crystals is >80% (Matthews coefficient of 6.3). Density modification was performed by using CNS with a solvent content of 80%. A polyalanine model of the archaeal Rpb4/Rpb7 homologs was placed in a map calculated from the solvent-flattened phases and rigid body refined by using CNS. The archaeal homolog model was then modified by using O to better fit the observed yeast density (19). Wherever possible the archaeal homolog was used, but, when necessary, additional polyalanine backbone was traced. A backbone model (Cα atoms only) of the complete 12-subunit pol II and structure factors have been deposited in the Protein Data Bank.
Fig. 1.
Complete, 12-subunit pol II electron density map. (A) Front view (as in refs. 10 and 11) of SigmaA-weighted Fobs – Fcalc electron density at 4.1-Å resolution (green) contoured at 3 σ, calculated from the initial placement of the pol II model (dark gray). The initial placement of archaeal RpoF (Rpb4 homolog) is shown in red, and placement of archaeal RpoE (Rpb7 homolog) in shown in blue. (B) Electron density map at 4.1-Å resolution (yellow) contoured at 1 σ, calculated by using observed amplitudes (Fobs) and phases after density modification. Superimposed is the final Cα Rpb4 (red) and Rpb7 (blue) model. The figure was generated by using O and POV-RAY (19).
Results and Discussion
The structure of complete, 12-subunit pol II was determined by molecular replacement with that of core pol II (Tables 1 and 2). All three previous structures, form 1, form 2, and transcribing complex, were used as search models. The transcribing complex structure gave the highest correlation coefficient and lowest initial R factor. Rigid body refinement with form 2, allowing the clamp to move, resulted in a position of the clamp essentially the same as that in the transcribing complex. We conclude that under the conditions analyzed here, the complete pol II is in the clamp-closed state. This conclusion is in agreement with results of electron microscopy (EM) and single-particle analysis of complete pol II, which also revealed the enzyme in the clamp-closed state, showing that this conformation was not induced by crystallization (20).
Table 1. Crystallographic data for complete pol II structure.
| Space group | C222(1) |
| Unit cell, Å | 224.0 by 394.5 by 284.3 |
| Molecules per asymmetric unit | 1 |
| Solvent content, % | 80 |
| Wavelength, Å | 0.98 |
| Mosaicity, degree | 0.43 |
| Resolution, Å | 40-4.1 (4.25-4.10) |
| Completeness, % | 98.8 (96.6) |
| Redundancy | 3.5 (3.0) |
| Unique reflections | 96,820 (9,357) |
| I/σI | 5.9 (1.06) |
| Rsym, % | 10.8 (61.4) |
Values in parentheses correspond to the highest-resolution shell. Rsym = Σi,h|I(i, h) - |I(h)〉|/Σi,h|I(i, h)|, where 〈I(h)〉 is the mean of the I observations of reflection h. Rsym was calculated with anomalous pairs merged; no σ cutoff was applied.
Table 2. Model data for complete pol II structure.
| Subunit | Residues in sequence | Residues in model | Identity to human | Model organism | Model PDB |
|---|---|---|---|---|---|
| Rpb4 | 221 | 151 | 32% | M. jannaschii | 1GO3 chain F |
| Rpb7 | 171 | 170 | 43% | M. jannaschii | 1GO3 chain E |
Difference density between the complete and core pol II structures clearly corresponded to the previously reported structure of archaeal Rpb4/Rpb7 (Fig. 1; ref. 21). As the crystals had a high solvent content (Table 1), density modification was performed to improve the map and help remove model bias. A backbone model could be built into the resulting map with the archaeal Rpb4/Rpb7 structure as a guide. The part of the model attributed to Rpb7 was virtually identical to the archaeal structure, in keeping with the sequence conservation between the yeast and archaeal proteins (25% identity, 34% similarity). The remainder of the model, attributed to Rpb4, was very similar to the structure of archaeal Rpb4. There is, however, no significant homology between yeast and archaeal Rpb4 sequences, and most homology between yeast and other eukaryotic Rpb4 sequences is located in the N-terminal 45 residues and C-terminal 75 residues (2, 21, 22). We therefore presume that the portion of the Rpb4 structure seen in the map is caused by the N- and C-terminal regions; a central, highly charged region of ≈70 residues, apparently unique to yeast, is not detected, because of motion or disorder.
Rpb7 interacts with both Rpb1 and Rpb6 (Fig. 2 and Fig. 4, which is published as supporting information on the PNAS web site, www.pnas.org). Based on alignment with the archaeal structure, a conserved region containing residues 15–20 (numbering scheme from Methanococcus jannaschii) appears to make a hydrophobic interaction with Ala-105 and Pro-106 of Rpb6. In archaeal Rpb7, conserved residues Gly-55, Gly-57, Gly-62, and Gly-64 (M. jannaschii numbering scheme) are located in a loop between two β-strands (21). In our map, residues corresponding to archeal 55, 57, and 59 appear to be in a β-strand that adds to a β-sheet region of Rpb1 around Val-1443 to Ile-1445, beneath the previously described “RNA exit groove 1” (10, 11). Residues 62 and 64 are in a loop penetrating the exit groove.
Again using the archaeal structure as a guide, we found that the N-terminal region of Rpb4 makes contact with the N-terminal region of Rpb1 around Ser-8 and Ala-9, located on the surface of the clamp above exit groove 1. Inasmuch as loops in Rpb1 that form the hinge for clamp movement are at the level of the exit groove, contacts of Rpb4 above the groove and Rpb7 below the groove would appear to bracket the clamp, constraining it in the closed state. It seems unlikely that the open conformations of the clamp seen in structures of free core pol II (10, 11) are possible in the presence of the Rpb4/Rpb7 heterodimer. As has been noted, the requirement for the heterodimer for the initiation of transcription and the effect of the heterodimer upon clamp closure suggest that promoter DNA binding and initiation occur in the clamp-closed state (20).
We previously considered the possibility of promoter DNA binding in the clamp-open state, which affords a straight path through the active center cleft for unbent promoter DNA (11). Binding in the cleft in the clamp-closed state requires bending the DNA to ≈90°, and such bending is likely to occur only after interaction with the polymerase and promoter melting. Interaction of straight promoter DNA with pol II in the clamp-closed state may occur as in the structure of the bacterial RNA polymerase holoenzyme–promoter DNA complex (23–25), in which the DNA passes above the clamp and adjacent protein “wall” [suggested for pol II by Craighead et al. (20)]. The DNA presumably descends into the active center region after melting and bending.
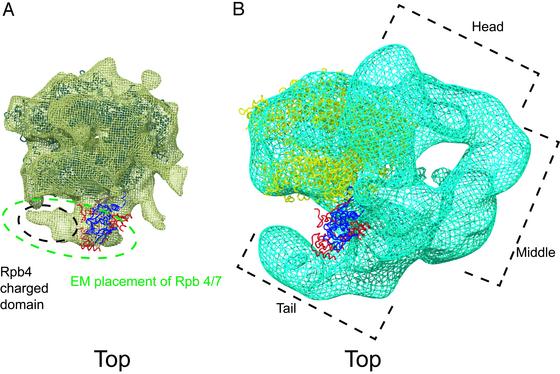
A second implication of the complete pol II structure for transcription concerns the possible involvement of Rpb7 in nucleic acid binding. Rpb7 contains a ribonucleoprotein (RNP) fold and an oligonucleotide-binding (OB) fold (dark and light blue, respectively, in Fig. 2) (2, 21). The Rpb4/Rpb7 heterodimer was shown to bind single-stranded DNA and RNA, and mutation of the OB fold abolished the binding (2). Previous structure determination of complete pol II by EM and single-particle analysis placed the heterodimer near RNA exit groove 1, leading to the suggestion that the heterodimer interacts with RNA emanating from the groove (20, 21). The location of the heterodimer in the x-ray structure agrees well with that determined by EM (Fig. 3A and Fig. 5, which is published as supporting information on the PNAS web site), although the orientation of the heterodimer differs from that previously proposed on the basis of the EM map (20). It is also consistent with results of immunoelectron microscopy on pol I (26), which led to the suggestion of heterodimer interaction with the “linker” domain near the C terminus of Rpb1 (see below). The volume occupied by the heterodimer in the EM map is sufficient to include not only the region of the heterodimer revealed in the x-ray structure, but also the central, charged domain of Rpb4 not seen in the x-ray map (Fig. 3A). Indeed a previous difference electron density map between EM structures of complete and core pol II may have been caused entirely by the charged domain (27).
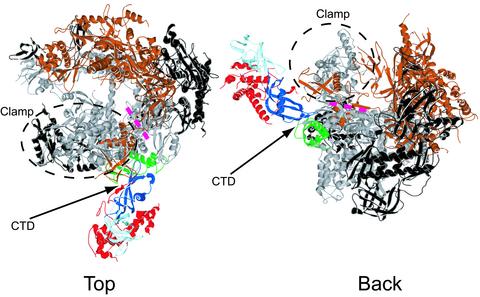
Fig. 2.
Backbone model of complete, 12-subunit pol II. Shown is a ribbon representation of the complete pol II structure (top and back views as in refs. 10 and 11). Rpb1 is gray, Rpb2 is bronze, Rpb4 is red, Rpb6 is green, the N-terminal half of Rpb7 that contains the RNP domain is dark blue, the C-terminal half of Rpb7 that contains the OB fold is light blue, and the remaining subunits are black. The locations of the clamp, the C-terminal repeat domain (CTD), and the previously proposed RNA exit groove 1 (pink dashed line) are indicated. The figure was generated with SWISS-PDB VIEWER and POV-RAY (40).
Fig. 3.
Relationship of complete pol II x-ray structure to EM structures of complete pol II (A) and Mediator–pol II complex (B). As this complex was prepared from exponentially growing yeast, it would have been largely deficient in Rpb4/Rpb7, accounting for the lack of density in this region of the EM map. The core pol II model is blue in A and yellow in B. Rpb4 is red and Rpb7 is dark blue. The figure was generated by using O and POV-RAY (19).
Details of the heterodimer in the x-ray structure further encourage speculation regarding RNA binding. The surface of the triple-stranded β-sheet of the RNP fold, involved in RNA binding in other examples of the fold, faces RNA exit groove 1 (28). As already mentioned, a loop containing residues 62 and 64, also involved in RNA binding in other instances, actually penetrates the groove. The question arises as to whether the RNP fold of Rpb7 has an affinity for RNA, because mutation of the OB fold abolished RNA binding in vitro (2). Binding was measured by gel electrophoretic mobility-shift analysis, and an affinity constant of a micromolar or less, which could significantly affect the stability of a transcribing complex, would not have been detected. It might be imagined that the RNP fold serves to guide the transcript toward the OB fold, which lies ≈50 Å from the exit of groove 1. A transcript length of 25–30 residues would be required to reach the OB fold, and both capping of the 5′ end and a transition to a stable transcribing complex occur at about this length (29–32).
The location of the Rpb4/Rpb7 heterodimer in the complete enzyme suggests a possible role in the assembly of the transcription initiation complex. The heterodimer is adjacent to the site of TFIIB binding in a pol II–TFIIB cocrystal (difference density attributable to TFIIB in the cocrystal is seen near RNA exit groove 1; unpublished work). Evidence for heterodimer–TFIIB interaction, stabilizing the transcription initiation complex, has come from surface plasmon resonance measurements, showing the greater affinity of a TFIIB–TATA box-binding protein–promoter DNA complex for complete pol II than for the core enzyme (27). Interaction of the heterodimer with TFIIB is also suggested by studies in the yeast pol III system, where the counterpart of Rpb4, termed C17, has been shown to bind the counterpart of TFIIB, termed Brf1, by two-hybrid and coimmunoprecipitation analyses (33). The location of the heterodimer in the complete enzyme in the vicinity of the CTD (Fig. 2) may be relevant to another reported interaction as well, that of Rpb4 with Fcp1, a phosphatase specific for the CTD (34).
Finally, the structure of complete pol II has possible implications for the mechanism of regulation by the multiprotein Mediator complex. Seven additional residues of Rpb1 could be traced in the complete structure beyond the N terminus seen in the core pol II structure. These additional residues, which appear to interact with Rpb7, form part of the linker between the CTD and the body of pol II (Fig. 2). The CTD is required for the binding of Mediator to pol II (35). The structure of a Mediator–pol II complex, determined at 35-Å resolution by EM and single-particle analysis, shows a crescent of Mediator density partly surrounding pol II (36). A gap between the “tail” region of the Mediator and the body of pol II, near the junction of the tail and “middle” regions, corresponds to the location of the Rpb4/Rpb7 and heterodimer in the x-ray structure (Fig. 3B), raising the possibility of direct Mediator–heterodimer interaction. There is genetic evidence for the involvement of both the heterodimer and Mediator in transcription control: Deletion of Rpb4 impairs the activating effect of Gal4 and other yeast regulatory proteins (37), and deletions of Mediator tail proteins have similar consequences (38, 39). The relevance of Mediator–heterodimer interaction to transcriptional control remains to be tested.
Supplementary Material
Acknowledgments
We thank the staff of the Berkeley Center for Structural Biology for help at the Advanced Light Source, Compaq for providing a Unix workstation, N. Thompson and R. Burgess for generously providing antibody for protein purification, R. Davis for the tandem affinity purification-tagged Rpb4 strain, and J. Craighead and F. Asturias for insightful discussion and access to maps from EM. This research is based in part on work done at the Stanford Synchrotron Radiation Laboratory, which is funded by the U.S. Department of Energy Office of Basic Energy Sciences. The structural biology program is supported by the National Institutes of Health National Center for Research Resources Biomedical Technology Program and the Department of Energy Office of Biological and Environmental Research. D.A.B. was supported by American Cancer Society Postdoctoral Fellowship PF-00-014-01-GMC. This research was supported by National Institutes of Health Grant GM49985 (to R.D.K.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: pol II, polymerase II; EM, electron microscopy; CTD, C-terminal repeat domain; OB, oligonucleotide binding; RNP, ribonucleoprotein.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1NIK).
See commentary on page 6893.
References
- 1.Edwards, A. M., Kane, C. M., Young, R. A. & Kornberg, R. D. (1991) J. Biol. Chem. 266, 71–75. [PubMed] [Google Scholar]
- 2.Orlicky, S. M., Tran, P. T., Sayre, M. H. & Edwards, A. M. (2001) J. Biol. Chem. 276, 10097–10102. [DOI] [PubMed] [Google Scholar]
- 3.Woychik, N. A. & Young, R. A. (1989) Mol. Cell. Biol. 9, 2854–2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McKune, K., Richards, K. L., Edwards, A. M., Young, R. A. & Woychik, N. A. (1993) Yeast 9, 295–299. [DOI] [PubMed] [Google Scholar]
- 5.Giaever, G., Chu, A. M., Ni, L., Connelly, C., Riles, L., Veronneau, S., Dow, S., Lucau-Danila, A., Anderson, K., Andre, B., et al. (2002) Nature 418, 387–391. [DOI] [PubMed] [Google Scholar]
- 6.Miyao, T., Barnett, J. D. & Woychik, N. A. (2001) J. Biol. Chem. 276, 46408–46413. [DOI] [PubMed] [Google Scholar]
- 7.Sheffer, A., Varon, M. & Choder, M. (1999) Mol. Cell. Biol. 19, 2672–2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan, Q., Li, X., Sadhale, P. P., Miyao, T. & Woychik, N. A. (2000) Mol. Cell. Biol. 20, 8124–8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pillai, B., Sampath, V., Sharma, N. & Sadhale, P. (2001) J. Biol. Chem. 276, 30641–30647. [DOI] [PubMed] [Google Scholar]
- 10.Cramer, P., Bushnell, D. A., Fu, J., Gnatt, A. L., Maier-Davis, B., Thompson, N. E., Burgess, R. R., Edwards, A. M., David, P. R. & Kornberg, R. D. (2000) Science 288, 640–649. [DOI] [PubMed] [Google Scholar]
- 11.Cramer, P., Bushnell, D. A. & Kornberg, R. D. (2001) Science 292, 1863–1876. [DOI] [PubMed] [Google Scholar]
- 12.Gnatt, A. L., Cramer, P., Fu, J., Bushnell, D. A. & Kornberg, R. D. (2001) Science 292, 1876–1882. [DOI] [PubMed] [Google Scholar]
- 13.Choder, M. & Young, R. A. (1993) Mol. Cell. Biol. 13, 6984–6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Puig, O., Caspary, F., Rigaut, G., Rutz, B., Bouveret, E., Bragado-Nilsson, E., Wilm, M. & Seraphin, B. (2001) Methods 24, 218–229. [DOI] [PubMed] [Google Scholar]
- 15.Myers, L. C., Leuther, K., Bushnell, D. A., Gustafsson, C. M. & Kornberg, R. D. (1997) Methods 12, 212–216. [DOI] [PubMed] [Google Scholar]
- 16.Bushnell, D. A., Bamdad, C. & Kornberg, R. D. (1996) J. Biol. Chem. 271, 20170–20174. [DOI] [PubMed] [Google Scholar]
- 17.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276, 307–326. [DOI] [PubMed] [Google Scholar]
- 18.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 19.Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47, 110–119. [DOI] [PubMed] [Google Scholar]
- 20.Craighead, J. L., Chang, W. H. & Asturias, F. J. (2002) Structure (Cambridge, Mass.) 10, 1117–1125. [DOI] [PubMed] [Google Scholar]
- 21.Todone, F., Brick, P., Werner, F., Weinzierl, R. O. & Onesti, S. (2001) Mol. Cell 8, 1137–1143. [DOI] [PubMed] [Google Scholar]
- 22.Siaut, M., Zaros, C., Levivier, E., Ferri, M. L., Court, M., Werner, M., Callebaut, I., Thuriaux, P., Sentenac, A. & Conesa, C. (2003) Mol. Cell. Biol. 23, 195–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vassylyev, D. G., Sekine, S., Laptenko, O., Lee, J., Vassylyeva, M. N., Borukhov, S. & Yokoyama, S. (2002) Nature 417, 712–719. [DOI] [PubMed] [Google Scholar]
- 24.Murakami, K. S., Masuda, S. & Darst, S. A. (2002) Science 296, 1280–1284. [DOI] [PubMed] [Google Scholar]
- 25.Murakami, K. S., Masuda, S., Campbell, E. A., Muzzin, O. & Darst, S. A. (2002) Science 296, 1285–1290. [DOI] [PubMed] [Google Scholar]
- 26.Peyroche, G., Levillain, E., Siaut, M., Callebaut, I., Schultz, P., Sentenac, A., Riva, M. & Carles, C. (2002) Proc. Natl. Acad. Sci. USA 99, 14670–14675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jensen, G. J., Meredith, G., Bushnell, D. A. & Kornberg, R. D. (1998) EMBO J. 17, 2353–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Varani, G. & Nagai, K. (1998) Annu. Rev. Biophys. Biomol. Struct. 27, 407–445. [DOI] [PubMed] [Google Scholar]
- 29.Woychik, N. A. & Hampsey, M. (2002) Cell 108, 453–463. [DOI] [PubMed] [Google Scholar]
- 30.Shatkin, A. J. & Manley, J. L. (2000) Nat. Struct. Biol. 7, 838–842. [DOI] [PubMed] [Google Scholar]
- 31.Proudfoot, N. J., Furger, A. & Dye, M. J. (2002) Cell 108, 501–512. [DOI] [PubMed] [Google Scholar]
- 32.Bentley, D. (2002) Curr. Opin. Cell. Biol. 14, 336–342. [DOI] [PubMed] [Google Scholar]
- 33.Ferri, M. L., Peyroche, G., Siaut, M., Lefebvre, O., Carles, C., Conesa, C. & Sentenac, A. (2000) Mol. Cell. Biol. 20, 488–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kimura, M., Suzuki, H. & Ishihama, A. (2002) Mol. Cell. Biol. 22, 1577–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Myers, L. C., Gustafsson, C. M., Bushnell, D. A., Lui, M., Erdjument-Bromage, H., Tempst, P. & Kornberg, R. D. (1998) Genes Dev. 12, 45–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davis, J. A., Takagi, Y., Kornberg, R. D. & Asturias, F. A. (2002) Mol. Cell 10, 409–415. [DOI] [PubMed] [Google Scholar]
- 37.Zhou, H. & Lee, K. A. (2001) Oncogene 20, 1519–1524. [DOI] [PubMed] [Google Scholar]
- 38.Myers, L. C. & Kornberg, R. D. (2000) Annu. Rev. Biochem. 69, 729–749. [DOI] [PubMed] [Google Scholar]
- 39.Jeong, C. J., Yang, S. H., Xie, Y., Zhang, L., Johnston, S. A. & Kodadek, T. (2001) Biochemistry 40, 9421–9427. [DOI] [PubMed] [Google Scholar]
- 40.Guex, N. & Peitsch, M. C. (1997) Electrophoresis 18, 2714–2723. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.