Abstract
Dengue virus is an emerging global health threat. Its major envelope glycoprotein, E, mediates viral attachment and entry by membrane fusion. A crystal structure of the soluble ectodomain of E from dengue virus type 2 reveals a hydrophobic pocket lined by residues that influence the pH threshold for fusion. The pocket, which accepts a hydrophobic ligand, opens and closes through a conformational shift in a β-hairpin at the interface between two domains. These features point to a structural pathway for the fusion-activating transition and suggest a strategy for finding small-molecule inhibitors of dengue and other flaviviruses.
Dengue virus, a member of the flavivirus family, imposes one of the largest social and economic burdens of any mosquito-borne viral pathogen (1). There is no specific treatment for infection, and control of dengue virus by vaccination has proved elusive (2). Several other flaviviruses are important human pathogens, including yellow fever, West Nile, tick-borne encephalitis virus (TBE), and Japanese encephalitis viruses (JE) (2).
Three structural proteins (“C,” “M,” and “E”) and a lipid bilayer package the positive-strand RNA genome of flaviviruses (3). The core nucleocapsid protein, C, assembles with RNA on the cytosolic face of the endoplasmic reticulum membrane. The assembling core buds through the endoplasmic reticulum membrane, thereby acquiring an envelope that contains the major envelope glycoprotein, E, and the so-called precursor membrane protein, prM. The particle passes through the secretory pathway, where a furin-like protease cleaves prM to M in a late trans-Golgi compartment. The cleavage, which removes most of the ectodomain of prM, releases a constraint on E and primes the particle for low-pH-triggered membrane fusion. Uncleaved, immature particles are not fusion competent (2, 3).
E, which mediates both receptor binding (4) and fusion (5), is a so-called “class II” viral fusion protein (6, 7). The more familiar class I fusion proteins, exemplified by the hemagglutinin of influenza virus and gp120/gp41 of HIV, have a “fusion peptide” at or near the N terminus of an internal cleavage point (8). This hydrophobic and glycine-rich segment, buried in the cleaved–primed trimer of the class I fusion protein, emerges when a large-scale conformational rearrangement is triggered by low pH (in the case of hemagglutinin), receptor binding (in the case of gp120/gp41), or other cell-entry related signal. The likely sequence of events that follows includes an interaction of the fusion peptide with the target-cell membrane and a refolding of the trimer. The latter step brings together the fusion peptide and viral-membrane anchor, thereby drawing together the cellular and viral membranes and initiating the bilayer fusion process (6). The class II proteins, found so far in flaviviruses and alphaviruses, have evolved a structurally different but mechanistically related fusion architecture (3, 7). As in class I proteins, a proteolytic cleavage (of prM to M in flaviviruses or pE2 to E2 in alphaviruses) yields mature virions, with the fusion proteins in a metastable conformation, primed for fusion. The fusion peptide, an internal loop at the tip of an elongated subdomain of the protein (5, 9), is buried at a protein interface and becomes exposed in the conformational change initiated by exposure to low pH (9, 10). Because only the prefusion structures of one flaviviral and one alphaviral envelope protein have previously been determined, we know rather little about the conformational rearrangements set in motion by exposure to low pH (in the early endosome after viral uptake). The structures do suggest that the conformational changes involve hinge motions about interdomain linkages (9), together with oligomeric rearrangements on the viral surface (11–13). In the case of the flaviviruses, the E dimers found on the surface of the virion recluster irreversibly into trimers when exposed to pH approximately <6.3 (11).
We report the structure of a soluble fragment (residues 1–394) of the E protein from dengue virus type 2. This fragment contains all but ≈45 residues of the E-protein ectodomain (Fig. 1A). It closely resembles the E protein from TBE in its dimeric structure and in the details of its protein fold, including a set of six disulfide bonds (9). Therefore, residues implicated as determinants of host range, tropism, or virulence in different flaviviruses and mutations that affect binding of neutralizing monoclonal antibodies map to the same locations in the dengue and TBE structures (see ref. 9 for a detailed analysis based on the TBE E structure). We have examined crystals grown in both the presence and the absence of the detergent n-octyl-β-D-glucoside (β-OG). The key difference between the two structures is a local rearrangement of the “kl” β-hairpin (residues 268–280) and the concomitant opening up of a hydrophobic pocket, occupied by a molecule of β-OG. Mutations affecting the pH threshold for fusion map to the hydrophobic pocket, which we propose is a hinge point in the fusion-activating conformational change. Detergent binding marks the pocket as a potential site for small-molecule fusion inhibitors.
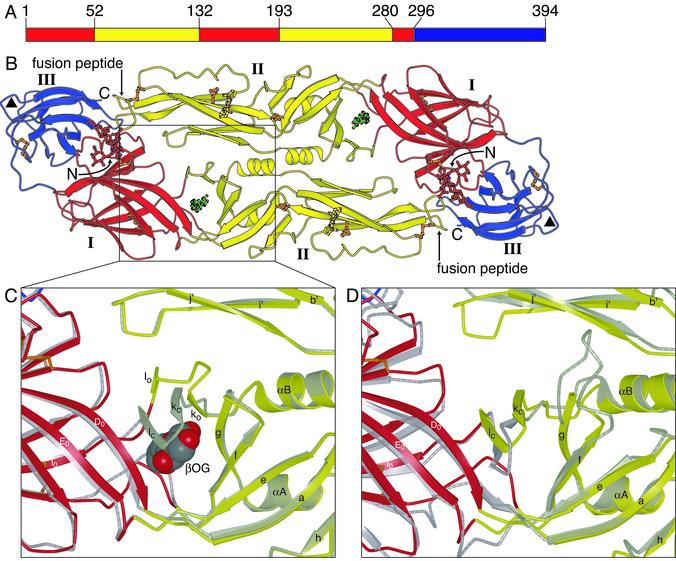
Fig. 1.
Dengue E protein and its ligand-binding pocket. (A) Domain definition of dengue E. Domain I is red, domain II is yellow, and domain III is blue. (B) The dengue E protein dimer, colored as in A, in complex with β-OG. The β-OG, shown in green, is bound in a hydrophobic pocket under the kl hairpin. A putative receptor-binding loop in domain III (residues 382–385) is marked with a triangle. The glycans in domains I and II are shown in a ball-and-stick representation in red and yellow, respectively. Disulfide bridges are shown in orange. (C) Enlargement of the kl hairpin region, with the structure of dengue E in the absence of β-OG (in gray) superimposed. The strands of the kl hairpin are labeled with “o” or “c” subscripts for the open (β-OG-bound) and closed forms, respectively. The β-OG molecule, shown in a space-filling representation, occupies the ligand-binding pocket. (D) Superposition of the structures of dengue E and TBE E, both in the absence of β-OG. Dengue E is colored as in B, and TBE E is in gray. The view is the same as in C. Figs. 1, 2, and 3 were generated with bobscript (33, 34) and raster 3d (35).
Materials and Methods
Expression, Purification, and Crystallization. E protein from dengue virus type 2 S1 strain (14) was supplied by Hawaii Biotech. The protein was expressed in Drosophila melanogaster Schneider 2 cells (American Type Culture Collection) from a pMtt vector (SmithKline Beecham) containing the dengue 2 prM and E genes (nucleotides 1–1185) as described by Ivy et al. (15). The resulting prM-E preprotein is processed during secretion to yield soluble E protein, which was purified from the cell culture medium by immunoaffinity chromatography (16). Crystals grow from a 10 g/liter solution at 4°C by hanging drop vapor diffusion in 11% polyethylene glycol 8000, 1 M sodium formate, 20% glycerol, and 0.1 M Hepes (pH 8). The addition of 0.5% β-OG before crystallization significantly improved the abundance and diffraction limit of the crystals. Dimensions of the primitive hexagonal cell were approximately a = b = 81 Å and c = 287 Å, with two molecules per asymmetric unit. An additional primitive hexagonal crystal form was observed, with cell dimensions a = b = 75 Å, c = 145 Å, and one molecule per asymmetric unit.
Data Collection and Processing. Crystals were derivatized by soaking in mother liquor containing 0.5 mM K2PtCl4, 0.5 mM Yb2(SO4)3, 0.5 mM KAu(CN)2, or 10 mM Me3PbAc for 24 h. Datasets were collected at 100°K on beamlines A1 and F1 of the Cornell High Energy Synchrotron Source (Cornell University, Ithaca, NY), except the Native1 dataset (Table 1), which was collected on beamline ID-19 at the Advanced Photon Source (Argonne National Laboratory, Argonne, IL). The data were processed with HKL (17). Statistics for data collection are summarized in Table 1.
Table 1. Crystallographic data statistics: Data collection and structure determination.
| Dataset | Native1 | Native2 | Me3PbAc | K2PtCl4 | YbSO4 | AuCN | Native3 | Native4 |
|---|---|---|---|---|---|---|---|---|
| Concentration of β-OG, mM | 17 | 17 | 17 | 17 | 17 | 17 | 0 | 0 |
| Resolution range, Å | 50-2.4 | 30-2.47 | 30-2.8 | 30-2.8 | 30-2.8 | 30-2.8 | 50-2.75 | 50-3.0 |
| Cell edges a(= b)/c | 81.6/287.4 | 81.2/286.6 | 81.3/286.8 | 81.3/286.9 | 81.1/286.5 | 81.2/285.5 | 81.5/288.6 | 74.6/144.7 |
| % completeness* | 97 (74) | 92 (45) | 99 (98) | 97 (88) | 99 (97) | 97 (99) | 90 (49) | 96 (82) |
| I/σ(I)* | 26.2 (3.3) | 15.1 (1.8) | 17.3 (4.0) | 17.8 (4.7) | 15.5 (2.7) | 11.9 (6.2) | 21.7 (2.5) | 13.6 (2.0) |
| Rmerge*†, % | 6.9 (28.9) | 6.1 (26.8) | 6.0 (27.4) | 8.2 (31.1) | 7.4 (39.2) | 5.3 (17.7) | 7.9 (40.9) | 8.4 (47.6) |
| No. of sites | 2 | 1 | 2 | 2 | ||||
| Phasing power‡ (centric/acentric; SHARP) | 0.85/1.3 | 0.43/0.52 | 0.25/0.49 | 0.36/0.54 | ||||
| Phasing power‡ (anomalous; SHARP) | 0.57 | 0.67 | 0.27 | 0.40 | ||||
| FOM§ centric (CNS/SHARP) | 0.68/0.24 | |||||||
| FOM§ acentric (CNS/SHARP) | 0.34/0.24 |
Numbers in parentheses are for the highest-resolution shells.
Rmerge = Σhkl|I - <I>|/ΣhklΣi(I).
Phasing power = (FH/lack of closure).
FOM = figure of merit ≈ cosine (phase error).
Structure Determination and Refinement. The pronounced anisotropy of the datasets was corrected by scaling each dataset anisotropically to a calculated dataset obtained from an arbitrary set of atomic coordinates. The datasets were scaled to the most isomorphous native dataset, Native2 (Table 1), and isomorphous difference Pattersons were calculated with SOLVE (18). Two initial heavy atom sites were identified by using the lead derivative. Additional sites were located in the three other derivative datasets by using cross-difference Fourier maps. Initial phases were optimized by refining the heavy atom parameters against maximum likelihood targets with SHARP (19). Phases were improved by solvent flattening and twofold noncrystallographic symmetry (NCS) averaging with DM (20) and RESOLVE (21). The solvent content was assumed to be 43%. The space group was determined as P3121, based on interpretable features in density-modified maps. An initial model was built into the maps with O (22). The atomic coordinates were refined against the best native dataset, Native1 (Table 1), first as a rigid body, then by simulated annealing using torsion angle dynamics with CNS (23). Further cycles also included restrained refinement of B-factors for individual atoms and energy minimization against maximum likelihood targets with CNS. Because the electron density for one of the molecules in the dengue E dimer was better defined than the other, the atomic coordinates of the two molecules were tightly restrained throughout refinement and therefore have very similar structures: the rms deviation is 0.34 Å (including side chain atoms). The B-factors were left unrestrained because of a large difference in overall B-factors for the two molecules in the asymmetric unit. The atomic model was completed by using 2Fo – Fc and Fo – Fc Fourier maps, and 137 water molecules were added by using an automated procedure in CNS and by visual inspection. The final model also includes two glycans, and one molecule of β-OG per protein molecule.
The structure of dengue E in the absence of β-OG was determined by refining the atomic coordinates against the Native3 dataset (Tables 1 and 2), which was collected from a crystal grown in the absence of β-OG. The protein atoms were first refined as six rigid bodies, corresponding to domains I, II, and III of each of the two chains in the asymmetric unit. The kl hairpin (residues 270–279) and residues 165–169 were completely rebuilt. Further refinement cycles consisted of simulated annealing with torsion angle dynamics, restrained B-factor refinement for individual atoms, and energy minimization against maximum-likelihood targets with CNS (23). The structure of dengue E without β-OG was also determined in a second crystal form (dataset Native4) by molecular replacement using a dengue E monomer as the search model in AMORE (24). The space group was identified in the translation search as P3221, with only one molecule per asymmetric unit. Rigid body refinement of domains I, II, and III resulted in substantial shifts, especially for domain II, which rotated ≈5° with respect to domains I and III. The axis of rotation passes through residue 193 and is roughly perpendicular to the dyad axis of the dimer. Further refinement cycles consisted of simulated annealing, restrained individual B-factor refinement, and energy minimization with CNS (23). The stereochemical quality of each atomic model was validated with PROCHECK (25). Statistics for data collection, phasing, and refinement are presented in Tables 1 and 2.
Table 2. Crystallographic data statistics: Model building and refinement.
| Dataset | Native1 (+β-OG) | Native3 (-β-OG) |
|---|---|---|
| Resolution range | 50-2.4 | 50-2.75 |
| Unique reflections | 44,435 | 24,851 |
| Rcryst* | 0.263 | 0.261 |
| Rfree† | 0.294 | 0.296 |
| Average B-factor, Å2 | ||
| Protein (chain A/B) | 88.7/64.5 | 79.1/72.8 |
| Solvent | 84.8 | 78.9 |
| rms deviation | ||
| Bond length, Å | 0.011 | 0.009 |
| Bond angle, o | 1.706 | 1.415 |
| Bonded B-factor, Å2 | ||
| Main chain | 4.37 | 3.30 |
| Side chain | 7.39 | 5.87 |
| Ramachandran plot, % | ||
| Favored | 82.3 | 73.2 |
| Allowed | 17.0 | 26.8 |
| Generous | 0.7 | 0.0 |
| Disallowed | 0 | 0 |
Rcryst = Σhkl∥Fo| -|<Fc>∥/Σhkl|Fo|.
Rfree = Rcryst using 5% of Fo sequestered before refinement.
Results and Discussion
Molecular Architecture of the Dengue E Dimer. Fig. 1B shows the three-domain structure of the dengue virus sE dimer. Domain I, the 8-stranded central β-barrel, organizes the structure. Its β-strands are denoted B0–I0, with the addition of a short amino-terminal strand (A0) parallel to strand C0 at one edge of the barrel. Insertions between strands D0 and E0 and strands H0 and I0 form the elongated domain II, which bears the fusion peptide at its tip (Fig. 1B). Domain II contains 12 β-strands, denoted a–l, and two α-helices, αA and αB. Domain III is an IgC-like module, with ten β-strands (A–G and a small extra sheet, AxCxDx; ref. 9). In all three domains, β-strands predominate. As expected from the 37% sequence identity between dengue and TBE sE, each domain of dengue sE has the same folded structure as its TBE counterpart, but several loops diverge in conformation. The relative domain orientations are also slightly different, consistent with the notion that the links between them might be flexible.
One consistent difference between E proteins from tick- and mosquito-borne flaviviruses is the presence in the latter of an additional four residues (382–385) between strands F and G of domain III. In our structure, these residues form a compact solvent-exposed bulge (Figs. 1B and 2 A). Their relatively high temperature factors suggest some degree of flexibility. This loop has been implicated in receptor binding in dengue virus (4).
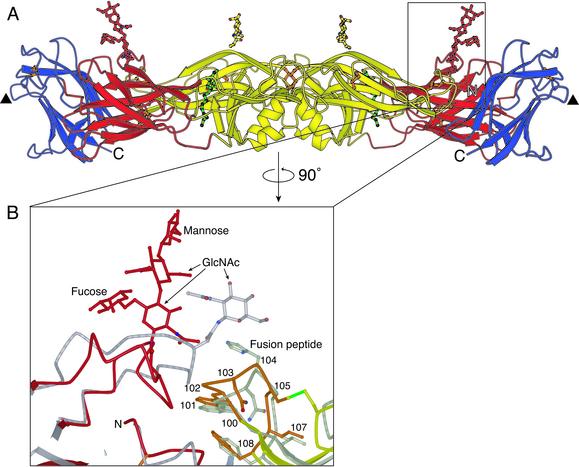
Fig. 2.
The glycan at residue 153 in dengue 2 virus E protein. (A) The E protein dimer, viewed perpendicular to the dyad axis (and the view in Fig. 1 A). Both glycans are approximately perpendicular to the viral surface. Domain I and the attached glycan are shown in red, domain II and the attached glycan are shown in yellow, and domain III is in blue. Disulfide bridges are shown in orange. The molecule of β-OG bound in the hydrophobic pocket underneath the kl hairpin is in green. A putative receptor-binding loop in domain III (residues 382–385) is marked with a triangle. (B) Enlargement of the area surrounding the glycan at residue 153 in domain I, with the structure of TBE envelope protein superimposed (gray) onto domain I of dengue virus E protein. The fusion peptide is highlighted in orange. The disulfide bridge between residues 92 and 105 is shown in green.
There are two glycosylated asparagines on each dengue E subunit: Asn-153 on domain I and Asn-67 on domain II (Fig. 2A). Asn-153, conserved in most flavivirus envelope proteins, bears a structure modeled here as a tetrasaccharide (Fig. 2B), although it contains additional, poorly ordered sugars. The fourth sugar is a mannose, which appears to be important for viral entry (26). The glycan projects outward from the surface of the protein, and somewhat discontinuous electron-density features suggest that it makes a crystal contact with the Asn-67 glycan of another sE dimer (Fig. 2). In TBE, its homolog extends laterally across the dimer interface and “covers” the fusion peptide (residues 100–108) on domain II of the dimer partner. In the absence of a crystal contact, the dengue Asn-153 oligosaccharide might do likewise. Indeed, stabilization of the dimer by the oligosaccharide would be consistent with the properties of mutants of dengue at position 153, which fuse with target membranes at a higher pH (27–29).
The Ligand-Binding Pocket. The most significant difference between the structures of dengue sE with and without β-OG is an altered conformation of the kl loop, which shifts toward the dimer contact in the presence of the detergent, forming a salt bridge and a hydrogen bond with i and j of the dimer partner. To effect this movement, strands k and l switch sheets, from F0E0D0lk to efgkl (Fig. 1C; also see figure 2 of ref. 9). The shift closes the “holes” along the dimer contact to either side of the twofold axis and opens a tapering, hydrophobic channel at the interface between domains I and II. This channel accepts a single β-OG molecule. The glucosyl head group of β-OG lies at the channel's mouth, with several hydrogen bonds fixing an ordered orientation; the hydrocarbon chain projects well into the channel's cavity. In TBE sE, which was studied in the absence of β-OG, the kl loop is in the “closed” position, and the hydrophobic residues are buried (Fig. 1D).
Mutations of residues that participate in the domain I/II interface just described alter the threshold pH for fusion (Fig. 3). Most of them involve side chains in the β-OG-binding pocket. We take this correlation as a strong indication that domains I and II indeed change orientation during the fusion-promoting conformational change. We propose that the shift in the kl hairpin opens the hydrophobic interface, allowing domain II to hinge away from its dimer partner and to project the fusion peptide at its tip toward the membrane of the target cell (Fig. 4A). Two crystallographic observations are consistent with such a hinge. When two different crystal forms of dengue sE obtained in the absence of β-OG are compared, domain II shifts by ≈5° with respect to domain I. The same is true for two different crystal forms of TBE sE (F. A. Rey and S.C.H., unpublished work). In both cases, the hinge angle is quite small, because a larger bend would disrupt the dimer contact at the tip of domain II and expose the fusion peptide. Indeed, it is just such a disruption that occurs at low pH.
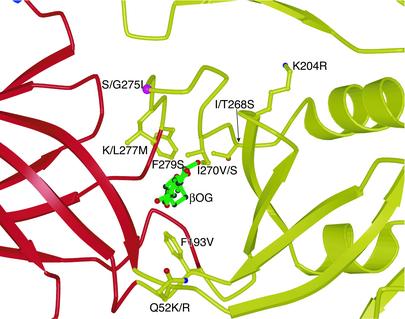
Fig. 3.
Mutations affecting the pH threshold of fusion (or virulence) in flaviviruses (36–41). The mutated residues line the interior of the ligand-binding pocket. For unconserved residues, the residue type in the virus in which the mutation was identified is listed first, followed by the residue type in dengue 2. The coloring is the same as in Fig. 1.
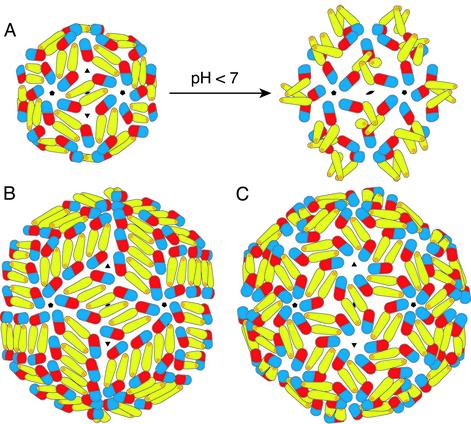
Fig. 4.
Proposed subunit packing interactions in various flaviviral icosahedral assemblies. (A) Suggested transition from the previously studied T = 1 subviral particles (12) to the fusion-competent T = 1 particle at low pH. On acidification, domain II is proposed to swing out about a hinge at the domain I/II interface, creating homotrimeric contacts at the threefold axis. Clusters of three fusion peptides are displayed at the tip of each trimer. (B) The packing in T = 3 virus-like particles deduced from image reconstructions of dengue virions (13). The 180 subunits are not related by local threefold symmetry. (C) Suggested T = 3 packing intermediate for the virion at low pH (13). E is shown in its native (high pH) conformation. Because all monomers are related by local threefold symmetry, the low-pH conformational change will result in the formation of trimers, as in A.
In the pH-threshold mutations, substitution of longer hydrophobic side chains by shorter ones generally lowers the maximum pH that triggers fusion (Fig. 3). We suggest that shorter side chains may allow a tighter and more stable closed form of the pocket, requiring a greater drop in pH to flip it open. Attenuated viruses with single mutations in the kl hairpin region have been obtained by passage in cell culture (30, 31). Accumulation of such mutations might result in even stronger attenuation.
Implications for Viral Assembly and Fusion. The outer surfaces of mature flavivirus particles contain 180 subunits each of E and M, in a compactly organized icosahedral array (3). Any conformational change in E is therefore likely to induce a concerted reorganization across the entire surface of the virion. The E proteins cluster into trimers when they undergo their conformational change induced by low pH (11). We do not yet know which domains contribute to the trimer contacts. Based on image reconstructions from electron cryomicroscopy of fusion-competent TBE recombinant subviral particles, which contain 60 subunits each of E and M (12), we propose that a hinge motion of domain II away from its dimer partner during the low-pH-induced transition could result in the formation of trimer contacts by domain II, with only a modest reorientation of domains I and III within the surface lattice (Fig. 4A). The resulting trimer would display three fusion-peptide loops at its tip. The packing of E deduced from image reconstructions of dengue virions (13) is at odds with this simple view, however, because the 90 dimers are not related by local threefold symmetry (Fig. 4B). It has been suggested that the surface proteins might rearrange to the structure shown in Fig. 4C as part of the low-pH-induced reorganization (13). Note the similarities between the structures shown in Figs. 4 A Left and C. As domain II bends out from the viral surface, it will release many of the surface-lattice packing constraints, giving individual E subunits (or groups of subunits) considerable lateral freedom. The very tight packing of subunits in the surface of the virion at neutral pH may therefore not, in practice, be a hindrance to the postulated rearrangement. The greater lateral freedom of E trimers on the viral surface is presumably also necessary for the fusion step, in which a further and more dramatic conformational change in E may be required to drive the cellular and viral membranes together.
In conclusion, we have identified the kl hairpin as a key structural element for initiating the low-pH conformational change that leads to formation of fusion-competent trimers. The opening up of a ligand-binding pocket just at the locus of a likely hinge suggests that compounds inserted at this position might hinder further conformational change and hence inhibit the fusion transition. In the context of the virion surface, their action might resemble that of some of the well studied antipicornaviral compounds, which block a concerted structural transition in the icosahedral assembly (32). Alternatively, small molecules that pry open the kl hairpin on binding in the ligand-binding pocket may inhibit infection by facilitating the low-pH conformational change, causing premature triggering. Our structural observations suggest direct ways to search for such inhibitors.
Acknowledgments
We thank George Gao, William Clemons, and the staff of Cornell High Energy Synchrotron Source beamlines F1 and A1 and Advanced Photon Source ID-19. This work was supported by a long-term fellowship to Y.M. from the Human Frontier Science Program Organization and by National Institutes of Health Grant CA13202 (to S.C.H.). S.C.H. is a Howard Hughes Medical Institute Investigator.
Abbreviations: TBE, tick-borne encephalitis virus; β-OG, n-octyl-β-D-glucoside.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID codes 1OAM and 1OAN).
See commentary on page 6899.
References
- 1.Gubler, D. J. (2002) Trends Microbiol. 10, 100–103. [DOI] [PubMed] [Google Scholar]
- 2.Burke, D. S. & Monath, T. P. (2001) in Fields Virology, eds. Knipe, D. M. & Howley, P. M. (Lippincott Williams & Wilkins, Philadelphia), pp. 1043–1125.
- 3.Lindenbach, B. D. & Rice, C. M. (2001) in Fields Virology, eds. Knipe, D. M. & Howley, P. M. (Lippincott Williams & Wilkins, Philadelphia), pp. 991–1041.
- 4.Crill, W. D. & Roehrig, J. T. (2001) J. Virol. 75, 7769–7773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allison, S. L., Schalich, J., Stiasny, K., Mandl, C. W. & Heinz, F. X. (2001) J. Virol. 75, 4268–4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weissenhorn, W., Dessen, A., Calder, L. J., Harrison, S. C., Skehel, J. J. & Wiley, D. C. (1999) Mol. Membr. Biol. 16, 3–9. [DOI] [PubMed] [Google Scholar]
- 7.Heinz, F. X. & Allison, S. L. (2001) Curr. Opin. Microbiol. 4, 450–455. [DOI] [PubMed] [Google Scholar]
- 8.Skehel, J. J. & Wiley, D. C. (2000) Annu. Rev. Biochem. 69, 531–569. [DOI] [PubMed] [Google Scholar]
- 9.Rey, F. A., Heinz, F. X., Mandl, C., Kunz, C. & Harrison, S. C. (1995) Nature 375, 291–298. [DOI] [PubMed] [Google Scholar]
- 10.Lescar, J., Roussel, A., Wien, M. W., Navaza, J., Fuller, S. D., Wengler, G. & Rey, F. A. (2001) Cell 105, 137–148. [DOI] [PubMed] [Google Scholar]
- 11.Allison, S. L., Schalich, J., Stiasny, K., Mandl, C. W., Kunz, C. & Heinz, F. X. (1995) J. Virol. 69, 695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferlenghi, I., Clarke, M., Ruttan, T., Allison, S. L., Schalich, J., Heinz, F. X., Harrison, S. C., Rey, F. A. & Fuller, S. D. (2001) Mol. Cell 7, 593–602. [DOI] [PubMed] [Google Scholar]
- 13.Kuhn, R. J., Zhang, W., Rossmann, M. G., Pletnev, S. V., Corver, J., Lenches, E., Jones, C. T., Mukhopadhyay, S., Chipman, P. R., Strauss, E. G., et al. (2002) Cell 108, 717–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hahn, Y. S., Galler, R., Hunkapiller, T., Dalrymple, J. M., Strauss, J. H. & Strauss, E. G. (1988) Virology 162, 167–180. [DOI] [PubMed] [Google Scholar]
- 15.Ivy, J., Nakano, E. & Clements, D. (1997) U.S. Patent 6,136,561.
- 16.Cuzzubbo, A. J., Endy, T. P., Nisalak, A., Kalayanarooj, S., Vaughn, D. W., Ogata, S. A., Clements, D. E. & Devine, P. L. (2001) Clin. Diagn. Lab. Immunol. 8, 1150–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276, 307–326. [DOI] [PubMed] [Google Scholar]
- 18.Terwilliger, T. C. & Berendzen, J. (1999) Acta Crystallogr. D 55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.La Fortelle, E. & Bricogne, G. (1997) Methods Enzymol. 276, 472–494. [DOI] [PubMed] [Google Scholar]
- 20.Collaborative Computational Project, Number 4 (1994) Acta Crystallogr. D 50, 760–763.15299374 [Google Scholar]
- 21.Terwilliger, T. C. (1999) Acta Crystallogr. D 55, 1863–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones, T. A., Zou, J.-Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47, 110–119. [DOI] [PubMed] [Google Scholar]
- 23.Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 24.Navaza, J. (2001) Acta Crystallogr. D 57, 1367–1372. [DOI] [PubMed] [Google Scholar]
- 25.Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993) J. Appl. Crystallogr. 26, 283–291. [Google Scholar]
- 26.Hung, S. L., Lee, P. L., Chen, H. W., Chen, L. K., Kao, C. L. & King, C. C. (1999) Virology 257, 156–167. [DOI] [PubMed] [Google Scholar]
- 27.Guirakhoo, F., Hunt, A. R., Lewis, J. G. & Roehrig, J. T. (1993) Virology 194, 219–223. [DOI] [PubMed] [Google Scholar]
- 28.Pletnev, A. G., Bray, M. & Lai, C. J. (1993) J. Virol. 67, 4956–4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kawano, H., Rostapshov, V., Rosen, L. & Lai, C. J. (1993) J. Virol. 67, 6567–6575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, E., Weir, R. C. & Dalgarno, L. (1997) Virology 232, 281–290. [DOI] [PubMed] [Google Scholar]
- 31.Monath, T. P., Arroyo, J., Levenbook, I., Zhang, Z. X., Catalan, J., Draper, K. & Guirakhoo, F. (2002) J. Virol. 76, 1932–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith, T. J., Kremer, M. J., Luo, M., Vriend, G., Arnold, E., Kamer, G., Rossmann, M. G., McKinlay, M. A., Diana, G. D. & Otto, M. J. (1986) Science 233, 1286–1293. [DOI] [PubMed] [Google Scholar]
- 33.Kraulis, P. J. (1991) J. Appl. Crystallogr. 24, 946–950. [Google Scholar]
- 34.Esnouf, R. M. (1997) J. Mol. Graphics 15, 132–134. [DOI] [PubMed] [Google Scholar]
- 35.Merritt, E. A. & Bacon, D. J. (1997) Methods Enzymol. 277, 505–524. [DOI] [PubMed] [Google Scholar]
- 36.Cecilia, D. & Gould, E. A. (1991) Virology 181, 70–77. [DOI] [PubMed] [Google Scholar]
- 37.Hasegawa, H., Yoshida, M., Shiosaka, T., Fujita, S. & Kobayashi, Y. (1992) Virology 191, 158–165. [DOI] [PubMed] [Google Scholar]
- 38.Lee, E., Weir, R. C. & Dalgaro, L. (1997) Virology 232, 281–290. [DOI] [PubMed] [Google Scholar]
- 39.Beasley, D. W. & Aaskov, J. G. (2001) Virology 279, 447–458. [DOI] [PubMed] [Google Scholar]
- 40.Hurrelbrink, R. J. & McMinn, P. C. (2001) J. Virol. 75, 7692–7702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Monath, T. P., Arroyo, J., Levenbook, I., Zhang, Z. X., Catalan, J., Draper, K. & Guirakhoo, F. (2002) J. Virol. 76, 1932–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]