Abstract
The crystal structure of the bifunctional enzyme 4-hydroxy-2-ketovalerate aldolase (DmpG)/acylating acetaldehyde dehydrogenase (DmpF), which is involved in the bacterial degradation of toxic aromatic compounds, has been determined by multiwavelength anomalous dispersion (MAD) techniques and refined to 1.7-Å resolution. Structures of the two polypeptides represent a previously unrecognized subclass of metal-dependent aldolases, and of a CoA-dependent dehydrogenase. The structure reveals a mixed state of NAD+ binding to the DmpF protomer. Domain movements associated with cofactor binding in the DmpF protomer may be correlated with channeling and activity at the DmpG protomer. In the presence of NAD+ a 29-Å-long sequestered tunnel links the two active sites. Two barriers are visible along the tunnel and suggest control points for the movement of the reactive and volatile acetaldehyde intermediate between the two active sites.
Enzymatic channeling is a process by which intermediates are moved directly between active sites in a sequential reaction pathway without equilibrating with the bulk phase (1, 2). Channeling processes are particularly advantageous over the free diffusion of reaction products through the bulk solvent because they can protect chemically labile intermediates from breakdown, prevent loss of nonpolar intermediates by diffusion across cell membranes, or protect the cell from toxic intermediates. Crystallographic studies on a number of different enzyme systems involved in substrate channeling (3–11) have revealed important structural factors that mediate intersubunit or inter-domain communication and facilitate the efficient transfer of intermediates between distant active sites.
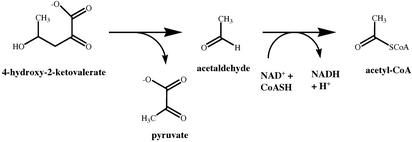
A bifunctional aldolase–dehydrogenase catalyzes the final two steps of the meta-cleavage pathway for catechol, an intermediate in many bacterial species in the degradation of phenols, toluates, naphthalene, biphenyls and other aromatic compounds (reviewed in ref. 12). Thus, 4-hydroxy-2-ketovalerate aldolase (DmpG; EC 4.1.3.-) and acetaldehyde dehydrogenase (acylating) (DmpF; EC 1.2.1.10) from a methylphenol-degrading pseudomonad convert 4-hydroxy-2-ketovalerate to pyruvate and acetyl-CoA by way of the intermediate acetaldehyde (Scheme 1).
Scheme 1.
The two enzymes are tightly associated with each other (13, 14). Whereas the aldolase appears to be inactive when expressed without the dehydrogenase, the dehydrogenase retains some activity when expressed in the absence of aldolase (14), suggesting that at least the dehydrogenase active site is distinct from that of the aldolase. Several lines of evidence are consistent with channeling of acetaldehyde between the active sites. For example, the conversion of 4-hydroxy-2-ketovalerate to acetyl-CoA occurs ≈20 times faster than that of acetaldehyde to acetyl-CoA, and the Km for acetaldehyde exceeds 50 mM, a physiologically irrelevant concentration (J.P., unpublished data). In addition, it has been shown that the aldolase activity is stimulated by the addition of the dehydrogenase cofactor to the assay mixture, suggesting that the reactivity of aldolase is allosterically modulated by the binding of the cofactor at the active site of DmpF (13).
To investigate the organization of the two active sites, the possible existence of a substrate channel connecting them, and how protein–protein interactions might facilitate efficient transfer of the intermediate acetaldehyde, we have determined the crystal structure of the bifunctional enzyme, DmpFG, from Pseudomonas sp. strain CF600, complexed with the cofactor NAD+. The structural results provide a unique view of the possible molecular events that facilitate the efficient transfer of acetaldehyde, a toxic and reactive intermediate, between two distant active sites without release to the cellular medium. Furthermore, the structures of a CoA-dependent aldehyde dehydrogenase and an aldolase from a subgroup of enzymes distinct from the existing structurally well characterized aldolases have not been determined previously.
Methods
Crystals of DmpFG were grown as described earlier (15). A slight modification in the procedure involved equilibrating the microseeded drops against 18% (wt/vol) polyethylene glycol rather than 15%, which was used to obtain the first multiple crystals used for seeds. Three-wavelength anomalous dispersion data to 2.2-Å resolution were collected from a selenomethionine (SeMet)-containing DmpFG crystal by using an ADSC Quantum 4 charge-coupled device detector at the Stanford Synchrotron Radiation Laboratory (Beamline 9.2). The native data were collected to 1.7-Å resolution from a single crystal at the Advanced Light Source, Lawrence Berkeley Laboratory (Beamline 5.0.2). The crystals were cryoprotected by using mother liquor with 20% (vol/vol) 2-methyl-2,4-pentanediol. The data collection and data processing statistics are shown in Table 1.
Table 1. Data collection and refinement statistics.
| Native | Inflection | Peak | Remote | |
|---|---|---|---|---|
| Collection | ||||
| Resolution, Å | 1.7 | 2.2 | 2.2 | 2.2 |
| Wavelength, Å | 1.000 | 0.97926 | 0.97907 | 0.97699 |
| Total reflections | 1,829,280 | 935,878 | 932,806 | 926,912 |
| Unique reflections | 299,052 | 260,811 | 260,928 | 261,232 |
| Completeness, % | 99.7 (99.3) | 98.8 (91.9) | 98.8 (92.0) | 98.8 (92.5) |
| Average I/α | 18.7 (2.0) | 12.9 (4.2) | 13.0 (3.7) | 14.2 (4.0) |
| Rmerge, % | 7.8 (56.5) | 7.5 (26.7) | 7.6 (30.1) | 7.0 (28.9) |
| Ranomalous | 0.072 | 0.091 | 0.065 | |
| Refinement | ||||
| Resolution range, Å | 20-1.7 | |||
| Total reflections used in refinement | 283,318 | |||
| R factor* | 0.189 | |||
| Free R factor† | 0.232 | |||
| rms deviation bond lengths, Å | 0.012 | |||
| rms deviation bond angles, ° | 1.5 | |||
| No. of nonhydrogen atoms | 22,429 | |||
| Average B factors, Å2 | 28.5 | |||
Statistics for the highest-resolution shells are given in parentheses.
R factor = ΣΣh∥Fo(h)| - k|Fc(h)∥/Σh|Fo(h)|.
Free R factor was calculated by using a 5% randomly selected subset of the total number of reflections.
The selenium atom substructure was located with the program Shake-and-Bake (SNB version 2.0) (16, 17), using the anomalous differences from the peak dataset. The experimental phases were extended to 1.7-Å resolution by using the free atom refinement method in ARP/WARP (18), and automatic chain tracing was carried out. The model was subjected to refinement using the program REFMAC (19). The final refinement statistics are shown in the lower part of Table 1. The stereochemical quality of the protein model was analyzed with PROCHECK (20). The figures have been made with MOLSCRIPT (21), RASTER3D (22), and SPOCK (23).
Results and Discussion
Structure Solution. The active biological unit of DmpFG is a 140-kDa tetramer consisting of two DmpFG heterodimers (DmpF is 32.5 kDa and DmpG is 37.5 kDa). The asymmetric unit of the orthorhombic crystals contains two DmpFG tetramers. The presence of 112 methionine residues in the asymmetric unit posed a formidable challenge to the application of multiwavelength anomalous dispersion phasing strategies for structure solution using a SeMet mutant form of the protein. The anomalous differences for the peak data from a SeMet mutant of the enzyme were used to successfully locate 86 of the total 112 selenium anomalous scatterers with the program SNB (16, 17). The successful substructure solution presented here constitutes one of the largest substructures that has been determined and represents a significant achievement in the application of direct methods to macromolecular structure determination.
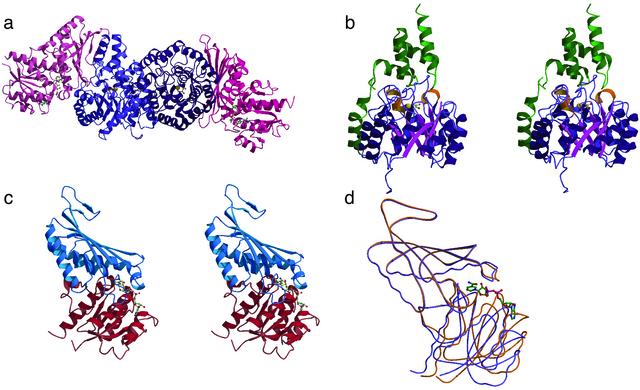
Overall Structure. Eight polypeptide chains, assigned as A–H, are present in the asymmetric unit of the crystal. The chains ACEG and BDFH represent DmpG and DmpF protomers, respectively; the four DmpFG heterodimers are named AB, CD, EF, and GH. The biological tetramers, consisting of chains A–D and E–H, are made up of two DmpFG heterodimers assembled such that the DmpG subunits from each heterodimer contact each other to form the core and the DmpF subunits are located at the peripheral ends of the tetrameric structure (Fig. 1a).
Fig. 1.
(a) Tetrameric assembly of the bifunctional enzyme DmpFG. The two DmpG molecules are shown in different shades of blue and the two DmpF protomers, located at the peripheral ends of the oligomer, are represented in different shades of red. The Mn2+ bound to DmpG is shown in rust and the oxalate and NAD+ bound to the active sites of DmpG and DmpF, respectively, are shown as ball-and-stick representations. (b) Stereo representation of the DmpG protomer. The TIM barrel domain is magenta and the helical communication domain is green. The helices that have been inserted into the barrel domain are shown in yellow. (c) Stereoview of the secondary structure of DmpF. The NAD+ domain is shown in red and the dimerization domain is in blue. (d) Worm representation showing the superposition of the holo form of DmpF (yellow) and the apo form of DmpF (purple). The α carbon atoms from the dimerization domains of DmpF (residues 131–285) for the holo and apo structures were used for the superposition. The NAD+ cofactor bound to the apoenzyme in c and d is represented as a ball-and-stick model.
DmpG is composed of two domains: an N-terminal (αβ)8 TIM barrel domain (residues 1–250) and a C-terminal helical communication domain (residues 251–345). The barrel exhibits a twofold repeat arrangement, (βα)1–4 and (βα)5–8, with two symmetrically related helical insertions between β1 and a1 and between β5 and α5 (Fig. 1b) that suggest this β-barrel protein may have evolved by gene duplication similar to what has been suggested for the barrel structures of HisA and HisF (24). The communication domain consists of five α-helices. Helix α9 moves the chain from the N terminus of the barrel to the C terminus, where the remaining four helices form an antiparallel helical bundle that plugs the C terminus of the barrel. This domain provides a significant number of intersubunit contacts with DmpF and thus appears to play a key role in the formation of the heterodimer.
The overall fold for DmpF resembles that of the glyceraldehyde-3-phosphate dehydrogenase enzyme family (25–27). The structure is organized into two domains: an NAD+-binding domain (residues 1–130 and 286–312) and a dimerization domain (residues 131–285) (Fig. 1c). Despite the fact that the enzyme had been crystallized in the presence of 2 mM NAD+, the structure reveals both apo and holo forms of DmpF. Three of the four DmpF protomers in the asymmetric unit are found in the holo form and one protomer is in the apo form. This mixture within a single crystal structure provides a unique opportunity to observe structural differences between the two forms of the enzyme and how these differences might contribute to activation of the aldolase in the presence of the pyridine nucleotide cofactor, bound to the dehydrogenase. A superposition of the holo and apo forms of DmpF reveals a significant movement of the main chain of the cofactor-binding domain relative to the dimerization domain (Fig. 1d). In the holo form of DmpF, the cofactor-binding domain rotates toward the dimerization domain, resulting in a more closed conformation relative to the apo form of the enzyme. Further structural comparisons between these two forms of the dehydrogenase reveal not only domain movements but also movements of specific amino acid side chains that may be important for the catalytic activity of the enzyme (described in more detail below). No insight is provided into the location of the CoA-binding site; an intriguing possibility is that it is shared with the NAD+-binding site if NADH dissociates before CoA binding, as in other CoA-dependent aldehyde dehydrogenases (28, 29).
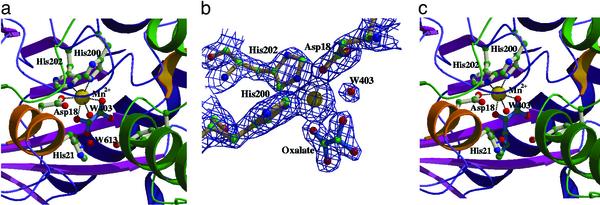
Active Sites. The active site of DmpG is located at the C terminus of the (α/β)8 barrel (Fig. 2a). The electron density within the active site clearly indicates the presence of a tightly bound metal ion. Biochemical analysis has shown that purified DmpG is partially active in the absence of added metal ions and that catalytic activity is stimulated upon addition of Mn2+ (13): preliminary studies using inductively coupled plasma–mass spectrometry detect significant quantities of Mn2+, but not Zn2+, in purified preparations of the enzyme (J.P. and L. Sahlman, unpublished results). On this basis, we have assigned the electron density in the active site as a Mn2+ ion. The metal ion ligands are Asp-18, His-200, His-202, and Wat-403.
Fig. 2.
(a) Active site of DmpG. The Mn2+ ion is shown as a yellow sphere. Red spheres correspond to bound water molecules. The oxalate ligand is shown with blue bonds. The dotted lines represent coordinating interactions between active site residues and the bound metal ion. (b)2Fo – Fc electron density in the active site of DmpG. The density is contoured at a 1.5-σ level. (c) Model of the substrate-bound complex of DmpG. The modeled substrate, 4-hydroxy-2-ketovalerate, is shown in blue bonds. The secondary structure elements are colored as described for Fig. 1.
In addition to the metal ion electron density at the active site, further electron density was found that completed octahedral coordination of the metal (Fig. 2b). This extra electron density was clearly observed for all four DmpG protomers in the asymmetric unit of the crystal. The identity of this bound ligand is unknown, and it is unlikely to be a known constituent of the crystallization solution, but either pyruvate (a product) or oxalate (a structural homologue to enolate pyruvate) can be successfully modeled into the observed electron density. The current model has oxalate modeled into the density with the two carboxylate oxygen atoms (O1 and O3) of oxalate forming equatorial ligands to the metal ion.
As mentioned above, a well-ordered water molecule (Wat-403) provides an axial ligand to the bound Mn2+. A second water molecule (Wat-613) is hydrogen bonded to the histidine residue (His-21) located in the insertion helix α1′ and to O4 of the bound oxalate molecule. Based on the positions of Wat-613 and the oxalate molecule we have modeled the substrate, 4-hydroxy-2-ketovalerate, in the active site of DmpG (Fig. 2c). The model was obtained from the HIC-Up (Hetero-compound Information Centre–Uppsala) web site (http://xray.bmc.uu.se/hicup/) (30) as 2-oxo-4-methylpentanoic acid. The 4-methyl group was changed to a hydroxyl group, the carboxylate and ketone moieties were superposed with those of the bound oxalate ligand and the hydroxyl group at Wat-613. In this model, the 4-hydroxyl group of the substrate lies within hydrogen bond distance of the side chains of His-21, Tyr-291, and Arg-17 and the main-chain oxygen atom of Gly-52.
The active site of DmpF is located at the interface between the NAD+-binding domain and the dimerization domain, where Cys-132 is adjacent to the nicotinamide ring of NAD+. Dehydrogenase activity, but not aldolase activity, is sensitive to inhibition by a thiol-modifying reagent, iodoacetate (31). Furthermore, a PSI-BLAST (32) search of the NCBI nonredundant protein sequence database (www.ncbi.nlm.nih.gov) revealed that the cysteine at position 132 of DmpF is the only strictly conserved cysteine within the protein sequence. In the apo structure, a water molecule is bound between the side chains of Cys-132 and Asp-209, suggesting that activation of Cys-132 may occur by means of base attack from the bound water, which is activated by Asp-209. Interestingly, the three holo structures of DmpF reveal no bound water between Cys-132 and Asp-209 and show two conformations for the side chain of Cys-132: one identical to that of the apo form and a second conformation where the thiol group is turned toward the bound NAD+ cofactor. This may indicate that once Cys-132 has been deprotonated by base attack from the bound water, the resulting hydronium ion is released and the thiolate side chain turns toward the bound cofactor in preparation for nucleophilic attack on the aldehyde intermediate. Such a mechanism has been postulated for CoA-independent aldehyde dehydrogenases (33).
Comparison of DmpG with Other Aldolases. Aldolase enzymes fall into two classes: those that rely on an active site lysine residue to assist in catalysis (class 1), and those in which aldol cleavage is assisted by divalent metal ions (class II). Structures are available for four class II aldolases: L-fuculose-1-phosphate aldolase (34, 35), fructose-1,6-bisphosphate aldolase (36, 37), tagatose-1,6-bisphosphate aldolase (38), and 2-dehydro-3-deoxygalactarate aldolase (18). The three latter enzymes display an (α/β)8 fold, whereas fuculose-1-phosphate aldolase does not. On this basis, it has been suggested that the class II aldolases fall into two distinct categories (37). On the basis of sequence identity and metal-binding preferences, class II aldolases exhibiting an (α/β)8 fold can be further subdivided into (i) fructose-1,6-bisphosphate and tagatose-1,6-bisphosphate aldolases, which share 23% sequence identity and bind Zn2+, and (ii) 2-dehydro-3-deoxygalactarate aldolase, unrelated in sequence to either of the above enzymes and preferentially binding Mg2+. The lack of significant sequence identity between DmpG and any of the above types of class II aldolases exhibiting an (α/β)8 fold (data not shown), and its unique metal-binding environment at the active site, suggest that DmpG represents a third subclass of (α/β)8 fold class II aldolases.
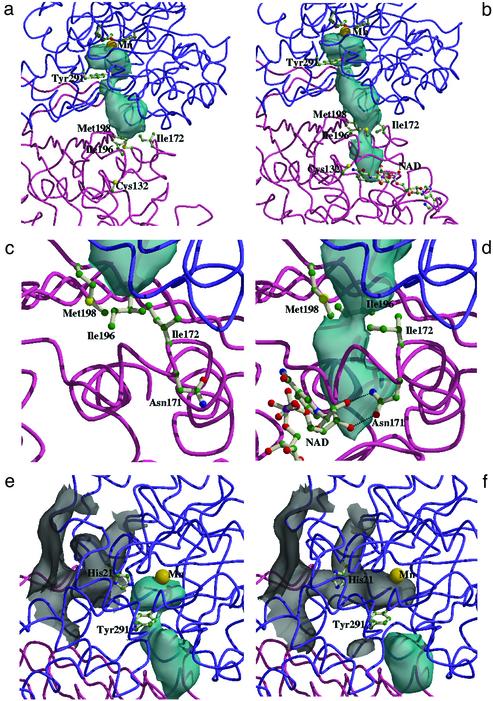
Molecular Channels Controlling Substrate and Intermediate Movement. An investigation of the solvent-accessible surfaces within the heterodimeric enzyme reveals an intriguing internal channel linking the two active sites (Fig. 3). Furthermore, significant differences in channel architecture are observed upon comparison of the apo and holo enzyme structures. In the presence of NAD+ the channel is 29 Å long, linking the aldolase and dehydrogenase active sites (Fig. 3b). The residues that line this “intermediate channel” are predominantly hydrophobic, providing an unreactive environment in which to move the highly reactive acetaldehyde intermediate. In the holoenzyme structure the two ends of the intermediate channel are blocked from accessing each of the active sites. The side chain of Tyr-291 obstructs entrance into the channel from the aldolase active site, and the side chains of Ile-172, Ile-196, and Met-198 block the exit from the channel to the dehydrogenase active site.
Fig. 3.
(a and b) Worm representation of DmpFG showing the apo form with the buried surface of the intermediate channel (a) and the holo form with the buried surface of the intermediate channel extending completely between the two active sites (b). (c and d) Worm representation showing a close-up view of the buried surface of the intermediate channel around the second barrier for the apo form (c) and the holo form (d) of the enzyme. The side chains of the residues that adopt multiple conformations at this barrier point are shown in a single conformation. In the holo form (d) this conformational arrangement results in an opening of the second barrier point from the intermediate tunnel to the active site of the DmpF molecule. (e and f) Worm representation showing the substrate entrance tunnel in the closed conformation blocked by the side chain of His-21 (e) and the open substrate entrance channel with His-21 in an alternative side-chain conformation (f). The DmpG chain is shown in purple and the DmpF chain is in magenta. Specific amino acid residues and the NAD cofactor are represented as ball-and-stick models, and the Mn2+ cofactor is shown as a yellow sphere. The intermediate channel is blue and the substrate entrance channel is gray. Hydrogen bonding interactions are shown in dotted lines. The solvent-accessible surfaces were computed with the program spock (23), using a probe radius of 1.4 Å.
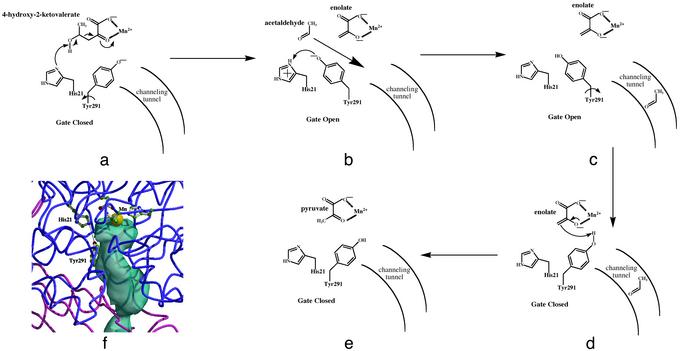
Close inspection of the active site of the aldolase and the position of Tyr-291 suggests that this residue may participate in two correlated functional events: (i) proton transfer and (ii) gating access of the acetaldehyde intermediate into the channel. These events are depicted in Fig. 4. The hydroxyl oxygen atom of Tyr-291 is hydrogen bonded to one of the carbonyl oxygen atoms of oxalate, which mimics the methylene group of the enolate intermediate, formed after aldolase cleavage. The position of His-21 in the active site of aldolase relative to the modeled substrate suggests that it is the base for abstraction of the 4-hydroxy proton (Fig. 4a). However, the side chain of His-21 is positioned too far from the methylene group of the enolate intermediate for direct protonation. Thus, the proton must be transferred to the enolate via either another residue or a water molecule. The structure shows the tyrosine side chain in contact with the bound oxalate ligand and access to the intermediate channel blocked. However, a torsional movement of Tyr-291 positions the phenolic oxygen within hydrogen bonding distance of His-21, from which it can accept the proton (Fig. 4b). This alternative conformation opens the “gate” to the intermediate channel, facilitating the movement of the intermediate into the channel (Fig. 4c). Tyr-291 is then repositioned to the gate-closed orientation (Fig. 4d), where it can protonate the enolate to form pyruvate (Fig. 4e). A model showing Tyr-291 in the alternative conformation with the intermediate channel open is shown in Fig. 4f, supporting the proposed reaction mechanism for aldolase activity and its role in gating the channeling activity. An alternative order of events may be envisioned in which the oxygen of Tyr-291 is initially in the protonated form and thus, in the gate-closed conformation, can protonate the enolate intermediate to form pyruvate. The tyrosine could then move to the gate-open conformation to reprotonate the phenolic oxygen by His-21. This gate-opening would result in channeling of the acetaldehyde intermediate after formation of pyruvate.
Fig. 4.
(a–e) Schematic representation of the postulated reaction mechanism for DmpG and the opening of the intermediate channel by Tyr-291. (f) Model of the Tyr-291 residue in the alternative conformation resulting in the channel open state. The protein chains are shown in worm representation with DmpG blue and DmpF magenta.
The second barrier, from the channel to the dehydrogenase active site, is made up of the side chains of three hydrophobic residues in the dehydrogenase protomer, Ile-172, Ile-196, and Met-198, all of which adopt multiple conformations. In the apoenzyme Ile-172 and Met-198 adopt multiple conformations, whereas in the holoenzyme Ile-196 and Met-198 adopt multiple conformations. The combinations of multiple conformations were analyzed in the context of how they affect the channel. In the apo form four conformational pairs were identified, all of which give a channel-closed structure (Fig. 3c). In contrast, in the holo forms, one of the four conformational pairs results in an opening of the channel (Fig. 3d). In addition, a unique conformation for Ile-172 is observed, which facilitates interaction between Asn-171 and NAD+. Thus the movement of Ile-172, induced by the interactions between the cofactor and Asn-171 and the multiple conformations of Ile-196 and Met-198, creates a structural arrangement that maintains the channel in an open state, allowing the intermediate to enter the dehydrogenase active site. This opening of the channel in the holo form may explain the mechanism by which cofactor binding at DmpF enhances DmpG activity (13). Clearly, if the exit from the intermediate channel remains closed, the aldehyde becomes trapped in the channel. The presence of the cofactor effectively enables complete channeling between the two active sites.
The structure of DmpFG does not reveal a solvent-accessible entrance into the active site of the aldolase protomer for the substrate. However, inspection of the surface of the molecule reveals a channel, positioned between the barrel domain and the communication domain, extending from the external aqueous environment toward the DmpG active site (Fig. 3e). The solvent-accessible path through this channel is obstructed by the side chains of His-21 and Ser-293; however, a torsional movement of the side chain of His-21 results in an opening of the channel, suggesting an entry point by which the substrate may access the DmpG active site (Fig. 3f). Once the substrate is positioned in the active site the side chain of His-21 may swing into the correct position to catalyze the bond cleavage reaction. Furthermore, the movement of the side chain of His-21 sequesters the aldolase active site such that the deleterious aldehyde is not able to escape through the entrance route; rather, it must move through the intermediate channel directly to the dehydrogenase active site.
In conclusion, these studies have provided us with a unique view of the role protein structure plays in maintaining a sequestered environment for the reactive and volatile aldehyde intermediate. Furthermore, the structure reveals mechanisms by which communication between the two proteins could control the movement of the intermediate between distant active sites. Further structural and kinetic studies of this bifunctional enzyme are needed to establish the role of specific residues in the protein in mediating subunit communication and substrate channeling.
Acknowledgments
We thank N. Croteau, R. Coulombe, and N. Skouris for technical assistance, P. Lario for helpful discussions, and C. Weeks for assistance with the program SnB and for access to the SnB server computational facilities at the Woodward–Hauptmann Medical Foundation. In addition, we thank A. Deacon and L.-W. Huang for assistance with data collection. This work was financially supported by research grants from the National Institutes of Health (to A.V.) and the Natural Sciences and Engineering Research Council (to J.P.).
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1NVM).
References
- 1.Ovadi, J. (1991) J. Theor. Biol. 152, 1–22. [PubMed] [Google Scholar]
- 2.Srere, P. A. (1987) Annu. Rev. Biochem. 56, 89–124. [DOI] [PubMed] [Google Scholar]
- 3.Chaudhuri, B. N., Lange, S. C., Myers, R. S., Chittur, S. V., Davisson, V. J. & Smith, J. L. (2001) Structure 9, 987–997. [PubMed] [Google Scholar]
- 4.Hyde, C. C., Ahmed, S. A., Padlan, E. A., Miles, E. W. & Davies, D. R. (1988) J. Biol. Chem. 263, 14925–14931. [PubMed] [Google Scholar]
- 5.Kim, J. H., Krahn, J. M., Tomchick, D. R., Smith, J. L. & Zalkin, H. (1996) J. Biol. Chem. 271, 15549–15557. [DOI] [PubMed] [Google Scholar]
- 6.Knighton, D. R., Kan, C.-C., Howland, E., Janson, C. A., Hostomska, Z., Welsh, K. M. & Matthews, D. A. (1994) Nat. Struct. Biol. 1, 186–194. [DOI] [PubMed] [Google Scholar]
- 7.Krahn, J. M., Kim, J. H., Burns, M. R., Parry, R. J., Zalkin, H. & Smith, J. L. (1997) Biochemistry 36, 11061–11068. [DOI] [PubMed] [Google Scholar]
- 8.Muchmore, C. R. A., Krahn, J. M., Kim, J. H., Zalkin, H. & Smith, J. L. (1998) Protein Sci. 7, 39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith, J. L., Zaluzec, E. J., Wery, J. P., Niu, L., Switzer, R. L., Zalkin, H. & Satow, Y. (1994) Science 264, 1427–1433. [DOI] [PubMed] [Google Scholar]
- 10.Larsen, T. M., Boehlein, S. K., Schuster, S. M., Richards, N. G., Thoden, J. B., Holden, H. M. & Rayment, I. I. (1999) Biochemistry 38, 16146–16157. [DOI] [PubMed] [Google Scholar]
- 11.Drennan, C. L., Heo, J., Sintchak, M. D., Schreiter, E. & Ludden, P. W. (2001) Proc. Natl. Acad. Sci. USA 98, 11973–11978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Powlowski, J. & Shingler, V. (1994) Biodegradation 5, 219–236. [DOI] [PubMed] [Google Scholar]
- 13.Powlowski, J., Sahlman, L. & Shingler, V. (1993) J. Bacteriol. 175, 377–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shingler, V., Powlowski, J. & Marklund, U. (1992) J. Bacteriol. 174, 711–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manjasetty, B. A., Croteau, N., Powlowski, J. & Vrielink, A. (2001) Acta Crystallogr. D 57, 582–585. [DOI] [PubMed] [Google Scholar]
- 16.Miller, R., Gallo, S. M., Khalak, H. G. & Weeks, C. M. (1994) J. Appl. Crystallogr. 27, 613–621. [Google Scholar]
- 17.Weeks, C. M. & Miller, R. (1999) J. Appl. Crystallogr. 32, 120–124. [Google Scholar]
- 18.Izard, T. & Blackwell, N. C. (2000) EMBO J. 19, 3849–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997) Acta Crystallogr. D 53, 240–255. [DOI] [PubMed] [Google Scholar]
- 20.Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993) J. Appl. Crystallogr. 26, 283–291. [Google Scholar]
- 21.Kraulis, P. (1991) J. Appl. Crystallogr. 24, 946–950. [Google Scholar]
- 22.Merritt, E. A. & Bacon, D. J. (1997) Methods Enzymol. 277, 505–525. [DOI] [PubMed] [Google Scholar]
- 23.Christopher, J. A. (1998) spock, The Structural Properties Observation and Calculation Kit (Center for Macromolecular Design, Texas A&M Univ., College Station).
- 24.Lang, D., Thoma, R., Henn-Sax, M., Sterner, R. & Wilmanns, M. (2000) Science 289, 1546–1550. [DOI] [PubMed] [Google Scholar]
- 25.Cobessi, D., Tete-Favier, F., Marchal, S., Azza, S., Branlant, G. & Aubry, A. (1999) J. Mol. Biol. 290, 161–173. [DOI] [PubMed] [Google Scholar]
- 26.Hadfield, A., Kryger, G., Ouyang, J., Petsko, G. A., Ringe, D. & Viola, R. (1999) J. Mol. Biol. 289, 991–1002. [DOI] [PubMed] [Google Scholar]
- 27.Vellieux, F. M. D., Hajdu, J., Verlinde, C., Groendijk, H., Read, R. J., Greenhough, T. J., Campbell, J. W., Kalk, K. H., Littlechild, J. A., Watson, H. C. & Hol, W. G. J. (1993) Proc. Natl. Acad. Sci. USA 90, 2355–2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shone, C. C. & Fromm, H. J. (1981) Biochemistry 20, 7494–7501. [DOI] [PubMed] [Google Scholar]
- 29.Smith, L. T. & Kaplan, N. O. (1980) Arch. Biochem. Biophys. 203, 663–675. [DOI] [PubMed] [Google Scholar]
- 30.Kleywegt, G. J. & Jones, T. A. (1998) Acta Crystallogr. D 54, 1119–1131. [DOI] [PubMed] [Google Scholar]
- 31.Monastariokas, S. (1998) M.Sc. thesis (Concordia Univ., Montreal).
- 32.Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hempel, J., Perozich, J., Chapman, T., Rose, J., Boesch, J. S., Liu, Z.-J., Lindahl, R. & Wang, B.-C. (1999) Adv. Exp. Med. Biol. 463, 53–59. [DOI] [PubMed] [Google Scholar]
- 34.Dreyer, M. K. & Schulz, G. E. (1993) J. Mol. Biol. 231, 549–553. [DOI] [PubMed] [Google Scholar]
- 35.Dreyer, M. K. & Schulz, G. E. (1996) J. Mol. Biol. 259, 458–466. [DOI] [PubMed] [Google Scholar]
- 36.Blom, N. S., Tetrault, S., Coulombe, R. & Sygusch, J. (1996) Nat. Struct. Biol. 3, 856–862. [DOI] [PubMed] [Google Scholar]
- 37.Cooper, S. J., Leonard, G. A., McSweeney, S. M., Thompson, A. W., Naismith, J. H., Qamar, S., Plater, A., Berry, A. & Hunter, W. N. (1996) Structure 4, 1303–1315. [DOI] [PubMed] [Google Scholar]
- 38.Hall, D. R., Bond, C. S., Leonard, G. A., Watt, C. I., Berry, A. & Hunter, W. N. (2002) J. Biol. Chem. 277, 18–24. [DOI] [PubMed] [Google Scholar]