Abstract
In response to elicitors in the oral secretions of caterpillars, plants produce and release volatile chemicals that attract predators and parasitoids of the caterpillar while it feeds. The most prevalent elicitors are fatty acid amides consisting of 18-carbon polyunsaturated fatty acids coupled with l-glutamine. We demonstrate rapid CoA- and ATP-independent in vitro biosynthesis of the fatty acid amide elicitor, N-linolenoyl-l-glutamine, by microsomal fractions of several alimentary tissues in Manduca sexta. N-linolenoyl-l-glutamine is a structural analog of several other elicitors including volicitin, the first fatty acid amide elicitor identified in caterpillars. The enzyme(s) that catalyzed biosynthesis of N-linolenoyl-l-glutamine was localized within the integral membrane protein fraction extracted from microsomes by Triton X-114 detergent phase partitioning and had maximum activity at alkaline pH. We found no evidence suggesting microbial or tissue-independent biosynthesis of N-linolenoyl-l-glutamine in M. sexta. The in vitro biosynthesis of N-linolenoyl-l-glutamine by membrane-associated enzyme(s) in M. sexta represents direct evidence of fatty acid amide synthesis by caterpillar tissues.
Keywords: fatty acid amide, linolenic acid, plant–insect interactions
Plants actively produce and release volatile chemical signals in response to elicitors present in the oral secretions of foraging lepidopterous herbivores (1–6). These volatile chemicals play a major role in enabling natural enemies of the herbivores such as insect parasitoids to locate host caterpillars (1, 7). Compounds found in the oral secretions of several species of caterpillars that have been reported to elicit volatile emission by plants include β-glucosidase (8) and a variety of fatty acid amides (FAAs) (4, 5, 9–11). The FAA elicitors comprise a family of compounds consisting of 18-carbon polyunsaturated fatty acids coupled to L-glutamine (5, 12) or L-glutamic acid (6, 13). Three structurally similar amides of linolenic acid, N-[17-hydroxylinolenoyl]-L-glutamine (volicitin), N-linolenoyl-L-glutamine, and N-linolenoyl-L-glutamic acid, are responsible for the majority of elicitor activity associated with the oral secretions of the caterpillar species analyzed thus far (4, 6, 10, 11). Two of these FAA elicitors, N-linolenoyl-L-glutamic acid and N-linolenoyl-L-glutamine, are present in the oral secretions of the tobacco hornworm, Manduca sexta (Fig. 1A), and their biological activities have been determined (6, 13).
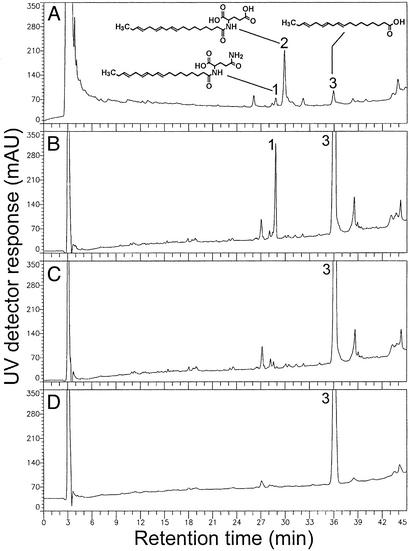
Fig. 1.
HPLC chromatograms of linolenic acid and related FAAs (structures shown) detected by UV analysis (200 nm) in M. sexta oral secretions (regurgitant) (A), M. sexta anterior midgut microsomes after in vitro incubation for 8hat4°C with sodium linolenate and either glutamine (B) or glutamic acid (C), and microbial suspensions derived from M. sexta regurgitant after in vitro incubation for8hat4°C with sodium linolenate and glutamine (D). Numbers represent the following: 1, N-linolenoyl-l-glutamine; 2, N-linolenoyl-l-glutamic acid; 3, linolenic acid. mAU, milliabsorbance units.
Despite numerous studies dealing with the induced emission of volatiles produced by plants in response to insect herbivory and the effect these volatile chemicals have on parasitoids of herbivores, little research has addressed the recurring question of how and where FAA elicitors are synthesized. Paré et al. (12) showed that the linolenic acid moiety of volicitin and N-linolenoyl-L-glutamine was derived from the diet of the caterpillar and suggested that the hydroxylation of linolenic acid (in volicitin) and its coupling to glutamine occurred within caterpillars via a hitherto uncharacterized biosynthetic mechanism(s). Research thus far has focused on gut contents or regurgitant collected from caterpillars as the most likely source of enzyme(s) involved with the biosynthesis of FAA elicitors. Spiteller et al. (14) showed that the regurgitant of Spodoptera exigua was capable of significantly delayed in vitro synthesis of an N-acylamino acid conjugate with general similarity to FAAs found in caterpillar oral secretions and postulated that gut microbes in the regurgitant might play a role in elicitor biosynthesis. Later, Mori et al. (15) showed rapid and complete enzymatic decomposition of FAA elicitors within regurgitant and, to a lesser degree, by gut tissues isolated from Heliothis virescens and Helicoverpa zea. Involvement of caterpillar tissues in FAA elicitor biosynthesis and the subcellular localization of putative enzyme(s) responsible for coupling linolenic acid with glutamine have not been reported. The objective of this study was to determine the role alimentary tissues and their subcellular fractions play in the biosynthesis of FAAs commonly found in the oral secretions of M. sexta and many other lepidopterous caterpillars. We demonstrate CoA-, ATP-, and NADPH-independent in vitro biosynthesis of N-linolenoyl-L-glutamine, an FAA elicitor of plant volatile emission, by M. sexta salivary gland, crop, and anterior midgut microsomes. An enzyme(s) that catalyzes rapid biosynthesis of N-linolenoyl-L-glutamine was localized within the integral membrane protein fraction extracted from microsomes by Triton X-114 detergent phase partitioning and exhibited optimal activity at alkaline pH, similar to the measured pH of the crop and midgut lumen.
Materials and Methods
Insect Rearing and Tissue Preparation. Tobacco hornworm, M. sexta, larvae were obtained from the Department of Entomology, North Carolina State University (Raleigh) and reared at 28°C, 74% relative humidity, and 14-h photoperiod on tobacco hornworm artificial diet (Southland Products, Lake Village, AR). M. sexta oral secretions (regurgitant) were collected as described (7). Labial salivary gland, crop, and anterior midgut tissues were removed from early fifth-instar M. sexta larvae that were anesthetized by immersion in tepid water for 15 min. The anterior midgut (mesenteron) was defined as the tissue extending from the posterior end of the crop to midway between attachment points for the dorsal tracheae originating from the second and third abdominal spiracles (16). Hemolymph, gut contents, peritrophic matrix (membrane), and crop contents were isolated from larval tissues and retained for further analysis. The pH of the crop and anterior midgut lumen were determined by measuring 100-μl aliquots of crop and midgut contents with a Horiba model B-213 twin pH tester (Horiba Instruments, Irvine, CA). The exterior of all tissues and the interior (lumen) of gut and crop tissues were rinsed thoroughly with a stream of sterile PBS/50 mM K2HPO4 (pH 7.5) containing 150 mM NaCl before microsome isolation.
Microsome Isolation. Rinsed M. sexta salivary gland, crop, and midgut tissues were homogenized in a Duall ground-glass tissue grinder (3 ml capacity, Kontes) with ice-cold homogenization buffer (3 ml/g fresh tissue) consisting of 50 mM K2HPO4 (pH 7.5) and 250 mM sucrose. To avoid possible interaction with the target enzyme activity, protease inhibitors were not added to homogenization buffer. The homogenates were centrifuged for 15 min at 9,000 × g, and the recovered supernatants were subsequently centrifuged for 90 min at 150,000 × g in an ultracentrifuge (Beckman L8–70M equipped with an SW 60 rotor) cooled to 4°C. The supernatants, containing cytosolic components, were retained for further analysis, and the microsome pellets were washed by resuspension in 2 ml of fresh homogenization buffer containing 150 mM KCl. After recentrifugation for 90 min at 150,000 × g, the washed microsome pellets were suspended in 200 μl of homogenization buffer and sonicated for 30 min by using a Bransonic 5210 ultrasonic bath (Branson) just before in vitro N-linolenoyl-L-glutamine biosynthesis assays. Protein quantities were determined with the detergent-compatible Bio-Rad DC protein assay (17) by using BSA fraction V as a standard. SDS (0.5% wt/vol) was included to enhance microsomal protein solubility before assays.
Triton X-114 Phase Partitioning of Microsomes. Phase partitioning of integral (intrinsic), peripheral (extrinsic), and extremely hydrophobic proteins from freshly prepared tissue microsomes was performed by using Triton X-114, based on the method of Bordier (18). Anterior midgut microsomes from early fifth-instar M. sexta larvae were suspended in ice-cold 25 mM imidazole-HCl (pH 7.5) to a final protein concentration of 1.9 mg/ml, and Triton X-114 (reduced form) (Sigma) was added to a final concentration of 2% (vol/vol). After incubation for 20 min on ice with occasional mixing, the temperature of the mixture was increased to 35°C in a water bath for 5 min to initiate phase separation of detergent micelles. The upper layer (detergent-depleted) containing peripheral membrane proteins and the lower layer (detergent-enriched) containing integral membrane proteins were collected separately for in vitro N-linolenoyl-L-glutamine biosynthesis assays. The pellet remaining after phase partitioning was dissolved in 200 μl of 25 mM imidazole-HCl (pH 7.5) containing 2% SDS (wt/vol) and centrifuged at 14,000 × g to yield a supernatant containing extremely hydrophobic proteins. Integral, peripheral, and extremely hydrophobic protein quantities were determined as they were for microsomes by using the Bio-Rad DC protein assay.
Microbial Suspensions. Aliquots (50 μl) of fresh regurgitant and each subcellular fraction (microsomal and cytosolic) isolated from M. sexta salivary gland, crop, or midgut tissues were plated separately on nutrient agar (catalog no. 78-5300, Carolina Biological Supply) and incubated overnight at the larval rearing temperature (28°C). All bacterial colonies growing on individual nutrient agar plates were lifted by using a sterile inoculation loop and combined as crude suspensions to an optical density (OD600) of 1.0–1.5 in 200 μl of sterile 50 mM K2HPO4 (pH 7.5) containing 250 mM sucrose. Microbial suspensions were used immediately for N-linolenoyl-L-glutamine biosynthesis assays.
Substrates for in Vitro N-linolenoyl-l-glutamine Biosynthesis Assays. Sodium linolenate was prepared by mixing 860 mg of linolenic acid (Sigma) with 120 mg of powdered sodium hydroxide (Fisher Scientific) and 2 ml of water in a screw-capped container. The container was filled with nitrogen, capped, and stirred in a water bath at 60°C overnight. After cooling to room temperature, the solidified product (sodium linolenate) was dissolved in 5 ml of water to a final concentration of 185 mg/ml. Free L-glutamine and L-glutamic acid monosodium salt were purchased (Sigma).
Assays for in Vitro Biosynthesis of N-linolenoyl-l-glutamine by Subcellular Fractions of Alimentary Tissues. Microsomal membrane (fresh or boiled for 3 min) and cytosolic fractions from each M. sexta tissue were diluted with 25 mM imidazole-HCl (pH 7.5) to contain 100 μg of protein in a total sample volume of 150 μl. The substrates sodium linolenate (5 μg/μl) and l-glutamine (20 μg/μl) were added to each sample to initiate the in vitro biosynthesis of N-linolenoyl-L-glutamine. In separate assays, L-glutamine was replaced by L-glutamic acid (monosodium salt) (20 μg/μl) to test for in vitro biosynthesis of N-linolenoyl-L-glutamic acid. After incubation for 8 h at 4°C, all assay mixtures were analyzed by high-performance liquid chromatography (HPLC). The incubation temperature of 4°C was chosen to minimize the likelihood of microbial growth or enzyme degradation in the absence of protease inhibitors. However, duplicate samples were incubated concurrently at 21°C for comparison. All assays were performed at pH 7.5–8.0 to simulate an alkaline environment that was a compromise between the pH of the crop and midgut lumen measured during tissue preparation (described above). By including ATP (10 mM), CoA (1 mM), and NADPH (5 mM) with assay buffers, we tested the effect of exogenous cofactors on in vitro biosynthesis of N-linolenoyl-L-glutamine by midgut microsomes. Assays consisting of glutamine (20 μg/μl) and sodium linolenate (5 μg/μl) in 25 mM imidazole-HCl (pH 7.5) buffer without microsomes and assays consisting of microsomes from each tissue incubated (as described above) in the absence of one or both substrates were included as negative controls.
Assays for in Vitro Biosynthesis of N-linolenoyl-l-glutamine by Microbial Suspensions, Regurgitant, Hemolymph, Crop Contents, and Gut Contents. Microbial suspensions isolated from regurgitant and subcellular fractions of each tissue were incubated with the substrates sodium linolenate (5 μg/μl) and L-glutamine (20 μg/μl) for 8 h at 4°C and for 72 h at the larval rearing temperature (28°C). Samples of regurgitant, hemolymph, crop contents, gut contents, and peritrophic matrix (membrane) each were diluted with an equal volume (100 μl) of 25 mM imidazole-HCl (pH 7.5) and incubated with substrates as described above for 8 h at 4°C. After incubation, all assay mixtures were analyzed by HPLC for compounds formed in vitro.
Assays for in Vitro Biosynthesis of N-linolenoyl-l-glutamine by Triton X-114-Partitioned Microsomes. To determine the membrane association of the enzyme(s) responsible for N-linolenoyl-L-glutamine biosynthesis, Triton X-114 phase-partitioned microsomes were assayed. Integral, peripheral, and extremely hydrophobic protein fractions were each diluted to final volumes of 200 μl with 25 mM imidazole-HCl (pH 7.5) and incubated with sodium linolenate (5 μg/μl) and L-glutamine (20 μg/μl) at 4°C. Samples (50 μl) were withdrawn for HPLC analysis at 1.5, 8, 23, and 46 h.
Optimum pH and Effect of Temperature on N-linolenoyl-l-glutamine Biosynthesis. The influence of pH on cumulative N-linolenoyl-L-glutamine biosynthesis was determined (by HPLC) for equal quantities of Triton X-114-partitioned integral membrane protein (200 μg) incubated in pH 5.0–11.0 buffers (final volume 200 μl) with sodium linolenate (5 μg/μl) and L-glutamine (20 μg/μl) for 8 h at 4°C. In other experiments, incubation time and microsomal protein quantities were altered to establish conditions of N-linolenoyl-L-glutamine biosynthetic reaction linearity at 21°C. The observed biosynthesis was linear over a 60-min time course when microsomal protein concentration was 1.2 μg/μl. The amount of N-linolenoyl-L-glutamine synthesized at 5, 10, 30, 45, and 60 min was determined (by HPLC) for multiple pH conditions (7.5–11.0) by using fixed substrate concentrations (glutamine, 18.9 μg/μl; sodium linolenate, 8.14 μg/μl) at 21°C. To terminate biosynthesis at each time point enzyme reactions were boiled for 3 min. From a plot of N-linolenoyl-L-glutamine synthesized versus time, the initial reaction velocities (Vo) and corresponding pH optimum (highest Vo) were determined (from slope). Buffers used for incubations were 25 mM Mes-Tris (pH 5.0–6.0), 25 mM Tris·HCl (pH 7.0–9.5), and 25 mM CAPS-NaOH (pH 10.0–11.0). The influence of temperature on cumulative N-linolenoyl-L-glutamine biosynthesis was determined by incubating midgut microsomes (1.5 μg/μl protein) in pH 9.5 buffer (optimum pH) with sodium linolenate (5 μg/μl) and L-glutamine (20 μg/μl) for 1 h.
HPLC and Mass Spectral Analyses of Compounds Synthesized in Vitro. Assay mixtures were boiled for 3 min to terminate enzyme catalysis (FAAs are heat-stable) and centrifuged at 14,000 × g for 5 min, and supernatants were analyzed by HPLC with a C18 reverse-phase column (YMC-Pack ODS-AMQ, 250 × 4.6-mm i.d.; YMC, Kyoto) maintained at 60°C and eluted with a 20–95% acetonitrile (in water) gradient as described by Mori et al. (15). Chromatographic separation was monitored by UV detection at 200 nm. Acid methanolysis and GC-MS analysis (HP-5 column, isobutane chemical ionization) of compounds resolved by HPLC was also performed as described (15). Compound identities were confirmed by comparison with chromatographic retention times and/or mass spectra of synthetic FAA standards. All quantitative analyses were performed after adding 10 μl of internal standard, N-palmitoleoyl-L-glutamine [1 μg/μl in acetonitrile/water 8:2 (vol/vol)], to 50 μl of each sample and then injecting 10 μl of the mixture onto the HPLC column. The mean quantity of N-linolenoyl-L-glutamine synthesized was analyzed by ANOVA followed by a Tukey multiple comparison test (α = 0.05) (19) by using MINITAB 12.1 (Minitab, State College, PA).
Results
N-linolenoyl-l-glutamine Biosynthesis by Subcellular Fractions of Alimentary Tissues. The in vitro biosynthesis of N-linolenoyl-L-glutamine by the subcellular fractions of M. sexta alimentary tissues was initiated by adding L-glutamine and sodium linolenate as substrates and did not require CoA-activated linolenic acid (Fig. 1B). The quantity of N-linolenoyl-L-glutamine synthesized in vitro by midgut microsomes incubated for 8 h at 4°C was significantly less in assays that included the cofactors ATP, CoA, and NADPH (144.6 ± 17.6 ng/μg protein) compared with assays without cofactors (324.6 ± 51.0 ng/μg protein) (mean ± SEM, n = 7, P < 0.05). The amount of N-linolenoyl-L-glutamine synthesized in vitro by midgut microsomes at 4°C (324.6 ± 51.0 ng/μg protein) and 21°C (394.3 ± 68.9 ng/μg protein) did not differ significantly after 8 h of incubation with substrates at pH 7.5 (mean ± SEM, n = 7, P > 0.05). N-linolenoyl-L-glutamine biosynthesis was detected primarily within microsomes isolated from crop (460 ± 87 ng/μg protein), anterior midgut (317 ± 32 ng/μg protein), and salivary gland (176 ± 46 ng/μg protein) tissues (Table 1). However, there was some biosynthesis by the cytosolic fraction of each tissue. Biosynthesis of N-linolenoyl-L-glutamine by crop microsomes was greater than by salivary gland microsomes (P < 0.05) but was not significantly different from biosynthesis by anterior midgut microsomes (P > 0.05). Biosynthesis of N-linolenoyl-L-glutamine by salivary gland and anterior midgut microsomes did not differ significantly (P > 0.05). There was no N-linolenoyl-L-glutamine biosynthesis by microsomes that were boiled for 3 min before incubation with substrates, microsomes incubated without substrates, microsomes incubated with only a single substrate, or substrates incubated without microsomes. When L-glutamine was replaced by L-glutamic acid (monosodium salt) and incubated in vitro with sodium linolenate and anterior midgut microsomes, there was no detectable biosynthesis of N-linolenoyl-L-glutamic acid (Fig. 1C).
Table 1.
N-linolenoyl-l-glutamine biosynthesis in vitro at pH 7.5 by M. sexta-derived samples and controls after incubation with the substrates l-glutamine (20 μg/μl) and sodium linolenate (5 μg/μl) for 8 h at 4°C unless otherwise indicated
| Sample assayed for N-linolenoyl-L-glutamine biosynthesis | n | ng of N-linolenoyl-L-glutamine synthesized per μg of protein |
|---|---|---|
| Salivary gland microsomes | 9 | 176 ± 46b |
| Salivary gland cytosol | 9 | 58 ± 17c |
| Salivary gland microbial suspensions | 6 | ND |
| Salivary gland microbial suspensions (72 h at 28°C) | 3 | ND |
| Crop microsomes | 9 | 460 ± 87a |
| Crop cytosol | 9 | 80 ± 30c |
| Crop microbial suspensions | 6 | ND |
| Crop microbial suspensions (72 h at 28°C) | 3 | ND |
| Midgut microsomes | 9 | 317 ± 32ab |
| Midgut cytosol | 9 | 23 ± 16c |
| Midgut microbial suspensions | 6 | ND |
| Midgut microbial suspensions (72 h at 28°C) | 3 | ND |
| Tissue microsomes boiled for 3 min before assay | 3 | ND |
| Regurgitant | 3 | ND |
| Regurgitant microbial suspensions | 4 | ND |
| Regurgitant microbial suspensions (72 h at 28°C) | 4 | ND |
| Crop contents | 3 | ND |
| Midgut contents | 3 | ND |
| Peritrophic matrix | 3 | ND |
| Hemolymph | 3 | ND |
| Substrate control (glutamine and Na-linolenate only) | 3 | ND |
| Tissue microsome controls (no substrates) | 3 | ND |
| Tissue microsome controls (single substrate only) | 3 | ND |
Data represent mean ± SEM. Means followed by the same letter are not significantly different (P > 0.05, Tukey test). ND, not detected.
N-linolenoyl-l-glutamine Biosynthesis Was Not Detected in Microbial Suspensions, Regurgitant, Hemolymph, or the Contents of Crop and Midgut. N-linolenoyl-L-glutamine biosynthesis was not detected in any of the microbial suspensions that were incubated with substrates for 8 h at 4°C (Fig. 1D) or after incubation for 72 h at the larval rearing temperature (28°C) (Table 1). Similarly, there was no detectable N-linolenoyl-L-glutamine biosynthesis by samples of oral secretions (regurgitant), crop contents, midgut contents, peritrophic matrix, or hemolymph after their incubation with substrates for 8 h at 4°C.
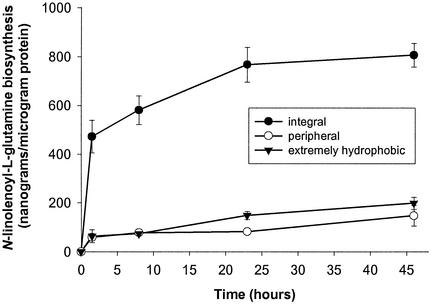
Rapid N-linolenoyl-l-glutamine Biosynthesis by Integral Membrane Protein Fraction. Rapid biosynthesis of N-linolenoyl-L-glutamine was associated with the integral membrane protein fraction isolated from M. sexta microsomes by Triton X-114 phase partitioning (Fig. 2). The quantity of N-linolenoyl-L-glutamine synthesized in vitro by the integral membrane protein fraction was 472 ± 67 ng/μg protein after just 90 min of incubation and 581 ± 59 ng/μg protein after 8 h (mean ± SEM, n = 7). N-linolenoyl-L-glutamine synthesized in vitro increased to 768 ± 71 and 806 ± 48 ng/μg protein after 23 and 46 h, respectively. By contrast, there was little in vitro N-linolenoyl-L-glutamine biosynthesis by extremely hydrophobic or peripheral protein fractions.
Fig. 2.
Time-dependent in vitro biosynthesis of N-linolenoyl-l-glutamine by Triton X-114-partitioned M. sexta midgut microsomes at pH 7.5. Integral, peripheral, and extremely hydrophobic protein fractions were each incubated with glutamine and sodium linolenate at 4°C. Data points represent mean ± SEM (n = 7).
N-linolenoyl-l-glutamine Biosynthesis Is Optimal at Alkaline pH. Cumulative N-linolenoyl-L-glutamine biosynthesis by the integral membrane protein fraction isolated from M. sexta midgut microsomes was greatest at alkaline pH (Table 2). The pH optimum of 9.0–9.5 for N-linolenoyl-L-glutamine biosynthesis by midgut microsomes was determined from analysis of initial reaction velocities (Vo) for assays performed at pH 7.5–11.0 (Table 2). The observed alkaline pH optimum was within the physiological pH range measured for the crop (7.9 ± 0.1) and midgut lumen (9.3 ± 0.1) of larvae (mean ± SEM, n = 8). A second apparent pH optimum of 11.0 was observed outside this physiological pH after a change in buffer salt.
Table 2. Influence of pH on cumulative N-linolenoyl-l-glutamine biosynthesis at 4°C by the integral protein fraction of Triton X-114-partitioned M. sexta midgut microsomes and initial velocity (Vo) of N-linolenoyl-l-glutamine biosynthesis at 21°C by midgut microsomes.
| pH | N-linolenoyl-L-glutamine biosynthesis at 4°C, ng/h per μg of protein (n = 4) | Initial velocity (Vo) of N-linolenoyl-L-glutamine biosynthesis at 21°C, pmol/min per mg of protein (n = 3) |
|---|---|---|
| 5.0 | 15.6 ± 1.1 | — |
| 6.0 | 21.6 ± 1.0 | — |
| 7.0 | 83.9 ± 8.2 | — |
| 7.5 | — | 0.90 ± 0.06 |
| 8.0 | 143.2 ± 12.7 | 1.7 ± 0.3 |
| 8.5 | — | 2.5 ± 0.2 |
| 9.0 | 178.1 ± 5.8 | 3.3 ± 0.4 |
| 9.5 | — | 3.4 ± 0.1 |
| 10.0 | 152.5 ± 9.1 | 2.2 ± 0.3 |
| 10.5 | — | 2.7 ± 0.3 |
| 11.0 | 189.9 ± 10.0 | 4.6 ± 0.3 |
Initial velocities were determined as described in Materials and Methods. Buffers included 25 mM MES-Tris (pH 5.0-6.0), 25 mM Tris·HCl (pH 7.0-9.5), and 25 mM CAPS (pH 10.0-11.0). Data are expressed as mean ± SEM.
N-linolenoyl-l-glutamine Biosynthesis Is Stable over a Wide Temperature Range. Cumulative N-linolenoyl-L-glutamine biosynthesis by midgut microsomes that were incubated with substrates at pH 9.5 was highest at 21–28°C (Table 3). Biosynthetic activity declined at incubation temperatures >40°C and was lost when microsomes were boiled for 3 min before incubation with substrates.
Table 3.
Influence of temperature on cumulative N-linolenoyl-l-glutamine biosynthesis at pH 9.5 by M. sexta midgut microsomes incubated with substrates l-glutamine (20 μg/μl) and sodium linolenate (5 μg/μl) for 1 h
| Temperature, °C | N-linolenoyl-L-glutamine biosynthesis at pH 9.5, ng/h per μg of protein (n = 3) |
|---|---|
| 4 | 35.3 ± 2.9 |
| 21 | 103 ± 1.7 |
| 25 | 96.7 ± 1.6 |
| 28 | 102 ± 0.6 |
| 35 | 87.3 ± 2.1 |
| 40 | 50.2 ± 2.8 |
| 45 | 10.2 ± 0.4 |
| 60 | 4.6 ± 0.4 |
| 21* | ND |
Data are expressed as mean ± SEM. ND, not detected.
M. sexta microsomes were boiled at 100°C for 3 min prior to incubation.
Discussion
We demonstrate in vitro biosynthesis of the FAA elicitor N-linolenoyl-L-glutamine by microsomes isolated from salivary gland, crop, and midgut tissues of M. sexta. The in vitro N-acylation of L-glutamine with sodium linolenate by microsomes proceeded significantly better when the cofactors ATP, CoA, or NADPH were omitted from assay buffers (P < 0.05) and did not require CoA-activated linolenic acid. Although unusual, ours is not the first report demonstrating N-acylation of a polar amide moiety with a free polyunsaturated fatty acid in the absence of exogenous ATP and CoA (20). However, our results demonstrate the potential involvement of insect alimentary tissues and their subcellular fractions in the enzymatic biosynthesis of N-linolenoyl-L-glutamine, an elicitor of plant volatile emission that is found in the oral secretions of M. sexta and many other Lepidoptera. We found no evidence to suggest that microbial or tissue-independent enzymes found within oral secretions (regurgitant) of M. sexta play a significant role in N-linolenoyl-L-glutamine biosynthesis.
Although caterpillar tissues have never been implicated in the biosynthesis of FAA elicitors, hydrolytic enzymes that are also capable of synthesizing biologically active FAAs at elevated substrate concentrations have been identified previously in microsomes isolated from mammalian tissues (20–22). We hypothesized that an enzyme(s) capable of catalyzing biosynthesis of the FAA elicitors found in caterpillar oral secretions might also be found in microsomes. N-linolenoyl-L-glutamine biosynthesis could indeed be catalyzed primarily by microsomes and to a lesser degree by cytosolic fractions isolated from M. sexta salivary gland, crop, and anterior midgut tissues (Table 1). Microbial involvement in the observed net biosynthesis was ruled out, because incubation of crude regurgitant-, microsome-, and cytosol-derived microbial suspensions with L-glutamine and sodium linolenate failed to produce N-linolenoyl-L-glutamine. Hemolymph, oral secretions (regurgitant), crop contents, gut contents, and peritrophic matrix also gave negative results when incubated with substrates and were eliminated as potential sources of tissue-independent enzyme(s) involved with N-linolenoyl-L-glutamine biosynthesis.
There was no significant difference (P > 0.05) with respect to the observed in vitro biosynthesis of N-linolenoyl-L-glutamine by M. sexta crop or anterior midgut microsomes at pH 7.5 (Table 1). Therefore, we used midgut microsomes to determine the optimum pH of N-linolenoyl-L-glutamine biosynthesis. The apparent physiological pH optimum for N-linolenoyl-L-glutamine biosynthesis was between 9.0 and 9.5 based on Vo values of 3.3 ± 0.4 and 3.4 ± 0.1 pmol/min per mg of protein, respectively (Table 2). This optimum pH was consistent with the alkaline environment we measured for the lumen of crop (7.9 ± 0.1) and midgut (9.3 ± 0.1) tissues in M. sexta larvae. A secondary pH optimum (11.0) outside the physiological range was measured when the buffer salt was changed from Tris to CAPS. Enzymatic biosynthesis of N-linolenoyl-L-glutamine was stable over a wide range of temperatures (Table 3). Midgut microsomes were also used for Triton X-114 phase partitioning to determine whether an integral, peripheral, or extremely hydrophobic enzyme(s) was involved with N-linolenoyl-L-glutamine biosynthesis. Rapid (≤90-min) biosynthesis of N-linolenoyl-L-glutamine was predominantly associated with the integral membrane protein fraction isolated from the microsomes (Fig. 2). Cumulative N-linolenoyl-L-glutamine biosynthesis by the integral protein fraction was highest at alkaline pH and coincided with the pH range in which Vo was the greatest (Table 2). Further study will be required to determine the kinetic parameters of the enzyme(s) involved with FAA biosynthesis and its precise membrane distribution.
The significance of our findings can be understood best in the context of previous research aimed at understanding the complex interactions between herbivore-produced FAA elicitors and the various biochemical pathways by which herbivore-damaged plants produce volatile chemical signals (6, 23–26). Beyond determining that elicitors are found in caterpillar oral secretions and that these compounds elicit the release of volatiles from damaged foliage, little research has addressed the questions of how and where FAA elicitors are made. Paré et al. (12) showed that the linolenic acid moiety of volicitin and other related elicitors was derived from leaves on which caterpillars had fed and suggested that the coupling of glutamine to linolenic acid during elicitor biosynthesis occurred within caterpillars. However, insect tissues were not fully explored as potential sources of the putative enzyme(s) involved with elicitor biosynthesis. Study of elicitors and their biosynthesis instead has been concentrated on gut contents or regurgitant collected from caterpillars. Spiteller et al. (14) reported that the regurgitant of S. exigua was capable of the strongly delayed coupling of 12-phenyldodecanoic acid with glutamine in vitro. The delayed (24–72 h) synthesis of this unique N-acylamino acid conjugate, with general similarity to caterpillar FAA elicitors, led to speculation that gut microbes within regurgitant might be involved somehow with elicitor biosynthesis. However, we did not detect in vitro N-linolenoyl-L-glutamine biosynthesis by microbial suspensions derived from fresh M. sexta regurgitant or by fresh crop and midgut contents that presumably contained a natural population of viable gut microbes (Table 1). More importantly, the slow (≥24-h) biosynthetic capability proposed by Spiteller et al. (14) cannot account for the rapid biosynthesis required for elicitor accumulation to overcome simultaneous and relatively rapid (≤8 h) enzymatic FAA hydrolysis inherent to the midgut lumen and regurgitant of caterpillars (15). Anaerobic microbes from tobacco hornworm regurgitant were not cultured or subjected to anaerobic assay conditions; therefore, we cannot exclude their potential role in elicitor biosynthesis. The rapid (≤90-min) biosynthesis of N-linolenoyl-L-glutamine by the enzyme(s) associated with the integral protein fraction of M. sexta tissues indeed could account for net elicitor accumulation in the lumen of the crop and midgut at sufficient precursor substrate concentrations.
Leaves contain an abundant supply of glutamine (27), and it has been suggested that after mechanical damage there is a relatively large release of free fatty acids, including linolenic, near the site of wounding (28). Such a release of fatty acids could account for the immediate availability of substrates used by caterpillars for FAA biosynthesis. Glutamine and linolenic acid both are constituents of the most biologically active FAA elicitors identified thus far (4, 6, 11). The FAA elicitors N-linolenoyl-L-glutamine and N-linolenoyl-L-glutamic acid both have been identified in the regurgitant of M. sexta (6), but only trace amounts of N-linolenoyl-L-glutamic acid have been reported in the regurgitant of caterpillar species other than M. sexta (10). Interestingly, N-linolenoyl-L-glutamic acid was not synthesized when anterior midgut microsomes were incubated in vitro with sodium linolenate and glutamic acid (Fig. 1C) or when incubations were repeated by using buffers containing the cofactors ATP (10 mM), CoA (1 mM), and NADPH (5 mM). It is possible that the glutamine moiety of N-linolenoyl-L-glutamine undergoes oxidative deamination in M. sexta, catalyzed by a separate enzyme(s) unique to M. sexta or derived from one of several known metabolic pathways responsible for the conversion of glutamine to glutamic acid (29–31). The enzyme(s) involved with this hypothetical conversion or specific cofactors necessary for their catalytic activities are probably absent from our in vitro assay mixtures.
The production of FAAs by an enzyme(s) intrinsic to caterpillar alimentary tissues is seemingly paradoxical, because some FAAs also elicit volatile emission by plants during herbivory that ultimately attract the caterpillar's natural enemies. It has been postulated that FAAs might simply serve as surfactants that somehow aid digestion in caterpillars (6, 14). From an evolutionary perspective, it is unclear why caterpillars would specifically synthesize conjugates of linolenic acid and glutamine as general surfactants when other compounds probably could serve the same function without eliciting a defensive response by plants. Although FAAs in general could potentially form micelles with detergent-like properties, we suggest that in caterpillars N-linolenoyl-L-glutamine could also play an important role in linolenic acid and/or glutamine metabolism in light of its hydrolysis by enzymes in the gut lumen (15) and the presence of enzyme(s) embedded in several tissues that are capable of its simultaneous biosynthesis. We must better understand the synthesis and physiological role of N-linolenoyl-L-glutamine and related FAA elicitors in light of the major roles their precursors, glutamine and linolenic acid, play in caterpillar metabolism and survival (32, 33). The fact that plants have evolved a mechanism for detecting and responding defensively to specific FAAs in caterpillar oral secretions and FAAs continue to be synthesized by caterpillars despite the threat they ultimately pose to their survival also supports the hypothesis that FAAs are a crucial component of caterpillar metabolism.
Acknowledgments
We thank P. Shirk for access to an ultracentrifuge and C. Briceño for assistance with rearing of caterpillars. We thank reviewers for helpful advice during the preparation of the manuscript.
Abbreviation: FAA, fatty acid amide.
References
- 1.Turlings, T. C. J., Tumlinson, J. H. & Lewis, W. J. (1990) Science 250, 1251–1253. [DOI] [PubMed] [Google Scholar]
- 2.Turlings, T. C. J. & Tumlinson, J. H. (1992) Proc. Natl. Acad. Sci. USA 89, 8399–8402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dicke, M., Van Baarlen, P., Wessels, R. & Dijkman, H. (1993) J. Chem. Ecol. 19, 581–599. [DOI] [PubMed] [Google Scholar]
- 4.Alborn, H. T., Turlings, T. C. J., Jones, T. H., Stenhagen, G., Loughrin, J. H. & Tumlinson, J. H. (1997) Science 276, 945–949. [Google Scholar]
- 5.Alborn, H. T., Jones, T. H., Stenhagen, G. S. & Tumlinson, J. H. (2000) J. Chem. Ecol. 26, 203–220. [Google Scholar]
- 6.Halitschke, R., Schittko, U., Pohnert, G., Boland, W. & Baldwin, I. T. (2001) Plant Physiol. 125, 711–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turlings, T. C. J., McCall, P. J. & Alborn, H. T. (1993) J. Chem. Ecol. 19, 411–425. [DOI] [PubMed] [Google Scholar]
- 8.Mattiacci, L., Dicke, M. & Posthumus, M. A. (1995) Proc. Natl. Acad. Sci. USA 92, 2036–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koch, T., Krumm, T., Jung, V., Engelberth, J. & Boland, W. (1999) Plant Physiol. 121, 153–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pohnert, G., Jung, V., Haukioja, E., Lempa, K. & Boland, W. (1999) Tetrahedron 55, 11275–11280. [Google Scholar]
- 11.Turlings, T. C. J., Alborn, H. T., Loughrin, J. H. & Tumlinson, J. H. (2000) J. Chem. Ecol. 26, 189–202. [Google Scholar]
- 12.Paré, P. W., Alborn, H. T. & Tumlinson, J. H. (1998) Proc. Natl. Acad. Sci. USA 95, 13971–13975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alborn, H. T., Brennan, M. M. & Tumlinson, J. H. (2003) J. Chem. Ecol. 29, 1333–1348. [DOI] [PubMed] [Google Scholar]
- 14.Spiteller, D., Dettner, K. & Boland, W. (2000) Biol. Chem. 381, 755–762. [DOI] [PubMed] [Google Scholar]
- 15.Mori, N., Alborn, H. T., Teal, P. E. A. & Tumlinson, J. H. (2001) J. Insect Physiol. 47, 749–757. [DOI] [PubMed] [Google Scholar]
- 16.Eaton, J. L. (1987) Lepidopteran Anatomy (Wiley, New York).
- 17.Alam, A. (1992) Anal. Biochem. 208, 121–126. [DOI] [PubMed] [Google Scholar]
- 18.Bordier, C. (1981) J. Biol. Chem. 256, 1604–1607. [PubMed] [Google Scholar]
- 19.Zar, J. H. (1984) Biostatistical Analysis (Prentice–Hall, Englewood Cliffs, NJ), 2nd Ed.
- 20.Kruszka, K. K. & Gross, R. W. (1994) J. Biol. Chem. 269, 14345–14348. [PubMed] [Google Scholar]
- 21.Ueda, N., Kurahashi, Y., Yamamoto, S. & Tokunaga, T. (1995) J. Biol. Chem. 270, 23823–23827. [DOI] [PubMed] [Google Scholar]
- 22.Sugiura, T., Kondo, S., Kodaka, T., Tonegawa, T., Nakane, S., Yamashita, A., Ishima, Y. & Waku, K. (1996) Biochem. Mol. Biol. Int. 40, 931–938. [DOI] [PubMed] [Google Scholar]
- 23.Korth, K. L. & Dixon, R. A. (1997) Plant Physiol. 115, 1299–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCloud, E. S. & Baldwin, I. T. (1998) Planta 203, 430–435. [Google Scholar]
- 25.Frey, M., Stettner, C., Paré, P. W., Schmelz, E. A., Tumlinson, J. H. & Gierl, A. (2000) Proc. Natl. Acad. Sci. USA 97, 14801–14806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen, B., Zheng, Z. & Dooner, H. K. (2000) Proc. Natl. Acad. Sci. USA 97, 14807–14812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Metzler, A., Izquierdo, M., Ziegler, A., Kockenberger, W., Komor, E., von Kienlin, M., Haase, A. & Decorps, M. (1995) Proc. Natl. Acad. Sci. USA 92, 11912–11915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ryu, S. B. & Wang, X. (1998) Biochim. Biophys. Acta 1393, 193–202. [DOI] [PubMed] [Google Scholar]
- 29.Stewart, G. R., Mann, A. F. & Fentem, P. A. (1980) in The Biochemistry of Plants, ed. Miflin, B. J. (Academic, London), pp. 271–327.
- 30.Seshachalam, R. V., Subramanyam, M. V. V. & Krishnamoorthy, R. V. (1992) Physiol. Entomol. 17, 281–287. [Google Scholar]
- 31.Hirayama, C., Konno, K. & Shinbo, H. (1997) J. Insect Physiol. 43, 959–964. [DOI] [PubMed] [Google Scholar]
- 32.Kutlesa, N. J. & Caveney, S. (2001) Pest Manag. Sci. 57, 25–32. [DOI] [PubMed] [Google Scholar]
- 33.Stanley-Samuelson, D. W. (1994) Adv. Insect Physiol. 24, 115–212. [Google Scholar]