Abstract
The use of DNA microarrays has revolutionized the manner in which mRNA populations are analyzed. One limitation of the current technology is that mRNAs are often purified on the basis of their 3′ poly(A) ends, which can be extremely short or absent in some mRNAs. To circumvent this limitation, we have developed a procedure for the purification of eukaryotic mRNAs using a mutant version of the mRNA 5′ cap-binding protein (eIF4E) with increased affinity for the m7GTP moiety of the cap. By using this procedure, we have compared the populations of mammalian mRNAs purified by oligo(dT) and 5′ cap selection with oligonucleotide microarrays. This analysis has identified a subpopulation of mRNAs that are present with short 3′ poly(A) ends at steady state and are missed or underrepresented after purification by oligo(dT). These mRNAs may respond to specific posttranscriptional control mechanisms such as cytoplasmic polyadenylation.
Initiation of eukaryotic mRNA translation requires the recruitment of many proteins, the initiator tRNA, and a 40S ribosomal subunit to the 5′ cap of mRNA. Eukaryotic initiation factor 4E (eIF4E or 4E) specifically recognizes the m7GDP moiety of the m7G(5′)ppp(5′)N cap present at the 5′ end of eukaryotic mRNAs (1–3). In addition to binding the 5′ cap of eukaryotic mRNAs, 4E also binds the ribosome adaptor protein (4G) and translational repressor proteins (4EBPs or PHAS I/II) and associates indirectly with poly(A)-binding protein to stimulate translation (4–9). 4E is a common point of growth regulation in both untransformed and cancer cells (10–13). Both structural and functional studies provide evidence that 4E binds the m7G moiety of the 5′ cap of mRNAs by a π-π stacking interaction between two tryptophan residues, as well as hydrogen bonds between m7G and acidic side chains of 4E (14–17). We have recently described mutants in the S4-H2 loop of 4E (N118A, K119A, and Q120A) that had an affinity for m7GTP several-fold increased relative to wild-type 4E (18).
DNA microarray technology has enabled new questions to be identified or answered in a wide variety of biological systems (19). Although total RNA can be analyzed by DNA microarrays to quantitate gene expression, purification of mRNA with oligo(dT) can frequently decrease problems with signal-to-noise ratios, and the analysis of specific subsets of mRNAs can provide useful information (20). Wild-type 4E fused to protein A has been used previously to isolate eukaryotic mRNAs by binding their 5′ caps (21). We tested the use of a high-affinity mutant of 4E to purify mRNA and determine how it compared with oligo(dT) and whether it might more effectively isolate a subset of eukaryotic mRNAs. One of these 4E mutants (4EK119A) enabled the high-yield 5′ cap-dependent purification of functional mRNAs from total RNA. Moreover, an unexpectedly large number of mRNAs were more efficiently isolated by using high-affinity 4E compared with oligo(dT), suggesting that they had short 3′ poly(A) ends that diminished their binding to oligo(dT). This mRNA phenotype may have biological regulatory implications, as has been demonstrated for Xenopus albumin mRNA, which has a 17-nt-long poly(A) tail and is rapidly degraded after estrogen treatment (22–24). In addition, examples of translationally silent mRNAs due to short 3′ poly(A) ends, which are rapidly activated after stimulation of cytoplasmic poly(A) polymerases, have been described during embryonic development, germline stem cell differentiation, and cell cycle regulation (25–29).
Materials and Methods
Expression and Purification of GST-4E Proteins. Escherichia coli BL21(DE3) expressing GST-4Ewt or GST-4EK119A were grown as previously described (17, 30). Cells were induced at an OD600 of 0.6 with 0.5 mM isopropyl β-d-thiogalactoside at 25°C and cultured overnight. Cells were resuspended in buffer (50 mM Tris·HCl, pH 7.4/150 mM NaCl/1 mM EDTA/1 mM DTT/1% Triton X-100) and sonicated or lysed with a French press. Solubilized proteins were loaded onto a 10-ml glutathione agarose column (Sigma), and protein was eluted with 20 ml of 50 mM Tris·HCl, pH 8.4, with 10 mM glutathione-reduced form. The eluted protein was further purified with a heparin sepharose FPLC column (5 ml of HiTrap; Amersham Pharmacia Biosciences). The approximate yield of purified GST-4E was 15 mg per 1 liter of E. coli culture.
Preparation of Radiolabeled 5′ Capped and Uncapped RNA. In vitro transcription with T7 RNA polymerase (Ambion, Austin, TX) was done to prepare capped and uncapped mRNA (31). The in vitro capping efficiency was estimated to be 10–30% (32). pCR 2.1 (Invitrogen) was digested with EcoR V and used as a template under conditions recommended by the manufacturer. m7GpppG was added to reactions to synthesize 5′ capped mRNA, and [α-32P]UTP (20 μCi) was used to prepare radiolabeled RNA. The template was removed by incubating with DNase I for 15 min at 37°C. RNA was isolated by acid phenol/chloroform (Ambion), precipitated, and analyzed by 7 M urea/polyacrylamide (6%) gel electrophoresis.
Tissue Samples and Total RNA. Normal human liver from unused sections of donor livers was the source of total RNA in these studies (Emory University Institutional Review Board approved). Total RNA was isolated by using TRI Reagent (Molecular Research Center, Cincinnati), as described by the manufacturer. Briefly, 0.8–0.9 g of human liver tissue in 10 ml of TRI Reagent was homogenized with a Polytron homogenizer (PowerGene 700, Fisher Scientific). The homogenates were incubated for 5 min at room temperature; and 2 ml of chloroform was added, mixed for 30 s, and incubated for 15 min at room temperature. After centrifugation at 12,000 × g for 15 min at 4°C, the aqueous phase was transferred to a new tube, and RNA was precipitated with an equal volume of isopropanol. The RNA was pelleted by centrifugation, washed with 10 ml of 75% ethanol, and resuspended in RNase-free water. Equal quantities of RNA from seven different liver specimens were pooled and used as starting material for the GST-4EK119A or oligo(dT) purification of mRNA.
Affinity Purification of 5′ Capped mRNA by Using High-Affinity GST-4E. GST-4EK119A was used to purify mRNA from total RNA. Purified GST-4EK119A (1 mg) or GST-4Ewt was mixed with glutathione agarose beads (1-ml packed volume) in PBS for 1 h at 4°C and washed with PBS. Glutathione agarose GST-4E beads (400 μl) were mixed with 315 μg of heat-denatured total RNA (70°C for 10 min) in 500 μl of binding buffer [10 mM KHPO4, pH 8.0/100 mM KCl/2 mM EDTA/5% glycerol/100 μg/ml E. coli tRNA (Roche, Indianapolis) or yeast tRNA (Sigma)/6 mM DTT/1.3% polyvinyl alcohol (Sigma)/0.005% Triton X-100/20 units RNasin (Roche)]. Mixing was end-over-end for 1 h in 1.5-ml nonstick hydrophobic microfuge tubes (Gene Mate, ISC BioExpress, Kaysville, UT) at room temperature. The resin was washed twice with 1 ml of binding buffer (minus tRNA) and three times with 1 ml of binding buffer (minus tRNA) containing 500 μM GDP. mRNA was eluted by mixing beads for 5 min with 1 ml of binding buffer containing 1 mM m7GDP. The beads were then extracted with equal volumes of acid phenol/chloroform (Ambion). Samples were precipitated with linear acrylamide (5 μg; Ambion), 3 M sodium acetate, and ethanol. RNA was analyzed by 7 M urea/polyacrylamide (6%) gel electrophoresis and autoradiography or Cerenkov counting. The Kd values for GST-4Ewild-type and GST-4EK119A were calculated as described (33). To quantitatively compare the GST-4EK119A purification with oligo(dT), 150 μg of total RNA was used as starting material with triplicate batch GST-4EK119A matrix (200 μl packed volume) or oligo(dT) (Qiagen, Chatsworth, CA) columns. The batch GST-4EK119A purification was done as described except that beads, not incubated with m7GDP, were washed twice with binding buffer without GDP before recovering mRNA by phenol/chloroform extraction. RNA was precipitated with ethanol and quantitated spectroscopically.
Oligo(dT) Purification of mRNA. The same total RNA used for 5′ cap-dependent mRNA purification was also used for the poly(A)-dependent purification of mRNA. The mRNA purification using oligo(dT) was done following the manufacturers instructions (Oligotex, Qiagen).
Cell-Free Translations of Purified mRNA. mRNA was isolated by 4EK119A or oligo(dT) purifications as outlined above. mRNA isolated by both procedures (1 μg) was translated in rabbit reticulocyte lysates with [35S]methionine (per conditions recommended by Promega), and the protein products were analyzed by 10% SDS/PAGE and autoradiography.
Preparation of cRNA. To synthesize cDNA, 5 μg of mRNA, from pooled m7GDP elutions and phenol/chloroform extractions, was incubated with reverse transcriptase (Superscript II, Invitrogen) for 1 h at 42°C with an oligo(dT)24 primer containing a T7 RNA polymerase promoter (34). Double-stranded cDNA was synthesized by using E. coli DNA polymerase, E. coli DNA ligase, and T4 DNA polymerase (16°C for 2 h). cRNA was synthesized by using cDNA and T7 RNA polymerase as recommended by the manufacturer (Enzo Diagnostics). The biotin-labeled cRNA was purified by using RNeasy spin columns (Qiagen). Twenty micrograms of cRNA was fragmented in buffer (40 mM Trisacetate, pH 8.1/100 mM potassium acetate/30 mM magnesium acetate) at 94°C for 35 min.
Oligonucleotide Array Hybridizations. Each cRNA sample was hybridized to a chip (Affymetrix HGU95AV2; 12,000 genes) at 45°C for 16 h in a rotisserie oven at 60 rpm following standard methods (Affymetrix). Microarrays were washed and stained by using an Affymetrix Fluid Station 400 and reagents provided by the manufacturer. Fluorescent images were scanned by using a Hewlett–Packard G2500A Gene Array Scanner and analyzed by using Affymetrix and Spotfire software.
Determining the Size of 3′ Poly(A) Ends. Poly(A) 3′ ends were analyzed by using the rapid amplification of cDNA ends poly(A) test (RACE-PAT) (35). To synthesize cDNA, 1 μg of total RNA was mixed with the following oligo(dT)12 primer: 5′-GCGAGCTCCGCGGCCG CG-T12 (200 ng) and heat denatured at 70°C for 10 min. The reaction mixture was incubated at 42°C for 60 min with Superscript II reverse transcriptase (20 units/μl, Invitrogen), Superscript RT II buffer (50 mM Tris·Cl, pH 8.3/75 mM KCl/3 mM MgCl2)/500 μM dNTP mix/10 mM DTT). The reaction was terminated by incubating at 70°C for 15 min. RNase H (2 units/μl) was added and incubated at 37°C for 30 min. PCR amplification was performed with PCR buffer (20 mM Tris·HCl, pH 8.4/50 mM KCl/1.5 mM MgCl2), a mRNA specific primer (0.5 μM), plus an oligo(dT)12 primer (0.5 μM), Taq DNA polymerase (2.5 units/μl), and 0.5 μl of cDNA template. mRNA specific primers used for RACE-PAT were as follows: H4 histone (X60484), 5′-GTGGCCAT TCACCCGGGGTCA; microsomal glutatione S-transferase 3 (AF026977), 5′-CTGGCATCAGCCTCATACCT; replication protein A 14-kDa subunit (L07493), 5′-GGTCAGATTAGATGCA AGA A; pre-mRNA processing factor 8 homolog (AB007510), 5′-CTCTGTCTGTGCTTGTGTTG; 1,4-α-glucan branching enzyme (L07956), 5′-GTAGCATTTCAGAAAATGTC; myeloid cell differentiation protein (L08246), 5′-AGGGAGTGGTGGGTTTATAGGG; multidrug resistance-associated protein 3B (AF085692), 5′-TCTGGGGTGCTGCCTGAATC; fau (X65923), 5′-GTACAACCGGCGCTTTGTCAACGTTGTGCC; U6 snRNA-associated Sm-like protein LSm7 (AA121509), 5′-GTCCTGGGCGCAGGGCCGCCC CAGCAT; prosomal protein-27 (X59417), 5′-CGTTAGTTTACCAGATCCGTGATGCCA C. The PCR was performed at 94°C for 5 min, followed by 30 cycles of 30 s at 94°C, 1 min at 50°C, 1 min at 72°C, and ending with a 10-min final extension at 72°C. PCR products were analyzed by either 6% nondenaturing polyacrylamide or 1% agarose gel electrophoresis.
Results
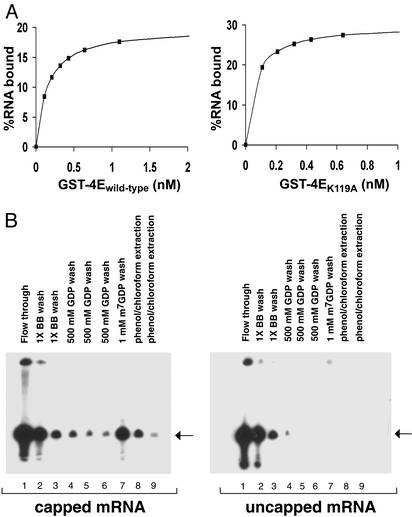
Use of 4EK119A to Isolate 5′ Capped mRNA. The following studies were done to determine whether a high-affinity mutant of 4E would enable the efficient 5′ cap-dependent purification of mRNA. 4Ewt and 4EK119A were expressed in E. coli as GST fusion proteins and purified to at least 95% purity as described in Materials and Methods (data not shown). The relative affinities of GST-4Ewt and GST-4EK119A for capped mRNAs were compared under similar buffer conditions to those used to isolate mRNA. GST-4Ewt and GST-4EK119A were linked to glutathione agarose beads and used to batch purify a model 5′ capped mRNA of 50 nt in length. Different concentrations of GST-4Ewt or GST-4EK119A were incubated with a fixed amount of 32P-labeled 5′ capped mRNA, and the quantity of mRNA bound to GST-4E agarose beads was quantified by Cerenkov counting. Saturation binding curves were obtained by plotting the percent of 5′ capped mRNA bound vs. the concentration of either GST-4Ewt or GST-4EK119A (Fig. 1A). On the basis of this analysis, the Kd for capped mRNA was estimated to be 0.15 nM for GST-4Ewt and 0.06 nM for GST-4EK119A, indicating that GST-4EK119A had an ≈2.5-fold higher affinity for 5′ capped mRNA under the test conditions. These data are in general agreement with the fluorescence quenching measurements used to determine the Kd of the nonfusion forms of wild-type and K119A 4E for m7GDP (18).
Fig. 1.
(A) Relative ability of GST-4Ewild-type and GST-4EK119A to bind 5′ capped mRNA. Batch mRNA-binding assays were performed to compare binding affinities of GST-4Ewild-type and GST-4EK119A. 5′ capped 32P-labeled mRNA was incubated in binding buffer with increasing amounts of GST-4Ewild-type and GST-4EK119A bound to agarose beads (see Materials and Methods). The quantity of mRNA bound to GST-4E agarose beads was measured by Cerenkov counts. The estimated dissociation constants (Kd) of GST-4Ewild-type and GST-4EK119A were 0.15 and 0.06 nM for capped mRNA, respectively. (B) Specificity of GST-4EK119A for 5′ capped mRNA. The batch purification of mRNA using GST-4EK119A was tested for its ability to bind both 5′ capped and uncapped mRNAs. Capped and uncapped mRNA synthesized in vitro using T7 polymerase were mixed with GST-4EK119A agarose beads (50 μl), washed with 1× binding buffer, 500 μM GDP, and eluted with 1 mM m7GDP (see Materials and Methods). mRNA that remained bound to GST-4E beads despite the m7GDP elution step was recovered by extraction with acid phenol/chloroform. mRNA isolated by using GST-4EK119A agarose beads are shown for purifications where 5′ capped (10–30%) and uncapped mRNA were used as starting material. mRNA present in each fraction was precipitated with ethanol and analyzed by 7 M urea/polyacrylamide (6%) gel electrophoresis and autoradiography. As determined by Cerenkov counts, the 1 mM m7GDP eluant (capped mRNA, lane 7) contained 85% of the total RNA recovered from the GST-4EK119A beads. The arrow indicates the size of the mRNA (50 nt) used as starting material.
To determine the specificity of this mRNA purification, both 5′ capped and uncapped radiolabeled mRNAs were tested as starting material. [32P]mRNA was batch purified by using GST-4EK119A agarose beads, eluted from the beads with m7GDP, precipitated with ethanol, and analyzed by denaturing polyacrylamide gel electrophoresis (Fig. 1B). Because the in vitro T7 polymerase synthesis of 5′ capped mRNA is relatively inefficient, a large amount of mRNA appeared in the flowthrough. To remove nonspecifically bound RNA, the GST-4EK119A beads were washed with binding buffer containing 500 μM GDP. Capped mRNA bound to GST-4EK119A beads was eluted with 1 mM m7GDP, whereas no mRNA was detected in the m7GDP eluant when uncapped RNA was used as starting material (Fig. 1B). These results provide evidence that GST-4EK119A enables a relatively specific 5′ cap-dependent purification of mRNA.
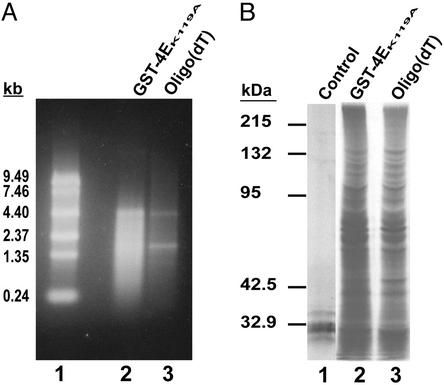
Purifying mRNA with GST-4EK119A and Comparison with Oligo(dT). To further test the use of GST-4EK119A in purifying mRNA, total RNA from liver was used as starting material and the GST-4EK119A and oligo(dT) purifications were compared. Equal quantities of total human liver RNA were mixed with GST-4EK119A agarose beads or applied to an oligo(dT) column (Oligotex, Qiagen). The recovery and purity of the mRNA isolated were compared by using spectrophotometric measurements and denaturing agarose gel analysis. The average recovery of mRNA from 150 μg of total RNA using oligo(dT) was 3.2 ± 0.9 μg (2.1% of total RNA), whereas the recovery with the GST-4EK119A batch purification was 10.1 ± 1.7 μg (6.7% of total RNA) (mean ± SEM). The capacity of the GST-4EK119A matrix was ≈10 μg of mRNA per 200-μl pack volume of beads. In agreement with the relative affinities of GST-4EK119A and wild-type GST-4E for 5′ capped mRNA, the GST-4EK119A matrix had an average 2-fold higher yield of mRNA than a GST-4Ewt matrix under the same conditions (not shown). In addition to having a higher yield, the mRNA purified by using GST-4EK119A had relatively less ribosomal RNA than the oligo(dT) isolated mRNA (Fig. 2A). To determine whether the mRNAs isolated using GST-4EK119A agarose beads were functional, they were incubated in rabbit reticulocyte lysate cell-free translations. Equal quantities of mRNA purified by the 5′ cap- and 3′ poly(A)-dependent methods were translated in vitro with [35S]methionine. The proteins synthesized were analyzed by 10% SDS/PAGE and autoradiography (Fig. 2B). The results demonstrated that functional mRNAs were purified by using the GST-4EK119A 5′ cap-dependent purification.
Fig. 2.
(A) Comparing 5′ cap- and 3′ poly(A)-dependent purifications of mRNA. Total RNA (315 μg) from normal liver was either mixed with GST-4EK119A beads as described in Materials and Methods or applied to an oligo(dC10T30) column. The GST-4EK119A matrix was washed, and mRNA was recovered by eluting with m7GDP (see Materials and Methods). An oligo(dT) column was used to purify mRNA as suggested by the manufacturer (Qiagen). Ten percent of the mRNA recovered from each purification was analyzed by formaldehyde agarose (1%) gel electrophoresis. An ethidium bromide-stained gel is shown: lane 1, 0.24- to 9.5-kb RNA ladder; lane 2, mRNA batch purified using GST-4EK119A agarose beads; lane 3, mRNA purified using an oligo(dT) column. (B) In vitro translation of mRNA purified using GST-4EK119A or oligo(dT). mRNA (1 μg) isolated by either GST-4EK119A or oligo(dT) was translated in a nuclease-treated rabbit reticulocyte lysate with [35S]methionine (see Materials and Methods). Protein products were analyzed by 10% SDS/PAGE and autoradiography: lane 1, control with no mRNA added; lane 2, mRNA purified with GST-4EK119A; and lane 3, mRNA purified with oligo(dT). Molecular mass standards are shown.
Recovery of Specific mRNAs with GST-4EK119A or Oligo(dT). We used high-density oligonucleotide gene arrays to determine the relative efficiency of the 5′ cap- and 3′ poly(A)-dependent purifications in isolating different species of mRNAs. Although the vast majority of genes present in microarrays have been cloned from cDNA libraries prepared from oligo(dT)-purified mRNAs, they do include several that are known to have no 3′ poly(A) end (e.g., histone H4). Total RNA from liver was used as starting material to purify mRNA by using GST-4EK119A or oligo(dT). mRNAs isolated by each method were used to prepare cRNA probes and were hybridized to high-density oligonucleotide microarrays (Affymetrix HGU95AV2; see Materials and Methods). Two independent sets of oligonucleotide microarrays were done by using this approach.
When the quantity of specific mRNAs isolated with GST-4EK119A was compared with the oligo(dT) purifications, we found that the 5′ cap-dependent purified group had at least 177 mRNAs present at ≥2× quantities (range 2–15×) as compared with the 3′ poly(A)-selected mRNA (Table 1; also see Table 2, which is published as supporting information on the PNAS web site, www.pnas.org). A smaller subset of 10 mRNAs was present in 5′ cap but not 3′ poly(A) selected mRNAs, suggesting they either had no 3′ poly(A) end or it was so short that it prevented any measurable binding with oligo(dT) (Table 3, which is published as supporting information on the PNAS web site). When mRNAs that were enriched by the 5′ cap-dependent purification were analyzed based on their function, we found a predominance of mRNAs encoding translational related proteins (e.g., ribosomal and RNA-binding proteins), nuclear-encoded mitochondrial proteins, and stress/growth-related proteins (Table 2). The clustering of biological functions of these mRNAs suggests a regulatory role for this mRNA phenotype. Several clusters of genes, such as protein kinases and cytoskeletal elements, were notably infrequent in this list.
Table 1. Relative quantities of specific mRNAs isolated by 5′- or 3′-dependent purification.
| Relative quantity
|
|||
|---|---|---|---|
| 4EK119A | Oligo(dT) | No. of mRNAs | Sequence characteristics |
| +++ | + | 177 | 2-15× relative increase; mean of 4.4× increase |
| + | Absent | 10 | Small average size; 816 nt; one exception |
| + | +++ | 26 | Large average size; 4,106* nt; large 3′ UTRs; mean of 1,024 nt |
| Absent | + | 9 | Large average size; 4,991 nt; large 3′ UTRs; mean of 1,870 nt |
The relative quantities of mRNAs are represented by plus signs. A detailed list of the numeric fold change for specific mRNAs in each category, GenBank numbers, and other features are provided in Tables 2-5. Several characteristics of mRNAs were determined by analysis by using the Celera and National Center for Biotechnology Information databases.
This does not include two exceptions, which were >10,000 nt.
A smaller subset of mRNAs were preferentially purified by oligo(dT) as compared with the GST-4EK119A purification. A total of 26 mRNAs were present at ≥2× quantities in 3′ poly(A) as compared with 5′ cap-dependent purified mRNA (Table 1 and Table 4, which is published as supporting information on the PNAS web site). An even smaller subset of nine mRNAs was detected in 3′ poly(A) but not in 5′ cap-dependent purified mRNA (Table 5, which is published as supporting information on the PNAS web site). These mRNAs were generally larger than the mRNAs identified in the other categories (Tables 2–4) and had large 3′ untranslated regions, which frequently encode sequence motifs that regulate mRNA stability or other biological properties.
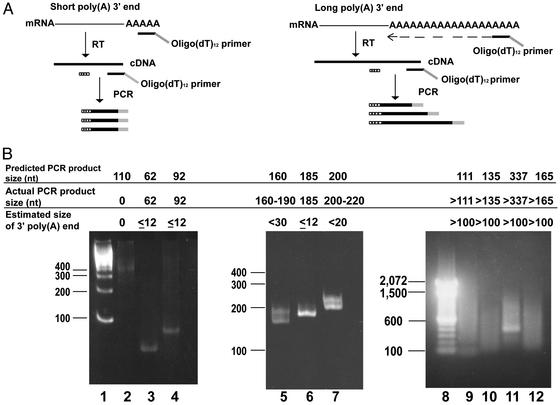
3′ Poly(A) Ends of Selected mRNAs. To determine the size of the 3′ poly(A) end of mRNAs with impaired oligo(dT) binding, we used RACE-PAT (Fig. 3A; see Materials and Methods) (35). RACE-PAT uses a 3′ end oligo(dT)12 primer with a 3′ G/C rich anchor sequence that permits a more accurate measurement of 3′ poly(A) ends than a homopolymeric oligo(dT) primer. The same total RNA used for the oligo(dT) and GST-4EK119A purifications of mRNA was used to synthesize cDNA with an oligo(dT)12 primer and reverse transcriptase. Hybridization of the oligo(dT)12 primer occurs at multiple sites along the 3′ poly(A) end of mRNAs. Subsequent PCR amplification of the heterologous pools of cDNA was done by using mRNA-specific primers and the RACE-PAT oligo(dT)12 primer, resulting in PCR products that included the length of the 3′ poly(A) end of the target mRNA.
Fig. 3.
Analysis of the 3′ poly(A) ends of mRNAs with impaired binding to oligo(dT) or GST-4EK119A. (A) Schematic diagram of the RACE-PAT method used to directly determine the size of the 3′ poly(A) ends. This analysis was done for specific mRNAs, which were preferentially purified with either GST-4EK119A or oligo(dT). Total RNA was reverse transcribed with an oligo(dT) primer with a G/C rich anchor sequence [designated oligo(dT)12 primer]. Hybridization of the oligo(dT) primer to the mRNA 3′ poly(A) end occurs along the entire length of the 3′ poly(A) end. Different-sized cDNAs primed at all possible positions along with the poly(A) end will be synthesized after reverse transcription. Subsequent PCR amplification using this pool of cDNAs with a message specific primer (dotted box) and an oligo(dT)12 primer produces a mixture of PCR products, which include the length of the 3′ poly(A) end of the target mRNA. (B) Results of RACE-PAT for selected mRNAs. PCR-amplified products were analyzed by either 6% nondenaturing polyacrylamide (lanes 1–7) or 1% agarose gel electrophoresis (lanes 9–12). The predicted minimum size of the PCR products was compared with the actual size observed to obtain an estimate of the size of the 3′ poly(A) ends of a specific mRNA. For example, the minimum expected PCR product for the mRNA analyzed in lane 4 (replication protein A/14-kDa subunit, L07493) was 92 nt [62 + 30 nt oligo(dT)12-G/C anchor primer]. The actual size of the PCR product produced was 92 nt, indicating that this mRNA had a 3′ poly(A) end of 12 nt or less. The ethidium bromide-stained gels show a 100-bp DNA ladder (Invitrogen) in lanes 1 and 8. The estimated size for 3′ poly(A) ends of other mRNAs are shown: present only in GST-4EK119A purified mRNA (lane 2, H4 histone mRNA/X60484; lane 3, microsomal glutatione S-transferase 3 mRNA/AF026977; lane 4, replication protein A 14-kDa mRNA/L07493; see Table 3); mRNAs preferentially isolated by GST-4EK119A (lane 5, fau mRNA/X65923; lane 6, U6 snRNA-associated Sm-like protein/AA121509; lane 7, PROS-27 mRNA/X59417; see Table 2); and present only in oligo(dT)-purified mRNA (lane 9, pre-mRNA splicing factor PRP8/AB007510; lane 10, 1,4-α-glucan branching enzyme mRNA/L07956; lane 11, myeloid cell differentiation protein mRNA/L08246; lane 12, multidrug resistance-associated protein mRNA/AF085692; see Table 5). The short size of the 3′ poly(A) ends of the mRNAs in lanes 3–7 was also found by RACE-PAT analysis of RNA isolated from cultured Huh7 hepatoma cells (data not shown).
We determined whether the impaired ability of some mRNAs to be purified with oligo(dT) was due to short poly(A) ends by directly analyzing their 3′ ends with RACE-PAT. mRNAs purified only by GST-4EK119A had 3′ poly(A) ends that were ≤12 nt (Fig. 3B, lanes 3 and 4), except for histone 4, which had no PCR product and has been reported to lack a 3′ poly(A) end (Fig. 3B, lane 2). mRNAs that were isolated preferentially by 5′ cap selection were found to have 3′ poly(A) ends between 12 and 30 nt (Fig. 3B, lanes 5–7). Direct sequencing of PCR products confirmed that their 3′ poly(A) ends were 11, 12, 20, 12, and 20 nt in length (Fig. 3B, lanes 3–7, respectively). On the other hand, mRNAs isolated only with oligo(dT) were found to have 3′ poly(A) ends that were generally >100 nt (Fig. 3B, lanes 9–12). These data provide direct evidence that the explanation for impaired binding to oligo(dT) exhibited by some of the mRNAs was a short 3′ poly(A) end.
Discussion
This study provides evidence that GST-4EK119A can be used in a practical high-yield purification that isolates biologically functional 5′ capped mRNA. Our results indicate there is likely to be a pool of mRNAs that persist at steady state in mammalian cells with very short 3′ poly(A) ends. Moreover, we may be under-representing this pool of mRNAs because we used oligo(dT) primers to synthesize cDNA used to produce cRNA hybridization probes for arrays. This method requires that the mRNA in question still have some poly(A) region to prime. In addition, the genes represented on the high-density oligonucleotide arrays used in these studies were generally identified by methods that included an oligo(dT) mRNA selection step.
The mRNAs that were purified with GST-4EK119A but not oligo(dT) were generally 1,000 nt in length (Table 3), and the 3′ poly(A) ends of representative mRNAs were ≤12 nt (Fig. 3B, lanes 2–4). On the other hand, the nine mRNAs that were purified only with oligo(dT) were all >2,900 nt in length, and their 3′ poly(A) ends were on average >100 nt. Many of these mRNAs were >4,000 nt in length, and most had lengthy 3′ UTRs, which frequently contain regulatory elements. Sequence analysis of these 3′ UTRs demonstrated the existence of potential PUF protein-binding sites as indicated by the UGUR tetranucleotide motif (36, 37). The regulation of gene expression by PUF proteins, which often enhance turnover or repress translation, are postulated to have the primordial function of sustaining mitotic proliferation of stem cells (38, 39). The inability of GST-4EK119A to bind these mRNAs may have been due to RNA structures that prevent recognition of the 5′ cap. An alternative possibility is that these mRNAs decap before deadenylation. Decapping represents an important step in mRNA degradation pathways, can be inhibited by 4E or poly(A)-binding protein, and usually follows deadenylation (40–42). An alternative mRNA degradation pathway, where decapping occurs first, may exist for this subset of mRNAs.
Using GST-4EK119A to purify mRNA may have some advantages over oligo(dT) for some studies. For example, our results suggest that purifying mRNA by 5′ cap selection would have advantages in gene profiling studies of eukaryotic cells that need to monitor the expression of genes involved in ribosome assembly, mRNA translation, mitochondrial functions, and cell stress/growth responses. Oligo(dT) purification of mRNA in some cases can misrepresent changes in mRNA pools if they have 3′ poly(A) ends shorter than 35 nt (39). The efficient isolation of mRNA by a 5′ cap-dependent method may prove particularly useful in organisms such as yeast where many mRNAs have 3′ poly(A) ends that are <40 nt (39).
The 5′ cap selection of mRNA with GST-4EK119A might be preferred for studies that are best done with full-length mRNA with intact 5′ ends. For example, preparing cDNA libraries, analyzing variant 5′ ends due to alternative transcriptional start sites, and preparing RNA based vaccines. In addition, isolating RNA with GST-4EK119A from pools of poly(A) depleted RNA may prove useful in studying noncoding RNAs (43, 44). Although cellular RNA-dependent RNA polymerases are required for aspects of RNA silencing in plants, the possibility that 5′ capped RNAs are involved in some RNA silencing pathways remains to be answered (44).
Why is the short 3′ poly(A) end mRNA phenotype more common than expected in mammalian liver and biased toward mRNAs encoding proteins with translation, mitochondrial, or growth functions? We propose that the explanation exists in a biological regulatory role for this mRNA phenotype. Regulation of gene expression by cytoplasmic polyadenylation of translationally silent mRNAs with short 3′ poly(A) ends has been well documented in Xenopus oocytes and plays a critical role in oocyte maturation (25, 45). cMos and several other prestored mRNAs in oocytes contain short poly(A) ends (≈20–40 nt). Translation of these mRNAs occurs when their 3′ poly(A) ends are elongated to ≈150 nt, requiring two elements in the 3′ UTR: the nuclear polyadenylation motif (AAUAAA) and an AU-rich cytoplasmic polyadenylation element, which is variable and includes UUUUAU, UUUUAACA, and UUUUUUAU (46, 47). The 3′ UTRs of mRNAs that were directly shown to have short 3′ poly(A) ends (Fig. 3) had both sequence elements indicating they are good candidates for cytoplasmic polyadenylation.
Increasing evidence in both invertebrate and vertebrate systems has suggested a more general role for the cytoplasmic polyadenylation of prestored mRNAs as a mechanism to rapidly stimulate gene expression. Studies in Caenorhabditis elegans have provided evidence for a cytoplasmic poly(A) polymerase that elongates the 3′ poly(A) end of specific mRNAs to regulate gene expression and germline developmental events (26). In Drosophila embryos, cytoplasmic polyadenylation of a subset of mRNAs is essential for initiation of development (27). In addition, studies in fission yeast have indicated that one cytoplasmic poly(A) polymerase (Cid1) targets mRNAs that include components of the S-M checkpoint and that another (Cid13) specifically targets the mRNA encoding a subunit of ribonucleotide reductase to regulate DNA replication (28, 29). Homologues of these polymerases are present in mammals. If the short 3′ poly(A) ends of specific mRNAs are elongated by such cytoplasmic poly(A) polymerases, these messages could be identified by the approach we have outlined. Protein synthesis represents a cellular function that has one of the greatest energy requirements. Regulation of critical elements of both the protein synthesis machinery and the mitochondrial respiratory system may permit these two systems to be linked by a rapidly responsive posttranscriptional rheostat. Specific tests of these hypotheses are now possible with the efficient 5′ cap-dependent purification of mRNA and classification of mRNAs into the functional groups presented.
Supplementary Material
Acknowledgments
We thank Stephen T. Warren and P. Jin for advice in the analysis of gene arrays, John LaVoy for excellent technical assistance, Tove Goldson for reading the manuscript, and Thomas Heffron for help with obtaining liver tissue. This work was supported by National Cancer Institute Grant CA63640 (to C.H.H.). Y.H.C. was supported by a Judith Graham Pool Postdoctoral Fellowship from the National Hemophilia Foundation.
Abbreviation: RACE-PAT, RACE poly(A) test.
References
- 1.Sonenberg, N., Rupprecht, K. M., Hecht, S. M. & Shatkin, A. J. (1979) Proc. Natl. Acad. Sci. USA 76, 4345–4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tahara, S. M., Morgan, M. A. & Shatkin, A. J. (1981) J. Biol. Chem. 256, 7691–7694. [PubMed] [Google Scholar]
- 3.Grifo, J. A., Tahara, S. M., Morgan, M. A., Shatkin, A. J. & Merrick, W. C. (1983) J. Biol. Chem. 258, 5804–5810. [PubMed] [Google Scholar]
- 4.Pause, A., Belsham G. J., Gingras, A. C., Donze, O., Lin, T. A., Lawrence, J. C., Jr., & Sonenberg, N. (1994) Nature 371, 747–748. [DOI] [PubMed] [Google Scholar]
- 5.Mothe-Satney, I., Brunn, G. J., McMahon, L. P., Capaldo, C. T., Abraham, R. T. & Lawrence, J. C., Jr., (2000) J. Biol. Chem. 275, 33836–33843. [DOI] [PubMed] [Google Scholar]
- 6.Borman, A. M., Michel, Y. M., Malnou, C. E. & Kean, K. M. (2002) J. Biol. Chem. 277, 36818–36824. [DOI] [PubMed] [Google Scholar]
- 7.Otero, L. J., Ashe, M. P. & Sachs, A. B. (1999) EMBO J. 18, 3153–3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Craig, A. W., Haghighat, A., Yu, A. T. & Sonenberg, N. (1998) Nature 392, 520–523. [DOI] [PubMed] [Google Scholar]
- 9.Gray, N. K., Coller, J. M., Dickson, K. S. & Wickens, M. (2000) EMBO J. 19, 4723–4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sonenberg, N. & Gingras, A. C. (1998) Curr. Opin. Cell. Biol. 10, 268–275. [DOI] [PubMed] [Google Scholar]
- 11.Donaldson, R. W., Hagedorn, C. H. & Cohen, S. (1991) J. Biol. Chem. 266, 3162–3166. [PubMed] [Google Scholar]
- 12.Zimmer, S. G., DeBenedetti, A. & Graff, J. R. (2000) Anticancer Res. 20, 1343–1351. [PubMed] [Google Scholar]
- 13.Crew, J. P., Fuggle, S, Bicknell, R, Cranston, D. W., de Benedetti, A. & Harris, A. L. (2000) Br. J. Cancer 82, 161–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marcotrigiano, J., Gingras, A. C., Sonenberg, N. & Burley, S. K. (1997) Cell 89, 951–961. [DOI] [PubMed] [Google Scholar]
- 15.Matsuo, H., Li, H., McGuire, A. M., Fletcher, C. M., Gingras, A. C., Sonenberg, N. & Wagner, G. (1997) Nat. Struct. Biol. 4, 717–724. [DOI] [PubMed] [Google Scholar]
- 16.McGuire, A. M., Matsuo, H. & Wagner, G. (1998) J. Biomol. NMR 12, 73–88. [DOI] [PubMed] [Google Scholar]
- 17.Hsu, P. C., Hodel, M. R., Thomas J. W., Taylor L. J., Hagedorn, C. H. & Hodel, A. E. (2000) Biochemistry 39, 13730–13736. [DOI] [PubMed] [Google Scholar]
- 18.Spivak-Kroizman, T., Friedland, D. E., De Staercke, C., Gernert, K. M., Goss, D. J. & Hagedorn, C. H. (2002) FEBS Lett. 516, 9–14. [DOI] [PubMed] [Google Scholar]
- 19.Arava, Y., Wang, Y., Storey, J. D., Liu, C. L., Brown, P. O. & Herschlag, D. (2003) Proc. Natl. Acad. Sci. USA 100, 3889–3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown, V., Jin, P., Ceman, S., Darnell, J. C., O'Donnell, W. T., Tenenbaum, S. A., Jin, X., Feng, Y., Wilkinson, K. D., Keene, J. D., et al. (2001) Cell 107, 477–487. [DOI] [PubMed] [Google Scholar]
- 21.Edery, I., Chu, L. L., Sonenberg, N. & Pelletier, J. (1985) Mol. Cell. Biol. 15, 3363–3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pastori, R. L., Moskaitis, J. E., Buzek, S. W. & Schoenberg, D. R. (1991) Mol. Endocrinol. 5, 461–468. [DOI] [PubMed] [Google Scholar]
- 23.Cunningham, K. S., Hanson, M. N. & Schoenberg, D. R. (2002) Nucleic Acids Res. 29, 1156–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gupta, J. D., Gu, H. & Schoenberg, D. R. (2001) RNA 7, 1034–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mendez, R. & Richter, J. D. (2001) Nat. Rev. Mol. Cell Biol. 2, 521–529. [DOI] [PubMed] [Google Scholar]
- 26.Wang, L., Eckmann, C. R., Kadyk, L. C., Wickens, M. & Kimble, J. (2002) Nature 419, 312–316. [DOI] [PubMed] [Google Scholar]
- 27.Juge, F., Zaessinger, S., Temme, C., Wahle, E. & Simonelig, M. (2002) EMBO J. 21, 6603–6613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Read, R. L., Martinho, R. G., Wang, S. W., Carr, A. M. & Norbury, C. J. (2002) Proc. Nat. Acad. Sci. USA 99, 12079–12084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saitoh, S., Chabes, A., McDonald, W. H., Thelander, L., Yates, J. R. & Russell, P. (2002) Cell 109, 563–573. [DOI] [PubMed] [Google Scholar]
- 30.Hagedorn, C. H., Spivak-Kroizman, T., Friedland, D. E., Goss, D. J. & Xie, Y. (1997) Protein Expression Purif. 9, 53–60. [DOI] [PubMed] [Google Scholar]
- 31.Baker, B. F., Miraglia, L. & Hagedorn, C. H. (1992) J. Biol. Chem. 267, 11495–11499. [PubMed] [Google Scholar]
- 32.Fuerst, T. R. & Moss, B. (1989) J. Mol. Biol. 206, 333–348. [DOI] [PubMed] [Google Scholar]
- 33.Weeks, K. M. & Crothers, D. M. (1992) Biochemistry 31, 10281–10287. [DOI] [PubMed] [Google Scholar]
- 34.Schnabl, B., Choi, Y. H., Olsen, J. C., Hagedorn, C. H. & Brenner, D. A. (2002) Lab. Invest. 82, 323–333. [DOI] [PubMed] [Google Scholar]
- 35.Salles, F. J., Darrow, A. L., O'Connell, M. L. & Strickland, S. (1992) Genes Dev. 6, 1202–1212. [DOI] [PubMed] [Google Scholar]
- 36.Zhang, B., Gallegos, M., Puoti, A., Durkin, E., Fields, S., Kimble, J. & Wickens, M. P. (1997) Nature 390, 477–484. [DOI] [PubMed] [Google Scholar]
- 37.Wharton, R. P., Sonoda, J., Lee, T., Patterson, M. & Murata, Y. (1998) Mol. Cell. 1, 863–872. [DOI] [PubMed] [Google Scholar]
- 38.Wickens, M., Bernstein, D. S., Kimble, J. & Parker, R. (2002) Trends Genet. 18, 150–157. [DOI] [PubMed] [Google Scholar]
- 39.Olivas, W. & Parker, R. (2000) EMBO J. 19, 6602–6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwartz, D. C. & Parker, R. (2000) Mol. Cell. Biol. 20, 7933–7942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tharun, S. & Parker, R. (2001) Mol. Cell 8, 1075–1083. [DOI] [PubMed] [Google Scholar]
- 42.Gao, M., Wilusz, C. J., Peltz, S. W. & Wilusz, J. (2001) EMBO J. 20, 1134–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McManus, M. T. & Sharp, P. A. (2002) Nat. Rev. Genet. 3, 737–747. [DOI] [PubMed] [Google Scholar]
- 44.Ahlquist, P. (2002) Science 296, 1270–1273. [DOI] [PubMed] [Google Scholar]
- 45.Dickson, K. S., Thompson, S. R., Gray, N. K. & Wickens, M. (2001) J. Biol. Chem. 276, 41810–41816. [DOI] [PubMed] [Google Scholar]
- 46.McGrew, L. L., Dworkin-Rastl, E., Dworkin, M. B. & Richter, J. D. (1989) Genes Dev. 3, 803–815. [DOI] [PubMed] [Google Scholar]
- 47.Fox, C. A., Sheets, M. D. & Wickens, M. P. (1989) Genes Dev. 3, 2151–2162. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.