Abstract
Human Hb, an α2β2 tetrameric oxygen transport protein that switches from a T (tense) to an R (relaxed) quaternary structure during oxygenation, has long served as a model for studying protein allostery in general. Time-resolved spectroscopic measurements after photodissociation of CO-liganded Hb have played a central role in exploring both protein dynamical responses and molecular cooperativity, but the direct visualization and the structural consequences of photodeligation have not yet been reported. Here we present an x-ray study of structural changes induced by photodissociation of half-liganded T-state and fully liganded R-state human Hb at cryogenic temperatures (25–35 K). On photodissociation of CO, structural changes involving the heme and the F-helix are more marked in the α subunit than in the β subunit, and more subtle in the R state than in the T state. Photodeligation causes a significant sliding motion of the T-state β heme. Our results establish that the structural basis of the low affinity of the T state is radically different between the subunits, because of differences in the packing and chemical tension at the hemes.
Allosteric transitions allow rapid regulation of protein function in biological systems. There is a wealth of structural data regarding the end states of allosteric transitions of various proteins (1), but very little is known about how they work. A typical example of allosteric regulation is the cooperative oxygen binding by human Hb. The Hb molecule is a heterotetramer consisting of two α subunits and two β subunits, α2β2, which are arranged as a dimer of αβ dimers. Each subunit contains one heme group to which one oxygen molecule binds reversibly. The oxygen affinity of each subunit rises as the other hemes in the same tetramer become saturated with oxygen. The binding of oxygen by Hb is therefore cooperative, allowing efficient transport of oxygen in the blood. Crystal structures of fully unliganded tense (T)-state and fully liganded relaxed (R)-state Hb reveal multiple differences at both tertiary and quaternary levels (2–5), but, as is the case for most other allosteric proteins, through what mechanism these different structures impact ligand reactivity is still not well understood (for review, see ref. 6). An essential part of this question is understanding the mechanism of restraints on ligand binding in the T state, because the oxygen affinity of the R state is close to that of isolated α and β subunits. In the early 1970s, Perutz suggested that the low affinity of the T state is caused by tension in the iron-proximal His(F8) bond in the liganded T state (7), but several lines of evidence show that the ligand affinity of the α and β subunits is regulated by different mechanisms (8–12). Despite the wealth of data from x-ray crystallography, a means of defining the stereochemical basis unambiguously for the difference in oxygen affinity between the T and R states, which is only a few kcal/mol, has not been determined (13). It is important to note that the large quaternary change that occurs on ligand (un)binding makes it difficult to determine to what extent the tertiary structure differences originate from a pure quaternary-linked change or from a ligation-induced difference. The problem is therefore the lack of detailed knowledge of how ligand (un)binding induces tertiary structural changes of an individual subunit and how these changes affect and can be affected by the quaternary state.
Insight into pure tertiary structural changes in Hb has been gained form time-resolved spectroscopic measurements with the use of the photodissociation of CO-liganded Hb (HbCO), to generate unliganded Hb (deoxyHb), which then evolves from the original R conformation toward the equilibrium T conformation before it fully recombines with the CO (14–17). These studies show that conformational changes that occur before 1 μs are purely tertiary, whereas those occurring later include quaternary changes as well (6, 14–17). Therefore, the ligation-linked tertiary changes can, in principle, be elucidated at the atomic level by determining the structure of a short-time photoproduct of HbCO by using x-ray crystallography. This type of experiment has recently been conducted on the CO photoproduct of myoglobin (Mb), a monomeric oxygen storage heme protein, either by x-ray crystallography at cryogenic temperatures (20–40 K) under continuous illumination of visible light (18–20) or time-resolved x-ray crystallography at room temperature after flash photolysis (21). It has been shown that cryogenic temperatures (20–40 K) slow the protein motions that permit ligand escape, so that a partially relaxed photoproduct intermediate, in which the dissociated CO still exists within the distal heme pocket, can be efficiently trapped under these conditions (18–20). This low-temperature photoproduct is essentially identical to the short-time photoproduct at room temperature (21).
Here we present a crystallographic study of CO complexes of the T and R states of Hb at cryogenic temperatures in both resting and photolysed states. Our current data show how differently the α and β subunits, within each allosteric form, respond to loss of ligand, where the free ligand lies, and what protein motions correlate with the regulatory mechanism of Hb. These findings may provide a deeper understanding of how Hb works than can be obtained from the static structures.
Materials and Methods
Sample Preparation. Cross-linked iron–nickel hybrid Hbs, XL[α(Fe-CO)β(Ni)]2 and XL[α(Ni)β(Fe-CO)]2, were prepared as described (22). Crystals of each half-liganded T-state hybrid Hb (space group P21) were grown from CO-saturated 1–2% (wt/vol) protein solution that contains 18% (wt/vol) polyethylene glycol (PEG) 3350, 0.15 M K2SO4, 5 mM homocysteine (Aldrich), 50 mM citrate-ammonium buffer (pH 6.6), and the submicroscopic fragments of the crystals of XL[α(Fe-CO)β(Ni)]2 as seeds. Glycerol (150 μl/ml) was added to the mother liquor to avoid crystal damage on cooling. Crystallization of R-state HbCO was carried out according to the protocol of Perutz (23), with slight modifications, to give a solution of 0.6% (wt/vol) HbCO in 2.2 M sodium/potassium phosphate buffer (pH 6.7), containing 10% (vol/vol) glycerol, 5 mM homocysteine (Aldrich), 2.0%(vol/vol) toluene, and the submicroscopic crystals as seeds. Crystals were mounted by using the loop technique and flash-cooled by a cold He gas stream.
Photolysis. Crystals were continuously photolysed 10 min before and during data collection (≈90 min) by red light alone in the case of the T-state Hb or three (red, green, and white) light beams at once in the case of the R-state Hb. Red, green, and white light were generated by a He–Ne laser (15 mW, 632.8 nm; Melles Griot, Irvine, CA), a He–Ne laser (5 mW, 543.5 nm; Melles Griot), and a fiber-optic illuminator equipped with a halogen-tungsten lamp (150 W), respectively. Using an open flow He cryostat (24), the temperatures were kept at ≈25 K for the T-state crystals and ≈35 K for the R-state crystals under illumination.
Data Collection and Refinement. All of the diffraction data were collected by using synchrotron radiation at RIKEN beamline BL44B2 (25) of SPring-8, Harima, Japan. To increase the signal-to-noise ratio of difference Fourier maps, both of the data sets on the photoproduct and the CO-bound structure were collected from the same crystal. The wavelength of the incident x-rays was 1.0 Å. Intensity data were collected with a MarCCD 165-mm detector (MarUSA, Evanston, IL). The diffraction data were integrated and scaled with HKL2000 and SCALEPACK (HKL Research, Charlottesville, VA). The initial models of all Hbs were obtained by molecular replacement using the CCP4 program MOLREP (26). The refinement was performed with the CCP4 programs REFMAC and CCP4 suite (26).
Results and Discussion
CO-Liganded T-State Hb. The problem with studying the photoproduct of T-state Hb by x-ray crystallography is that fully HbCO prefers the R state rather than the T state, making it difficult to produce stable crystals of CO-liganded T-state Hb. Two approaches have been introduced to attack this problem. One approach is to use “half-liganded” metal-substituted hybrid Hbs, in which one pair of subunits has an inert metal porphyrin group and the other a CO-reactive iron(II) (for review, see ref. 27), instead of thermodynamically less stable fully liganded T-state Hb. Several hybrid Hbs of this kind have been used as a model system to study T-state Hb (28–32). The other approach to further improving crystal quality is to use intramolecular crosslinking, which prevents the dissociation of Hb tetramer into αβ dimers (33–35). It has been shown that dissociation into αβ dimers is an obligatory step in thermal unfolding (36) and dissociation of heme from globin (37), both of which may interfere with crystallization (38).
By combining these two approaches, remarkably stable T-state crystals were obtained from two half-liganded human Hbs, XL[α(Fe-CO)β(Ni)]2 and XL[α(Ni)β(Fe-CO)]2, in which the iron in either the α or β subunits is replaced by nickel(II) and the two αβ dimers are cross-linked between the two Lys(EF6)82β residues by a fumaryl group (22). We chose these hybrid Hbs for the following reasons. First, nickel(II)-protoporphyrin IX binds neither oxygen nor CO and mimics a ferrous deoxy heme (30, 31), and thus the structural effects of ligand binding to the α or β subunits may be studied in isolation. Second, the fumaryl cross-link between the two Lys(EF6)82β residues little affects the structure and function of human Hb (22, 39). Finally, because of the cross-linking, both half-liganded hybrids yield well diffracting, optically thin isomorphous crystals.
We collected x-ray diffraction data sets from crystals of XL[α(Fe-CO)β(Ni)]2 and XL[α(Ni)β(Fe-CO)]2 at a temperature near 25 K under continuous illumination by visible laser light, as well as control data sets on the CO-bound states at both 25 and 100 K in the dark (Table 1). Note that because the temperature difference (25 vs. 100 K) was found to have no significant effect on the CO-bound structures, the data at 100 K with higher resolution are presented in Table 1. Our refined structural models of the CO-bound states of XL[α(Fe-CO)β(Ni)]2 and XL[α(Ni)β(Fe-CO)]2 show that both half-liganded hybrids assume a T-quaternary structure characteristic of native unliganded Hb (deoxyHb). The iron atoms of both α and β CO-liganded hemes are located at the center of the four-pyrrole nitrogen plane like HbCO, whereas both CO-liganded ferrous hemes remain domed like deoxyHb. The F-helices of both liganded subunits lie between the positions of HbCO and deoxyHb, and the conformations of His(E7)63β and Val(E11)67β in the liganded β subunit are very similar to those in HbCO.
Table 1. Data collection and refinement statistics.
| T-state α(Fe-CO) hybrid Hb
|
T-state β(Fe-CO) hybrid Hb
|
||||||
|---|---|---|---|---|---|---|---|
| Bound state | Photo-product | Difference map* | Bound state | Photo-product | Difference map* | R-state HbCO difference map* | |
| Data collection | |||||||
| Light | None | Laser, red | None (laser, red) | None | Laser, red | None (laser, red) | None (multiple beams) |
| Temperature, K | 100 | 25 | 25 (25) | 100 | 25 | 25 (25) | 25 (35) |
| Resolution range, Å | 20-1.60 | 20.0-1.55 | 20.0-1.70 (20.0-1.70) | 20.0-1.45 | 20.0-1.45 | 20.0-1.75 (20.0-1.75) | 20.0-2.50 (20.0-2.50) |
| Reflections | |||||||
| Measured | 437,187 | 600,432 | 403,926 (387,426) | 751,479 | 878,993 | 385,971 (381,959) | 50,436 (51,059) |
| Unique | 144,674 | 153,793 | 120,229 (119,562) | 200,565 | 197,252 | 110,529 (111,952) | 9,387 (9,363) |
| Completeness, %† | 94.4/83.9 | 90.7/51.8 | 94.4/82.0 (93.7/78.2) | 95.9/77.3 | 95.9/70.4 | 93.8/63.9 (94.7/66.3) | 97.1/87.5 (96.7/86.5) |
| Mean <I>/<σ(I)> | 26.0 | 5.8 | 7.2 (7.2) | 8.1 | 7.9 | 5.8 (6.2) | 7.0 (7.2) |
| Multiplicity | 3.0 | 3.9 | 3.4 (3.2) | 3.7 | 4.5 | 3.5 (3.4) | 5.4 (5.5) |
| Rmerge‡, %† | 3.5/9.6 | 6.0/27.6 | 5.6/22.6 (5.9/22.5) | 6.7/21.3 | 5.7/20.4 | 6.5/19.7 (6.3/19.0) | 5.9/29.8 (5.6/27.8) |
| Refinement statistics | |||||||
| Resolution range, Å | 20.0-1.60 | 20.0-1.55 | 20.0-1.45 | 20.0-1.45 | |||
| σcut-off | 0.0 | 0.0 | 0.0 | 0.0 | |||
| Reflections used | 144,674 | 153,793 | 200,565 | 197,252 | |||
| R-factor§, % | 17.4 | 18.4 | 19.1 | 18.7 | |||
| Free R-factor, % | 20.4 | 21.3 | 21.1 | 20.8 | |||
| Solvent | 715 | 937 | 929 | 969 | |||
| rms deviations from ideals | |||||||
| Bond lengths, Å | 0.012 | 0.013 | 0.010 | 0.009 | |||
| Bond angles, o | 1.312 | 1.480 | 1.310 | 1.350 | |||
To increase the signal-to-noise ratio of difference Fourier maps, both of the datasets on the photoproduct and the bound state were collected from the same crystal.
Completeness and Rmerge are given for overall data and for the highest-resolution shell (overall/the highest-resolution shell). The highest-resolution shells for the most leftward column to the right are 1.66-1.60 Å, 1.61-1.55 Å, 1.76-1.70 Å, 1.50-1.45 Å, 1.50-1.45 Å, 1.81-1.75 Å, and 2.59-2.50 Å, respectively.
Rmerge = Σ|Ii — 〈I〉|/Σ|Ii|; where Ii is the intensity of an observation and 〈I〉 is the mean value for that reflection and the summations are over all reflections.
R-factor = Σh∥Fo(h)| — |Fc(h)∥ /ΣhFo(h), where Fo and Fc are the observed and calculated structure factor amplitudes, respectively. The free R-factor was calculated with 5% of the data excluded from the refinement.
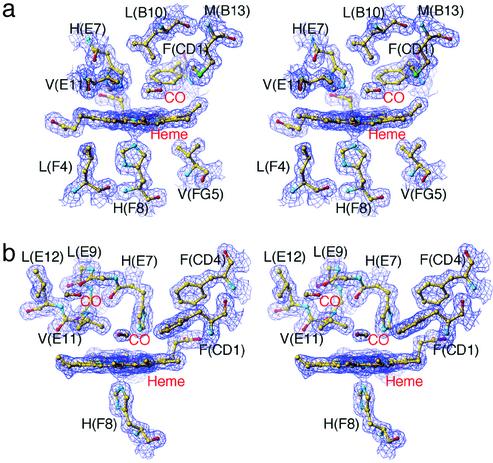
Location of Dissociated CO in the T State. We first note that the asymmetric unit of each T-state crystal structure contains two almost identical tetrameric molecules, designated molecules 1 and 2, and all of the structural features that will be described in this paper are highly significant in each of the crystallographically independent α and β subunits with different crystal contacts. Nevertheless, because the photolysis-induced tertiary structural changes in the α1 subunit of molecule 1 and in the β2 subunit of molecule 2 are slightly more visible than those in the other subunits, these two subunits are selected for presentation in the results and figures.
Electron density maps and structural models for the α and β heme surroundings of the photolysed T-state hybrid Hbs are shown in Fig. 1. Comparing the resting and photolysed states, the α and β irons move out of the heme plane toward the proximal His(F8) by 0.31 and 0.22 Å, respectively. As shown in Fig. 1, no electron density of the bound CO is seen in both photolysed states (Fig. 1), implying that complete photodissociation has occurred in both subunits. In the α(Fe) subunit, new density representing the CO appears in the distal heme pocket at a location above pyrrole ring C ≈3.5 Å from the iron, making the C-O molecular axis approximately parallel to the heme plane (Fig. 1a). This site corresponds to the primary docking site identified by previous x-ray studies on the photoproduct of CO-liganded Mb (MbCO) (18–21). In the β(Fe) subunit, photodissociated CO molecules are detected at two locations: a distal pocket docking site above pyrrole ring C ≈3.7 Å from the iron, and a cavity surrounded by hydrophobic residues, Leu(B10)28β, Val(E11)67β, Leu(E12)68β, and Leu(G8)106β, in the back of the distal pocket ≈8.5 Å from the iron (Fig. 1b). This hydrophobic cavity preexists in the β subunit and also in Mb, in which it is known to be a xenon-binding site (40), but not in the α subunit. Our data are consistent with predictions by molecular dynamics simulation, which show that in the T-state β subunit the ligands move rapidly toward Leu(B10)28β and Leu(G8)106β, whereas in the T-state α subunit they remain near the heme iron (41), and in agreement with the experimental observations that geminate recombination of oxygen and NO are markedly slower in the T-state β subunit than in the T-state α subunit (41, 42). Because we find no open channel between the two observed docking sites in the β subunit (Fig. 1b), migration of the CO to the secondary docking site is probably assisted by relatively large movements of Val(E11)67β, much larger than seen for the equivalent valine residue in the α subunit. In the T state, Val(E11)67β is pressed against any heme ligand. On photodissociation, the stress is relieved. We speculate that the released energy is dissipated by structural fluctuations that help the ligand escape. This suggestion is consistent with the photolysis-induced movements of the β heme and Val(E11)67β described below.
Fig. 1.
Stereoview of electron density maps (2Fo – Fc map) of the active-site structures of photolysed CO complexes of T-state hybrid Hbs contoured at 1.3σ. (a) The α1 heme region in photolysed XL[α(Fe-CO)β(Ni)]2 (molecule 1) at 25 K. (b) The β2 heme region in photolysed XL[α(Ni)β(Fe-CO)]2 (molecule 2) at 25 K.
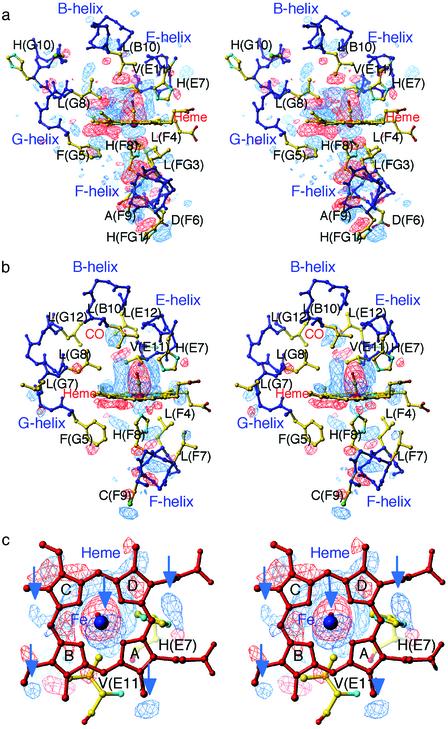
Structural Changes in the T State. Difference Fourier maps between the T-state photoproducts (laser on) and the CO-bound structures (laser off) give a uniquely clear picture of the dynamical responses of the hemes and protein moiety after photolysis (Fig. 2). They reveal three important differences between the α(Fe) and β(Fe) subunits. First, the movement of the F-helix is more marked in the α(Fe) subunit than in the β(Fe) subunit. In the α(Fe) subunit, the F-helix moves significantly away from the heme, with displacement of the proximal His(F8)87α toward the F-helix (Fig. 2a). This movement is propagated to the FG-helix corner. In the β(Fe) subunit, by contrast, the movement of the F-helix is relatively small and localized (Fig. 2b). This finding may be correlated with differences in the packing of the heme pocket between the α and β subunits: on ligation, the packing of the β heme pocket allows the iron to move into plane with very little strain on the proximal side. Second, pyrrole rings B and C move more significantly toward the distal side in the α heme than in the β heme. This motion of the α heme is coupled with movement of the phenyl ring of the Phe(G5)98α toward the heme plane (Fig. 2a), suggesting that steric repulsion between this group and the α heme contributes to strain energy in the liganded T state. Third, the β heme undergoes a translation in the direction from pyrrole ring C to B, as is evident from the difference Fourier map viewed along the iron-proximal His(F8)92β bond (Fig. 2c). As seen in Fig. 2c, the positive, iron-associated electron density below the heme is not located on the heme normal, but is displaced from it, indicating the direction of the heme motion. Several other features at the rim of the heme add support for the occurrence of the heme motion (arrows in Fig. 2c). Importantly, this heme motion is consistent in direction with that arising from the relief of steric interaction between the bound CO and the Cγ2 of Val(E11)67β. The side chain of Val(E11) in the β subunit, but not in the α subunit, moves significantly toward the site formerly occupied by the bound CO.
Fig. 2.
Stereoview of difference Fourier maps of the active-site structures between T-state photoproducts and CO-bound structures contoured at ±3σ.(a) The α1 heme region in XL[α(Fe-CO)β(Ni)]2 (molecule 1) at 25 K. (b) The β2 heme region in XL[α(Ni)β(Fe-CO)]2 (molecule 2) at 25 K. (c) The β2 heme region in XL[α(Ni)β(Fe-CO)]2 (molecule 2) at 25 K viewed along the β iron-proximal His(F8) bond. Arrows pointing to the positive densities at the iron and the rim of the heme indicate the direction of the sliding motion of the β heme. In this and all subsequent difference maps, negative and positive density are shown in red and blue, respectively.
The T-state α(Fe) and β(Fe) subunits also share several common features: motion of the distal His(E7) toward the location formerly occupied by the bound CO, slight rearrangements of the side chains of the distal pocket-forming residues, such as Leu(B10) and Leu(G8), that are in contact with the dissociated CO, and subtle movements of the helices B, E, and G (Fig. 2).
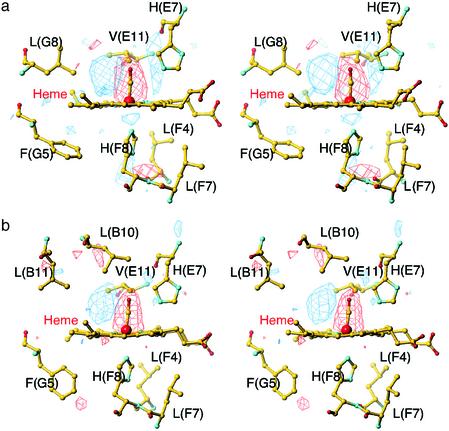
CO Location and Structural Changes in the R State. We next investigated the crystal structure of the photolysed R-state HbCO. To attain a maximal yield of the R-state photoproduct whose geminate rebinding rate is much faster than that of the T state (43), a small crystal (≈0.04 mm × 0.02 mm × 0.02 mm) was irradiated with three light beams at the same time (see Materials and Methods). The use of the multiple beams raised the temperature of the crystal to ≈35 K. Using this experimental setup, we obtained ≈50% photolysis, as judged from the tube-like electron density of the CO ligand previously observed in the photoproduct of CO-liganded Mb (MbCO) at 36 K (20). Difference Fourier maps between the R-state photoproduct and the CO-bound structure show that, in both subunits, the most prominent features are the loss of density of the bound CO and the appearance of new density representing the photodissociated CO on pyrrole ring C (Fig. 3). Photodissociation of CO induces small movements of the iron, proximal His(F8), and distal His(E7) in the α subunit but has little effect on the β heme surroundings. Neither the E-helix nor the F-helix moves significantly in either subunit. Overall, the protein conformational relaxation in the R state is very small, as in the case of the photoproduct of MbCO (18–21), implying that there is little ligation-linked strain. This finding is consistent with the view that the more plastic R state can accommodate equally well both liganded and unliganded heme (44).
Fig. 3.
Stereoview of difference Fourier maps of the active-site structures between the R-state photoproduct and the CO-bound structure contoured at ±3σ. (a) The α heme region in HbCO at 35 K. (b) The β heme region in HbCO at 35 K.
Conclusion
Comparison of the effects of ligand dissociation within a single crystal allows the tertiary changes of the α and β subunits to be observed without least-squares superimposition, but directly in terms of electron density movements. This study shows clearly that the structural basis of the low affinity of T-state Hb is radically different between the α and β subunits, even though these subunits have similar tertiary structures. Because all of the structural features described above are significant in each of the crystallographically independent α and β subunits with different crystal contacts (the asymmetric unit contains two tetramers), they must reflect intrinsic features of the protein in solution. In the T-state α subunit, the conformational relaxation on photolysis can be viewed as reflecting the release of strain energy stored in the proximal side, involving the α heme and the F-helix. In the T-state β subunit, by contrast, there is far less strain in the iron-proximal His(F8) bond. Instead, our structural analysis provides direct evidence of the unfavorable steric interaction between the liganded β heme and its surrounding protein structure. We find little evidence of structural changes transmitted beyond the E- and F-helices and FG-helix corner. The salt bridges formed by the terminal residues are left intact, and no significant movements are observed at the α1β2 contact, suggesting quaternary structure change. Altogether, these results suggest that the reduced ligand affinity of T-state human Hb is brought about by interactions close to the hemes. Changes in these interactions may be decoupled from breakage of the quaternary-linked salt bridges.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: HbCO, CO-liganded Hb; Mb, myoglobin; R, relaxed; T, tense.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID codes 1J3Z, 1J3Y, 1J40, and 1J41).
References
- 1.Perutz, M. F. (1989) Q. Rev. Biophys. 22, 139–237. [DOI] [PubMed] [Google Scholar]
- 2.Perutz, M. F. (1970) Nature 228, 726–739. [DOI] [PubMed] [Google Scholar]
- 3.Baldwin, J. & Chothia, C. (1979) J. Mol. Biol. 129, 175–220. [DOI] [PubMed] [Google Scholar]
- 4.Shaanan, B. (1983) J. Mol. Biol. 171, 31–59. [DOI] [PubMed] [Google Scholar]
- 5.Fermi, G., Perutz, M. F., Shaanan, B. & Fourme, R. (1984) J. Mol. Biol. 175, 159–174. [DOI] [PubMed] [Google Scholar]
- 6.Eaton, W. A., Henry, E. R., Hofrichter, J. & Mozzarelli, A. (1999) Nat. Struct. Biol. 6, 351–358. [DOI] [PubMed] [Google Scholar]
- 7.Perutz, M. F. (1972) Nature 237, 495–499. [DOI] [PubMed] [Google Scholar]
- 8.Sawicki, C. A. & Gibson, Q. H. (1977) J. Biol. Chem. 252, 7538–7547. [PubMed] [Google Scholar]
- 9.Nagai, K. & Kitagawa, T. (1980) Proc. Natl. Acad. Sci. USA 77, 2033–2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujii, M., Hori, H., Miyazaki, G., Morimoto, H. & Yonetani, T. (1993) J. Biol. Chem. 268, 15386–15393. [PubMed] [Google Scholar]
- 11.Barrick, D., Ho, N. T., Simplaceanu, V., Dahlquist, F. W. & Ho, C. (1997) Nat. Struct. Biol. 4, 78–83. [DOI] [PubMed] [Google Scholar]
- 12.Unzai, S., Eich, R., Shibayama, N., Olson, J. S. & Morimoto, H. (1998) J. Biol. Chem. 273, 23150–23159. [DOI] [PubMed] [Google Scholar]
- 13.Wyman, J. (1964) Adv. Protein Chem. 19, 223–286. [DOI] [PubMed] [Google Scholar]
- 14.Sawicki, C. A. & Gibson, Q. H. (1976) J. Biol. Chem. 251, 1533–1542. [PubMed] [Google Scholar]
- 15.Friedman, J. M. (1994) Methods Enzymol. 232, 205–231. [DOI] [PubMed] [Google Scholar]
- 16.Jayaraman, V., Rodgers, K. R., Mukerji, I. & Spiro, T. G. (1995) Science 269, 1843–1848. [DOI] [PubMed] [Google Scholar]
- 17.Björling, S. C., Goldbeck, R. A., Paquette, S. J., Milder, S. J. & Kliger, D. S. (1996) Biochemistry 35, 8619–8627. [DOI] [PubMed] [Google Scholar]
- 18.Teng, T.-Y., Šrajer, V. & Moffat, K. (1994) Nat. Struct. Biol. 1, 701–705. [DOI] [PubMed] [Google Scholar]
- 19.Schlichting, I., Berendzen, J., Phillips, G. N., Jr., & Sweet, R. M. (1994) Nature 371, 808–812. [DOI] [PubMed] [Google Scholar]
- 20.Hartmann, H., Zinser, S., Komninos, P., Schneider, R. T., Nienhaus, G. U. & Parak, F. (1996) Proc. Natl. Acad. Sci. USA 93, 7013–7016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Šrajer, V., Ren, Z., Teng, T. Y., Schmidt, M., Ursby, T., Bourgeois, D., Pradervand, C., Schildkamp, W., Wulff, M. & Moffat, K. (2001) Biochemistry 40, 13802–13815. [DOI] [PubMed] [Google Scholar]
- 22.Shibayama, N., Imai, K., Morimoto, H. & Saigo, S. (1995) Biochemistry 34, 4773–4780. [DOI] [PubMed] [Google Scholar]
- 23.Perutz, M. F. (1968) J. Crystal Growth 2, 54–56. [Google Scholar]
- 24.Nakasako, M., Sawano, M. & Kawamoto, M. (2002) Rev. Sci. Instrum. 73, 1318–1320. [Google Scholar]
- 25.Adachi, S., Oguchi, T., Tanida, H., Park, S.-Y., Shimizu, H., Miyatake, H., Kamiya, N., Shiro, Y., Inoue, Y., Ueki, T. & Iizuka, T. (2001) Nucl. Instrum. Methods Phys. Res. Sect. A 467/468, 711–714. [Google Scholar]
- 26.Bailey, S. (1994) Acta Crystallogr. D 50, 760–763.15299374 [Google Scholar]
- 27.Venkatesh, B., Manoharan, P. T. & Rifkind, J. M. (1998) Prog. Inorg. Chem. 47, 563–684. [Google Scholar]
- 28.Blough, N. V. & Hoffman, B. M. (1984) Biochemistry 23, 2875–2882. [DOI] [PubMed] [Google Scholar]
- 29.Simolo, K., Stucky, G., Chen, S., Bailey, M., Scholes, C. & McLendon, G. (1985) J. Am. Chem. Soc. 107, 2865–2872. [Google Scholar]
- 30.Shibayama, N., Morimoto, H. & Miyazaki, G. (1986) J. Mol. Biol. 192, 323–329. [DOI] [PubMed] [Google Scholar]
- 31.Shibayama, N., Morimoto, H. & Kitagawa, T. (1986) J. Mol. Biol. 192, 331–336. [DOI] [PubMed] [Google Scholar]
- 32.Park, S.-Y., Nakagawa, A. & Morimoto, H. (1996) J. Mol. Biol. 255, 726–734. [DOI] [PubMed] [Google Scholar]
- 33.Walder, J. A., Zaugg, R. H., Walder, R. Y., Steele, J. M. & Klotz, I. M. (1979) Biochemistry 18, 4265–4270. [DOI] [PubMed] [Google Scholar]
- 34.Chatterjee, R., Welty, E. V., Walder, R. Y., Pruitt, S. L., Rogers, P. H., Arnone, A. & Walder, J. A. (1986) J. Biol. Chem. 261, 9929–9937. [PubMed] [Google Scholar]
- 35.Benesch, R. E. & Kwong, S. (1988) Biochem. Biophys. Res. Commun. 156, 9–14. [DOI] [PubMed] [Google Scholar]
- 36.Bellelli, A., Ippoliti, R., Brancaccio, A., Lendaro, E. & Brunori, M. (1990) J. Mol. Biol. 213, 571–574. [DOI] [PubMed] [Google Scholar]
- 37.Benesch, R. E. & Kwong, S. (1995) J. Biol. Chem. 270, 13785–13786. [DOI] [PubMed] [Google Scholar]
- 38.McPherson, A. (1985) Methods Enzymol. 114, 112–120. [DOI] [PubMed] [Google Scholar]
- 39.Shibayama, N., Imai, K., Hirata, H., Hiraiwa, H., Morimoto, H. & Saigo, S. (1991) Biochemistry 30, 8158–8165. [DOI] [PubMed] [Google Scholar]
- 40.Tilton, R. F., Jr., Kuntz, I. D., Jr., & Petsko, G. A. (1984) Biochemistry 23, 2849–2857. [DOI] [PubMed] [Google Scholar]
- 41.Shibayama, N., Yonetani, T., Regan, R. M. & Gibson, Q. H. (1995) Biochemistry 34, 14658–14667. [DOI] [PubMed] [Google Scholar]
- 42.Gibson, Q. H., Ikeda-Saito, M. & Yonetani, T. (1985) J. Biol. Chem. 260, 14126–14131. [PubMed] [Google Scholar]
- 43.Murray, L. P., Hofrichter, J., Henry, E. R., Ikeda-Saito, M., Kitagishi, K., Yonetani, T. & Eaton, W. A. (1988) Proc. Natl. Acad. Sci. USA 85, 2151–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilson, J., Phillips, K. & Luisi, B. (1996) J. Mol. Biol. 264, 743–756. [DOI] [PubMed] [Google Scholar]