Abstract
We present a computational model for the mammalian circadian clock based on the intertwined positive and negative regulatory loops involving the Per, Cry, Bmal1, Clock, and Rev-Erb α genes. In agreement with experimental observations, the model can give rise to sustained circadian oscillations in continuous darkness, characterized by an antiphase relationship between Per/Cry/Rev-Erbα and Bmal1 mRNAs. Sustained oscillations correspond to the rhythms autonomously generated by suprachiasmatic nuclei. For other parameter values, damped oscillations can also be obtained in the model. These oscillations, which transform into sustained oscillations when coupled to a periodic signal, correspond to rhythms produced by peripheral tissues. When incorporating the light-induced expression of the Per gene, the model accounts for entrainment of the oscillations by light-dark cycles. Simulations show that the phase of the oscillations can then vary by several hours with relatively minor changes in parameter values. Such a lability of the phase could account for physiological disorders related to circadian rhythms in humans, such as advanced or delayed sleep phase syndrome, whereas the lack of entrainment by light-dark cycles can be related to the non-24h sleep-wake syndrome. The model uncovers the possible existence of multiple sources of oscillatory behavior. Thus, in conditions where the indirect negative autoregulation of Per and Cry expression is inoperative, the model indicates the possibility that sustained oscillations might still arise from the negative autoregulation of Bmal1 expression.
Keywords: circadian rhythms, computational biology, oscillations, entrainment, sleep-wake disorders
Most living organisms, from cyanobacteria to plants, insects, and mammals, are capable of displaying spontaneously sustained oscillations with a period close to 24 h. These circadian rhythms can occur in constant environmental conditions, e.g., constant darkness, and are therefore endogenous. Recent experimental advances have shed much light on the molecular mechanism of circadian rhythms. Although the most studied organisms were initially Drosophila (1) and Neurospora (2), molecular studies of circadian rhythms have since been extended to cyanobacteria (3), plants (4), and mammals (5). The picture that emerges from these experiments is that in all cases investigated so far, the molecular mechanism of circadian oscillations relies on negative autoregulation of gene expression (6–9).
A number of genes and their protein products involved in such a regulatory mechanism have been identified. Thus, in Drosophila (1, 7), the PER (period) and TIM (timeless) proteins form a complex that indirectly represses the activation of the per and tim genes. Expression of these genes is enhanced by the complex formed by the activators CYC (cycle) and CLOCK. Binding of the PER–TIM complex to CYC and CLOCK prevents the activation of per and tim expression. The situation in mammals (5, 9) resembles that observed in Drosophila, but instead of TIM, it is the cryptochrome (CRY) protein that forms a regulatory complex with a PER protein (9). Several forms of these proteins exist (PER1, PER2, PER3, CRY1, and CRY2). The PER–CRY complex inhibits the expression of the Per and Cry genes in an indirect manner, by binding to the complex CLOCK–BMAL1; the latter, formed by the products of the Clock and Bmal1 genes, activates Per and Cry transcription (5, 10).
Besides this negative regulation of gene expression, indirect positive regulation is also involved. In Drosophila, the PER–TIM complex derepresses the transcription of clock by binding to CLOCK, which exerts a negative autoregulation on the expression of its gene (11). This indirect autoinhibition of clock is likely mediated by the product of the vri gene (12). Similarly, in mammals, Bmal1 expression is subjected to negative autoregulation by BMAL1, through the product of the Rev-Erbα gene (13). The complex between PER2 and CRY1 or CRY2 enhances Bmal1 expression in an indirect manner (5) by binding to CLOCK–BMAL1, and thereby reducing the transcription of the Rev-Erbα gene (13).
Adaptation of biological organisms to their periodically varying environment is mediated through the entrainment of circadian rhythms by light-dark (LD) cycles. Light can entrain circadian rhythms by inducing degradation of the TIM protein in Drosophila (14), whereas in mammals (15) it acts by inducing the expression of the Per gene.
Mathematical models for circadian rhythms have so far been proposed for Drosophila (16–20) and Neurospora (20–22). These deterministic models, based on experimental observations, predict that in a certain range of parameter values, the genetic regulatory network undergoes sustained oscillations of the limit cycle type corresponding to circadian rhythmic behavior, whereas outside this range, the gene network operates in a stable steady state. The roles and advantages of such a computational approach to circadian rhythms have recently been reviewed (23, 24). Similar results have been obtained by means of stochastic simulations, which show that circadian rhythms remain robust with respect to molecular noise, even when the maximum numbers of mRNA and protein molecules involved in the oscillatory mechanism are in the order of tens and hundreds, respectively (25).
The purpose of this article is to present a deterministic model for the mammalian circadian clock. It incorporates the regulatory effects exerted on gene expression by the PER, CRY, BMAL1, CLOCK, and REV-ERBα proteins, as well as posttranslational regulation of these proteins by reversible phosphorylation, and light-induced Per expression. The model can account for autonomous, sustained circadian oscillations in conditions corresponding to continuous darkness, and for entrainment by LD cycles. In the latter conditions, the shift of the phase and the lack of entrainment observed for some parameter values can be related to syndromes associated with physiological disorders of the circadian system in humans. The analysis of the model uncovers the possibility of multiple sources of periodic behavior in the genetic regulatory network controlling circadian oscillations.
Computational Model for the Mammalian Circadian Clock. The model is schematized in Fig. 1. The list of molecular processes incorporated into the model and the kinetic equations governing its time evolution are given in Supporting Text. The model describes the regulatory interactions between the products of the Per, Cry, Bmal1, Clock, and Rev-Erbα genes. For simplicity, at this stage we do not distinguish between the Per1, Per2, and Per3 genes and represent them in the model by a single Per gene; similarly Cry1 and Cry2 are represented by a single Cry gene.
Fig. 1.
Model for circadian oscillations in mammals involving interlocked negative and positive regulations of Per, Cry, Bmal1, and Rev-Erbα genes by their protein products. We focus on the case where BMAL1 exerts a direct negative feedback on the expression of its gene. The role of the Rev-Erbα gene product in the indirect regulation of Bmal1 expression by BMAL1 (indicated in gray) is considered in a second stage (the gray loop then replaces the direct negative feedback exerted by BMAL1). The kinetic equations governing the time evolution of the model are listed in Supporting Text, together with the definition and values of the parameters, given in the legend to Fig. 5 and in Table 1, respectively, which are published as supporting information on the PNAS web site, www.pnas.org.
We shall first treat the regulatory effect of BMAL1 on Bmal1 expression as a direct, negative autoregulation. We will then extend the model by considering explicitly the action of REV-ERBα in the indirect negative feedback exerted by BMAL1 on the expression of its gene. Similar conclusions are reached in the two cases. The version of the model without REV-ERBα is governed by a set of 16 kinetic equations, whereas three more equations are needed in the model incorporating the Rev-Erbα mRNA and the REV-ERBα protein (see Supporting Text).
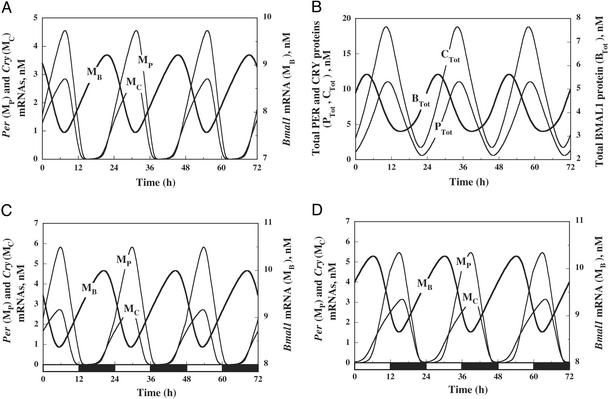
Circadian Oscillations in Continuous Darkness. In a certain range of parameter values the system of Eqs. 1–16 (see Supporting Text) produces sustained oscillations with a circadian period. Because they occur for parameter values that do not change in the course of time, these oscillations are endogenous, as observed for circadian rhythms that persist in continuous darkness or light. In agreement with experimental observations (5, 10), Bmal1 mRNA oscillates in antiphase with Per and Cry mRNAs (Fig. 2A). The proteins undergo similar oscillations and follow their mRNA by a few hours (Fig. 2B). Given that most parameter values remain to be determined experimentally, these oscillations were obtained for a semiarbitrary choice of parameter values, in a physiological range, so as to yield a period of oscillations in continuous darkness (DD) close to 24 h (see Table 1). Parameter values were also selected so as to satisfy other constraints set by experimental observations. Among these, a major constraint is that the model should allow entrainment of the oscillations by LD cycles (see below).
Fig. 2.
Circadian oscillations in DD (A and B) and entrainment by LD cycles (C and D). (A) The mRNA of Bmal1 oscillates in antiphase with respect to the mRNAs of Per and Cry.(B) Corresponding oscillations of the PER, CRY, and BMAL1 proteins. (C) Oscillations of the mRNAs after entrainment by 12:12 LD cycles. The peak in Per mRNA occurs in the middle of the light phase. (D) Oscillations are delayed by 9 h and the peak in Per mRNA occurs in the dark phase when the value of parameter KAC is decreased from 0.6 to 0.4 nM. Other parameter values correspond to the basal set of values listed in Table 1. In C and D, the maximum value of the rate of Per expression, vsP, varies in a square-wave manner such that it remains at a constant low value of 1.5 nM/h during the 12-h-long dark phase (black rectangle) and is raised up to the high value of 1.8 nM/h during the 12-h-long light phase (white rectangle). The curves have been obtained by numerical integration of Eqs. 1–16 (see Supporting Text) of the model without REV-ERBα.
Sustained oscillations only occur in an appropriate range of parameter values. Outside this range, rhythmic behavior disappears and the system evolves toward a stable steady state; such an evolution is often accompanied by damped oscillations. Given the large number of parameters considered in the model, it is difficult to thoroughly assess its sensitivity to changes in parameter values. Useful insights can nevertheless be obtained by determining, for each parameter, one at a time, the range of values producing sustained oscillations as well as the variation of the period over this range, while keeping for the other parameters the basal values used in Fig. 2A (see Supporting Text, Table 1, and Fig. 6, which are published as supporting information on the PNAS web site).
When damped oscillations occur in the model in DD, they can readily be entrained by the periodic variation in one of the parameters. Such a situation is illustrated in Fig. 7, which is published as supporting information on the PNAS web site, where damped oscillations in A become entrained in B by a periodic variation in parameter, vsP.
Entrainment by LD Cycles. To assess whether the model can account for entrainment of the circadian clock by LD cycles, we incorporated the effect of light on the maximum rate of Per expression, vsP. Rather than remaining constant as in DD, this parameter now varies periodically, e.g., as a square wave, going from a constant low value during the dark phase up to a higher constant value, vsPmax, during the light phase. In such conditions, entrainment by a 12:12 LD cycle (12 h of light followed by 12 h of darkness) can be obtained over an appropriate range of vsPmax values. The antiphase relationship between Per and Cry mRNAs on one hand, and Bmal1 mRNA on the other, is maintained during entrainment (Fig. 2 C and D) as in DD (Fig. 2 A), but the particular value of the phases depends on vsPmax.
The phase of the oscillations after entrainment is also sensitive to the choice of other parameter values. Different phases can even be obtained in LD for parameter values yielding comparable periods of circadian oscillations in DD. An example of this situation is illustrated in Fig. 2D, where the only difference with respect to Fig. 2C is a change in parameter KAC, the equilibrium constant describing the activating effect of CLOCK–BMAL1 on Cry expression. The autonomous period in DD is 23.85 h and 23.55 h in Fig. 2 C and D, respectively, whereas the phase of Per mRNA is delayed by ≈9 h in the latter case, so that Per mRNA reaches its maximum during the dark phase instead of peaking in the light phase.
Link with Physiological Disorders of the Circadian System. In searching for entrainment by an LD cycle, we did not succeed initially to find suitable conditions and often found, instead, quasi-periodic oscillations. Entrainment, if and when it occurred, was observed only over a reduced range of the maximum rate of light-induced Per expression, vsPmax. The reason for this lack of robust entrainment could be traced to the need for a sufficiently high level of CRY protein. Indeed, during the light phase, Per mRNA increases, and as a result, the level of PER protein also rises. If CRY is not present in adequate amounts, free PER will accumulate because there is not enough of CRY present to form a complex with PER. In such conditions, entrainment by the LD cycle fails to take place. Only when the light-independent maximum rate of Cry expression is sufficiently high can entrainment occur. The fact that the absence of CRY1 or CRY2 produces only a slight change in period (26), might suggest that each of the genes produces sufficient levels of CRY.
Lack of entrainment can occur as a function of other control parameters as well. Of particular interest is the effect of the maximum rate of PER phosphorylation, Vphos. The effect of progressively increasing Vphos is shown in Fig. 3A, both for DD and LD conditions. The period of oscillations in DD (upper curve) rises, then decreases to a minimum, and increases again as Vphos increases. The nonlinear nature of the dependence of the period on Vphos likely reflects the antagonistic effects played by the cytosolic and nuclear forms of the kinase acting on PER. Eventually, sustained oscillations disappear when the control parameter exceeds a critical value outside the range considered in Fig. 3A. In a 12:12 LD cycle, different types of dynamic behavior can be observed as a function of Vphos. A domain of entrainment (the white region in Fig. 3A) is flanked by two regions (in gray) in which entrainment fails to occur. The lower curve in Fig. 3A shows the phase of the peak in Per mRNA, defined with respect to the onset of the light phase, when entrainment occurs in LD.
Fig. 3.
Relating the model to syndromes associated with disorders of the circadian oscillatory system. (A) Effect of the maximum rate of PER phosphorylation on the free running period in DD and on the phase of the oscillations in LD. The phase corresponds to the time (in h) at which the maximum in Per mRNA occurs after the onset of the light phase. Situations 1 and 2 show that different values of the control parameter can produce different phases after entrainment, even though they correspond to the same free running period in DD. Situation 1 corresponds to the entrainment shown in Fig. 2C. The double-arrow lines show how to obtain the free running period and the phase in LD for a given value of the control parameter. Situations a–c indicate that decreasing (increasing) the rate of phosphorylation, Vphos, with respect to the basal situation b can produce a phase advance (delay) as well as a decrease (increase) in the free running period. The transition from b to a would correspond to the phase shift observed in FASPS (see text). The gray areas on the left and right refer to absence of entrainment. The data are obtained by integration of Eqs. 1–16 (see Supporting Text) of the model without REV-ERBα, for the basal parameter values listed in Table 1, with Vphos = V1P = V1PC = V3PC. (B) Quasi-periodic behavior and phase jump outside the range of entrainment. The phase of the circadian oscillations does not lock to a constant value with respect to the 24-h LD cycle, as occurs in the case of entrainment (see Fig. 2 C and D). Instead, the phase advances every day by <1 h. During 25 successive days the peak in Per mRNA falls within the light phase of the LD cycle, until it reaches the end of the preceding dark phase. Then, in only 4 successive days (horizontal arrows), it crosses the dark phase and reaches the end of the preceding light phase. Gray and white bars represent the dark and light phases of the LD cycles, respectively. The curve in B has been obtained as in Fig. 2C, for the same parameter values except KIB = 1.64 nM instead of 2.2 nM.
The phosphorylation status of PER has been related to disorders of the sleep-wake cycle in humans. Thus, a mutation of hPER2 that reduces its ability to be phosphorylated by casein kinase Iε has been linked with the familial advanced sleep phase syndrome, FASPS (27). The free running period in DD in a subject affected by FASPS is shorter than 24 h (28). In a 24-h LD cycle, sleep onset and sleep offset occur very early, and the phase of sleep is advanced by 3–4 h. This phenomenon can be accounted for by the results of Fig. 3A. For case b, which corresponds to an intermediate value of the maximum phosphorylation rate, Vphos, the peak in Per mRNA occurs 9 h after the onset of the light phase. For case a, which corresponds to a smaller value of Vphos, the autonomous period in DD is reduced and the phase in LD is advanced by a few hours. Case c indicates that a phase delay in LD could occur as a result of increased rate of PER phosphorylation, which corresponds to a longer period in DD. This result is of interest because an hPER3 gene polymorphism has recently been implicated in the delayed sleep phase syndrome (29), although hyperphosphorylation of the protein has not been demonstrated.
The results of Fig. 3A show, unexpectedly, that a phase advance in LD is not necessarily associated with a reduction in the free running period in DD. In a narrow range of Vphos values, the period in DD increases, whereas the phase in LD is advanced. The data further indicate that two distinct values of the control parameter that yield the same period in DD (dotted vertical lines marked by 1 and 2 in Fig. 3A) can produce different phases in LD (double-arrow lines).
The behavior outside the range of entrainment also bears on physiological disorders of the sleep-wake cycle. Lack of entrainment corresponds to the non-24-h sleep-wake syndrome in which the phase of the sleep-wake pattern constantly changes with respect to the LD cycle (30). Such free running circadian oscillations have been observed both in blind (31) and sighted subjects (32). An example of this behavior in the model is shown in Fig. 3B where quasi-periodic oscillations occur in LD. Although the phase of the peak in Per mRNA never settles to a constant value, it is generally located in the light part of the LD cycle. In the case shown in Fig. 3B, it falls in the light phase during 25 consecutive days, then falls in the dark phase during 4 consecutive days, before returning again to the light phase. The rapid passage of the peak through dark phase can be seen as a phase jump, of the kind observed in patients affected by a non-24-h sleep-wake syndrome (33).
Multiple Sources for Oscillations in the Genetic Regulatory Network. The existence of intertwined positive and negative regulations in the scheme of Fig. 1 raises the possibility that the mechanism producing sustained oscillations may not be unique. To test whether the oscillations rely primarily, as expected, on the indirect negative feedback loop involving the inactivation of the CLOCK–BMAL1 complex through its binding to PER–CRY, we may determine whether oscillations still occur when preventing this inactivation. When silencing the negative feedback loop involving the PER–CRY complex, e.g., by setting to zero the rate of synthesis of the PER protein, oscillations disappear and the system evolves toward a stable steady state (Fig. 4A), as observed in mice for the double mutants mPer1/mPer2 (34).
Fig. 4.
The possibility of oscillations due to multiple oscillatory mechanisms. (A) The oscillations in Fig. 2 A disappear in the absence of PER protein synthesis (ksP = 0). The curves show the asymptotic, stable steady state reached after transients have subsided. (B) Sustained oscillations are restored in these conditions when the degree of cooperativity, m, of repression of Bmal1 by CLOCK-BMAL1 increases from 2 to 4. The fact that oscillations can occur in the absence of PER protein indicates the existence of another oscillatory mechanism that relies only on CLOCK-BMAL1 autoregulation. The curves are obtained by numerical integration of Eqs. 1–16 of the model without REV-ERBα, for the basal parameter values listed in Table 1, except ksP = 0, m = 4, ksB = 0.5 h-1; for this choice of parameter values, the period is 19.83 h. The period varies from some 12 h to 40 h as the rate constant, ksB, measuring BMAL1 synthesis decreases from 1.4 h-1 to 0.1 h-1. (C) Oscillations occurring solely due to the PER–CRY negative autoregulation can occur in the absence of negative feedback of BMAL1 on Bmal1 expression. The curves, showing the time evolution of Per, Cry, and Bmal1 mRNAs, were obtained as in Fig. 2 A, for the same parameter values except KIB = 100 nM (the effect of the negative feedback of BMAL1 on Bmal1 transcription is negligible at such a high value of the inhibition constant), vmB = 0.96 nM/h. The period of the oscillations is 23.91 h.
Simulations performed for slightly different parameter values indicate that the possibility of oscillations due to a second oscillatory mechanism nevertheless exists. Thus, when the degree m of cooperativity of repression exerted on the Bmal1 gene by its product BMAL1 is raised from 2 (as in Figs. 2 A and 4A) to 4, sustained oscillations reappear (Fig. 4B). The period of these oscillations can be markedly different from 24 h; for the selected choice of parameter values, it is close to 19.8 h. Because the indirect negative feedback loop involving PER–CRY is inoperative, since no PER is made, the oscillations in Fig. 4B are solely due to the negative feedback exerted by CLOCK–BMAL1 on the expression of the Bmal1 gene. In the absence of this negative feedback, oscillations can originate from the PER–CRY negative feedback loop (Fig. 4C). This result agrees with the observation of circadian oscillations in the absence of REV-ERBα in mice (13).
Incorporation of REV-ERBα into the Model for the Mammalian Clock. Taking into account explicitly the role of REV-ERBα in the indirect negative feedback exerted by BMAL1 on the expression of the Bmal1 gene requires an extension of the model, which is now governed by 19 instead of 16 kinetic equations (see Supporting Text). Sustained oscillations with a circadian period can occur in this extended model in DD, much as in the model based on direct inhibition of Bmal1 expression by BMAL1. The mRNAs of Per, Cry, and Rev-Erbα oscillate in phase, and out of phase with respect to Bmal1 mRNA (see Fig. 8A, which is published as supporting information on the PNAS web site). The oscillations can be entrained by LD cycles, with the peak in Per mRNA falling in the light phase (see Fig. 8B).
Discussion
The model considered here represents a step toward a detailed computational model for the mammalian circadian clock. It incorporates the main clock components identified so far, but some additional components, such as the recently discovered Dec1 and Dec2 genes (35), are not considered. Over a sizeable range of parameter values, the model accounts for the occurrence of autonomous sustained oscillations, in conditions corresponding to DD. In agreement with experimental observations, the model predicts an antiphase relationship between the oscillations of Per and Cry mRNAs on the one hand, and Bmal1 mRNA on the other. When incorporated into the model, Rev-Erbα mRNA oscillates in phase with Per and Cry mRNAs. The model also accounts for entrainment by LD cycles.
One use of the model for mammalian circadian rhythms could be to test the effect of treatments affecting the levels of expression of the various clock genes, and the consequences of mutations affecting the clock gene products. Thus, in the conditions of Fig. 2 A, decreasing the level of CRY by halving the rate of CRY synthesis does not abolish the oscillations, but merely decreases the period in DD from 23.9 h to 22.1 h. This result agrees with the observation that circadian oscillations persist, with a roughly normal free running period, in Cry single-mutant mice (26). The model also predicts that oscillations persist when PER or CRY is expressed constitutively (see Table 1, for KAP = 0 and KAC = 0).
It is noteworthy that the model also predicts the possibility of sustained oscillations in the absence of Per mRNA or PER protein (as in Fig. 4B). Indeed, the model indicates that the negative autoregulatory feedback exerted by CLOCK–BMAL1 on the expression of the Bmal1 gene suffices to produce sustained oscillatory behavior in all of the other variables, although the period of these oscillations may not necessarily be circadian. At least two oscillators are thus coupled within the circadian control system. The first oscillator relies primarily on the indirect negative feedback exerted on the expression of Per and Cry through the binding of PER–CRY to the CLOCK-BMAL1 activating complex. As shown by experiments on Rev-Erbα knockout mice (13), and by the results of Fig. 4C, this mechanism suffices to produce circadian oscillations. The second mechanism capable of generating sustained oscillations is based on the negative feedback exerted by CLOCK-BMAL1, through REV-ERBα, on the expression of the Bmal1 gene. The latter mechanism should become unmasked only when the other feedback becomes inoperative, provided that parameter values are such that they allow for sustained oscillations. Parameter values may indeed be such that in the absence of the PER–CRY feedback loop, the system evolves, with or without damped oscillations, to a nonoscillatory state, as in the case illustrated in Fig. 4A. It is also possible that the second oscillatory mechanism is only capable of producing damped oscillations, which could become sustained when entrained by a periodic signal, such as temperature cycles. The possibility of a second oscillatory mechanism in mammals could explain the observation that in mPer1/mPer2-deficient mice, rhythmicity can be restored for several days by an extended light pulse (K. Bae and D. Weaver, personal communication). A major goal of future studies will be to use the model to clarify how the two mechanisms of oscillations interact to produce circadian rhythms.
The model schematized in Fig. 1 applies, with few modifications, to circadian rhythms in nonmammalian organisms such as Drosophila. Differences pertain to the effect of light, which is to trigger degradation of the TIM protein rather than Per expression, and to the partners of PER and CLOCK, which, instead of CRY and BMAL1, are TIM and CYC, respectively, whereas the role of REV-ERBα is played by VRILLE. The finding of multiple oscillatory mechanisms in the mammalian model could thus be related to experimental evidence for multiple oscillators in Drosophila (36), and might also bear on related observations in Neurospora (37) and plants (38).
In mammals, light pulses generally cause phase delays in early subjective night, phase advances in late subjective night, and no phase shift (dead zone) in subjective day (see, for example, ref. 39). Phase-response curves (PRCs) of this type can be obtained theoretically, as illustrated in Fig. 8C for the extended model incorporating REV-ERBα. The results of numerical simulations indicate, however, that the shape of the PRC is sensitive to parameter values and to the duration and amplitude of the perturbation.
Although the central pacemaker located in the suprachiasmatic nuclei (SCN) produces sustained circadian rhythms in an autonomous manner, peripheral tissues such as liver, kidney, or skeletal muscle can also give rise to circadian rhythms, with a phase in LD that differs from that observed for SCN rhythms (5, 40). Peripheral rhythms are damped unless they are driven by periodic signals received from the SCN (41, 42). The model indicates, accordingly, that damped oscillations can transform into sustained circadian rhythms when subjected to forcing by a periodic signal (see Fig. 7). Such a result can account for the observation that circadian oscillations produced by the “slave oscillators” present in isolated peripheral tissues are damped, in contrast to sustained oscillations generated in the master clock contained in the SCN. This finding corroborates the view (5) that the difference between the two situations is of quantitative, rather than qualitative nature. The model further shows that changes in parameter values that cause the transition from sustained to damped oscillations are accompanied by a change in the phase of the peripheral oscillators after entrainment. This result would explain why the phase of circadian oscillations in peripheral tissues differs from that observed in the SCN.
Because the effect of light is to induce Per expression, the peak in Per mRNA after entrainment in LD cycles often falls within the light phase (see Fig. 2C). In a counterintuitive manner, the model indicates that the phase of the oscillations markedly depends on other parameters of the model that are not affected by light. For example, in Fig. 2D, a decrease in the equilibrium constant characterizing the Cry gene activation by CLOCK–BMAL1 shifts the maximum in Per mRNA from the light to the dark phase. The model also indicates (see Fig. 3A), that different parameter values yielding the same free running period in DD can correspond to different phases of the oscillations after entrainment in LD.
The model could be used to explore syndromes or pathological conditions resulting from disorders of circadian rhythms. Of particular import is the observation that severe disruption of circadian rhythms can lead to accelerated growth of malignant tumors (43, 44). In regard to the sleep-wake cycle, we already showed here that the results of Fig. 3A bear on the molecular, dynamical origin of syndromes (27–33) associated with milder perturbations of the human circadian clock, such as FASPS (27), the delayed sleep phase syndrome (29), or the non-24-h sleep-wake syndrome (30). The model corroborates the view that these disorders of the circadian system may be viewed as “dynamical diseases” (45), i.e., physiological dysfunctions resulting from changes in dynamic behavior because of a shift outside the physiological range of some control parameter values.
Supplementary Material
Acknowledgments
We thank Dr. D. Weaver for fruitful comments on the manuscript. This work was supported by Grant 3.4607.99 from the Fonds de la Recherche Scientifique Médicale (Belgium) and by Defense Advanced Research Projects Agency Grant F30602-01-2-0554. J.-C.L. is Chargé de Recherches du Fonds National de la Recherche Scientifique (Belgium).
Abbreviations: PER, period; TIM, timeless; CYC, cycle; CRY, cryptochrome; LD, light-dark; DD, continuous darkness; SCN, suprachiasmatic nuclei.
References
- 1.Young, M. W. & Kay, S. A. (2001) Nat. Rev. Genet. 2, 702-715. [DOI] [PubMed] [Google Scholar]
- 2.Loros, J. J. & Dunlap, J. C. (2001) Annu. Rev. Physiol. 63, 757-794. [DOI] [PubMed] [Google Scholar]
- 3.Mori, T. & Johnson, C. H. (2001) Semin. Cell Dev. Biol. 12, 271-278. [DOI] [PubMed] [Google Scholar]
- 4.Roden, L. C. & Carre, I. A. (2001) Semin. Cell Dev. Biol. 12, 305-315. [DOI] [PubMed] [Google Scholar]
- 5.Reppert, S. & Weaver, D. (2002) Nature 418, 935-941. [DOI] [PubMed] [Google Scholar]
- 6.Hardin, P. E., Hall, J. C. & Rosbash, M. (1990) Nature 343, 536-540. [DOI] [PubMed] [Google Scholar]
- 7.Glossop, N. R. J., Lyons, L. C. & Hardin, P. E. (1999) Science 286, 766-768. [DOI] [PubMed] [Google Scholar]
- 8.Lee, K., Loros, J. J. & Dunlap, J. C. (2000) Science 289, 107-110. [DOI] [PubMed] [Google Scholar]
- 9.Shearman, L. P., Sriram, S., Weaver, D. R., Maywood, E. S., Chaves, I., Zheng, B., Kume, K., Lee, C. C., van der Horst, G. T., Hastings, M. H. & Reppert, S. M. (2000) Science 288, 1013-1019. [DOI] [PubMed] [Google Scholar]
- 10.Lee, C., Etchegaray, J. P., Cagampang, F. R., Loudon, A. S. & Reppert, S. M. (2001) Cell 107, 855-867. [DOI] [PubMed] [Google Scholar]
- 11.Bae, K., Lee, C., Sidote, D., Chuang, K.-Y. & Edery, I. (1998) Mol. Cell. Biol. 18, 6142-6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blau, J. & Young, M. W. (1999) Cell 99, 661-671. [DOI] [PubMed] [Google Scholar]
- 13.Preitner, N., Damiola, F., Lopez-Molina, L., Zakany, J., Duboule, D., Albrecht, U. & Schibler, U. (2002) Cell 110, 251-260. [DOI] [PubMed] [Google Scholar]
- 14.Zeng, H., Qian, Z., Myers, M. P. & Rosbash, M. (1996) Nature 380, 129-135. [DOI] [PubMed] [Google Scholar]
- 15.Zylka, M. J., Shearman, L. P., Weaver, D. R. & Reppert, S. M. (1998) Neuron 20, 1103-1110. [DOI] [PubMed] [Google Scholar]
- 16.Goldbeter, A. (1995) Proc. R. Soc. London Ser. B 261, 319-324. [Google Scholar]
- 17.Goldbeter, A. (1996) Biochemical Oscillations and Cellular Rhythms: The Molecular Bases of Periodic and Chaotic Behavior (Cambridge Univ. Press, Cambridge, U.K.).
- 18.Leloup, J.-C. & Goldbeter, A. (1998) J. Biol. Rhythms 13, 70-87. [DOI] [PubMed] [Google Scholar]
- 19.Ueda, H. R., Hagiwara, M. & Kitano, H. (2001) J. Theor. Biol. 210, 401-406. [DOI] [PubMed] [Google Scholar]
- 20.Smolen, P., Baxter, D. A. & Byrne, J. H. (2001) J. Neurosci. 21, 6644-6656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leloup, J.-C., Gonze, D. & Goldbeter, A. (1999) J. Biol. Rhythms 14, 433-448. [DOI] [PubMed] [Google Scholar]
- 22.Ruoff, P., Vinsjevik, M., Monnerjahn, C. & Rensing, L. (2001) J. Theor. Biol. 209, 29-42. [DOI] [PubMed] [Google Scholar]
- 23.Leloup, J.-C. & Goldbeter, A. (2000) BioEssays 22, 83-92. [DOI] [PubMed] [Google Scholar]
- 24.Goldbeter, A. (2002) Nature 420, 238-245. [DOI] [PubMed] [Google Scholar]
- 25.Gonze, D., Halloy, J. & Goldbeter, A. (2002) Proc. Natl. Acad. Sci. USA 99, 673-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van der Horst, G. T., Muijtjens, M., Kobayashi, K., Takano, R., Kanno, S., Takao, M., de Wit, J., Verkerk, A., Eker, A. P., van Leenen, D., et al. (1999) Nature 398, 627-630. [DOI] [PubMed] [Google Scholar]
- 27.Toh, K. L., Jones, C. R., He, Y., Eide, E. J., Hinz, W. A., Virshup, D. M., Ptacek, L. J. & Fu, Y.-H. (2001) Science 291, 1040-1043. [DOI] [PubMed] [Google Scholar]
- 28.Jones, C. R., Campbell, S. S., Zone, S. E., Cooper, F., DeSano, A., Murphy, P. J., Jones, B., Czajkowski, L. & Ptacek, L. J. (1999) Nat. Med. 5, 1062-1065. [DOI] [PubMed] [Google Scholar]
- 29.Ebisawa, T., Uchiyama, M., Kajimura, N., Mishima, K., Kamei, Y., Katoh, M., Watanabe, T., Sekimoto, M., Shibui, K., Kim, K., et al. (2001) EMBO Rep. 2, 342-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richardson, G. S. & Malin, H. V. (1996) J. Clin. Neurophysiol. 13, 17-31. [DOI] [PubMed] [Google Scholar]
- 31.Lockley, S. W., Skene, D. J., James, K., Thapan, K., Wright, J. & Arendt, J. (2000) J. Endocrinol. 164, R1-R6. [DOI] [PubMed] [Google Scholar]
- 32.McArthur, A. J., Lewy, A. J. & Sack, R. L. (1996) Sleep 19, 544-553. [DOI] [PubMed] [Google Scholar]
- 33.Uchiyama, M., Okawa, M., Ozaki, S., Shirakawa, S. & Takahashi, K. (1996) Sleep 19, 637-640. [DOI] [PubMed] [Google Scholar]
- 34.Zheng, B., Albrecht, U., Kaasik, K., Sage, M., Lu, W., Vaishnav, S., Li, Q., Sun, Z. S., Eichele, G., Bradley, A. & Lee, C. C. (2001) Cell 105, 683-694. [DOI] [PubMed] [Google Scholar]
- 35.Honma, S., Kawamoto, T., Takagi, Y., Fujimoto, K., Sato, F., Noshiro, M., Kato, Y. & Honma, K.-I. (2002) Nature 419, 841-844. [DOI] [PubMed] [Google Scholar]
- 36.Yoshii, T., Sakamoto, M. & Tomioka, K. (2002) Zool. Sci. 19, 841-850. [DOI] [PubMed] [Google Scholar]
- 37.Merrow, M., Brunner, M. & Roenneberg, T. (1999) Nature 399, 584-586. [DOI] [PubMed] [Google Scholar]
- 38.Green, R. M. & Tobin, E. M. (1999) Proc. Natl. Acad. Sci. USA 96, 4176-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yan, L. & Silver, R. (2002) Eur. J. Neurosci. 16, 1531-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stokkan, K. A., Yamazaki, S., Tei, H., Sakaki, Y. & Menaker, M. (2001) Science 291, 490-493. [DOI] [PubMed] [Google Scholar]
- 41.Brown, S., Zumbrunn, G., Fleury-Olela, F., Preitner, N. & Schibler, U. (2002) Curr. Biol. 12, 1574-1583. [DOI] [PubMed] [Google Scholar]
- 42.Balsalobre, A. (2002) Cell Tissue Res. 309, 193-199. [DOI] [PubMed] [Google Scholar]
- 43.Fu, L., Pelicano, H., Liu, J., Huang, P. & Lee, C. C. (2002) Cell 111, 41-50. [DOI] [PubMed] [Google Scholar]
- 44.Filipski, E., King, V. M., Li, X., Granda, T. G., Mormont, M. C., Liu, X., Claustrat, B., Hastings, M. H. & Levi, F. (2002) J. Natl. Cancer Inst. 94, 690-697. [DOI] [PubMed] [Google Scholar]
- 45.Mackey, M. C. & Glass, L. (1977) Science 197, 287-289. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.