Abstract
A frequent characteristic of human papillomavirus (HPV)-positive cervical cancers is the loss of viral E2 gene expression in HPV-infected cervical epithelial cells as a consequence of viral DNA integration into the cellular genome. The expression of E2 in HPV-positive cancer cells results in the repression of the viral E6/E7 oncogenes, activation of the p53 and pRB pathways, and a G1 cell cycle arrest, followed by induction of cellular senescence. The transcriptional consequences of E2-mediated cell cycle arrest that lead to senescence currently are unknown. Using conditional senescence induction in HeLa cells and microarray analysis, we describe here the expression profile of cells irreversibly committed to senescence. Our results provide insight into the molecular anatomy of senescence pathways and its regulation by HPV on-coproteins. These include the induction of the RAB vesicular transport machinery and a general down-regulation of chromatin regulatory molecules. The repression of tumor-specific G antigens during E2 senescence supports a reversal of the tumorigenic phenotype by E2 and the potential approach of tumor-specific G antigen-specific immunotherapy for cervical cancer.
High-risk human papillomaviruses (HPVs) are a group of small DNA tumor viruses that are associated with cancers of the uterine cervix (1). More than 95% of cervical cancers contain DNA from one of the high-risk HPV types and express the viral E6 and E7 oncogenes. The major transforming E6 and E7 activities include the targeted degradation of p53 and transcriptional induction of the cellular enzyme telomerase by E6 and the inactivation of the cellular retinoblastoma protein pRB by E7 (2).
Progression of HPV-positive precursor lesions to cancer usually is associated with the integration of the HPV DNA into the cellular genome with the disruption of the viral E2 ORF (3, 4). E2 can repress the transcription of the viral oncogenes E6 and E7 (5–11) and requires a functional E2 DNA-binding and transcriptional activation domain for this activity (12–14). Disruption of the E2 gene as a consequence of integration results in the deregulated expression of E6 and E7 mRNAs.
Exploiting the functions of E2 represents one possibility for the treatment of HPV-associated cancers. The reexpression of E2 proteins in HPV-positive, but not HPV-negative, cervical cancer cells results in a G1 cell cycle arrest (11, 12, 15, 16). E2-mediated G1 growth arrest is accompanied by the reactivation of known cellular targets for E6 and E7 and is followed by the induction of cellular senescence (17–19). As had been observed for E2-mediated growth arrest, the induction of senescence is specific for HPV-positive cancer cells and requires E6/E7 promoter repression by E2 (19, 20). E2 expression also has been shown to induce apoptotic cell death under some circumstances (15). Direct interactions between E2 and yet unidentified cellular apoptotic regulators have been implicated in this process (21). Recent data from the DiMaio laboratory further confirm the ability of E2 to cause both apoptosis and senescence and highlight distinct mechanisms that are governed by E6 and E7, respectively (22).
The term cellular or replicative senescence defines the finite replicative capacity of most somatic cells in culture, which eventually results in the complete and irreversible cessation of cellular division (for reviews, see refs. 23 and 24). To explore in detail the molecular pathways involved in E2 senescence, we used a genomic approach to analyze the transcriptional programs invoked within HeLa cells irreversibly committed to senescence. Adenovirus delivery of temperature-sensitive E2 resulted in temperature-dependent regulation of known E2 targets and the induction of senescence. Using temperature-shift experiments, we determined that senescence was irreversibly established 3 days after E2 expression. We selected this early time point for the transcriptional profiling of E2-expressing HeLa cells and have identified genes that either reflect or contribute directly to the senescence phenotype.
Materials and Methods
Cell Culture. The HeLa cervical cancer cell line was obtained from the American Type Culture Collection. The 293 cell line was a gift from Patrick Hearing (Stony Brook University, Stony Brook, NY). Monolayer HeLa and 293 cells were maintained in DMEM supplemented with 10% FBS.
Recombinant Vectors. The pRC-CMV-tsE2 expression plasmid encoding a temperature-sensitive bovine E2 protein has been described (12, 25). The E2ts gene was excised by using XbaI and HindIII digestion and subcloned into the pAdTrack-CMV shuttle plasmid (26). Recombinant adenoviral vectors were a generous gift from the Vogelstein laboratory (The Johns Hopkins University, Baltimore), and virus production was performed as described (26). Viruses were amplified and titers were determined by plaque assays on 293 cells.
Luciferase Assay. HeLa cells (1 × 106) were plated in 6-well dishes and were transfected with 500 ng of 2x2xE2BS-luc luciferase reporter plasmid (27). The cells were superinfected after 12 h with increasing multiplicities of infection of either AdE2ts or empty Ad. Infected cell aliquots were incubated at 32°C and 39.5°C, respectively, and lysates were prepared and analyzed 48 h postinfection as described (20).
Senescence Assay. Staining for perinuclear senescence assay–β-galactosidase (SAβ-gal) activity was performed as described (28).
Gene Array Hybridization and Analysis. Three separate experimental trials were performed. In each, a single culture was divided into four treatment groups that consisted of Ad vector or AdE2ts-infected cells incubated at 39.5°C or 32°C. From each group, total RNA was extracted and cRNA target was prepared and labeled according to the manufacturer's instructions. Target quality was ensured by hybridization with a Test3 Array. The target subsequently was hybridized to the Affymetrix (Santa Clara, CA) U95Av2 oligonucleotide arrays, representing ≈10,000 full-length human genes, and scanned. Initial analyses were performed with MICROARRAY SUITE 5.0 (MAS 5.0; Affymetrix), and subsequent analyses were carried out with GENE-SPRING (Silicon Genetics, Redwood City, CA).
To maximize relative gene expression changes caused by AdE2ts expressed at the 32°C permissive temperature, we performed a two-step normalization procedure. First, the signal intensity for each gene was divided by the median signal intensity of all genes on the GeneChip not flagged as absent by MAS 5.0. Each of the three trials then was normalized independently by dividing the signal intensity of each gene by its average signal intensity in the Ad and AdE2ts-infected cell samples that were incubated at 39.5°C. The resulting normalized data from all 12 samples were filtered for genes that were flagged as present by the MAS 5.0 algorithm in two or more chips, resulting in an initial pool of 6,475 genes. The relative gene expression changes between Ad and AdE2ts at 32°C in the three trials then were analyzed by using three different statistical group comparisons: Student's t test identified 193 genes (P = 0.025), Welch t test identified 266 genes (P = 0.05), and a nonparametric (Wilcoxon–Mann–Whitney) test identified 703 genes (P = 0.0215). All genes that were identified by Student's t test and Welch t test also were contained in the nonparametric gene list. Therefore, the 703 genes from the nonparametric list were clustered into a hierarchical gene and experiment tree by using a Pearson correlation. Replicate experiments were found to cluster together as expected.
RT-PCR. Total RNA was isolated from infected HeLa cells by using Trizol reagent. Synthesis of cDNA was from 5 μg of RNA by using the Superscript II Reverse Transcriptase system from Invitrogen. PCR subsequently was performed with 35 cycles and an annealing temperature of 50°C by using the following primers: for GAPDH, 5′-CCA CCC ATG GCA AAT TCC-3′ and 5′-GTC TTA CTC CTT GGA GGC CAT G-3′; for bovine papillomavirus E2, 5′-ATC GCC CAG ACG GAG TCT G-3′ and 5′-GGG AGC CGA GCA AAG AAG A-3′.
Western Blot Analysis. The cells were washed with PBS and lysed in RIPA buffer (1% Triton X-100/1% sodium deoxycholate/ 0.1% SDS/160 mM NaCl/10 mM Tris, pH 7.4/5 mM EDTA). Aliquots containing 50 μg of total protein were boiled in SDS sample buffer and resolved on a 12.5% SDS/PAGE gel. Proteins were transferred to poly(vinylidene difluoride) membranes from NEN, according to standard procedures, and membranes were probed with either p53 antibody (Ab-6) from Oncogene Science or p21CIP antibody (15091A) from PharMingen.
Northern Blot Analysis. Total RNA was isolated from infected HeLa cells by using Trizol extraction. RNA samples (10 μg) were separated on a 1.2% agarose gel containing formaldehyde, transferred to a Duralon charged nylon membrane from Stratagene, and UV cross-linked. The membrane was hybridized with randomly primed 32P-labeled DNA probe, washed, and exposed according to standard protocols. A STORM PhosphorImager from Molecular Dynamics was used to quantitate p21CIP and GAPDH-specific signals for subsequent normalization.
Results
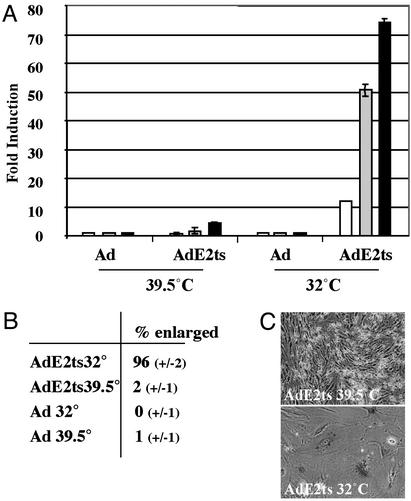
Adenovirus-Mediated Expression of a Temperature-Sensitive Bovine Papillomavirus E2 Protein. To generate an optimally controlled E2 senescence system and to circumvent background problems that usually are associated with transfection experiments, we constructed an adenovirus vector encoding a temperature-sensitive bovine papillomavirus E2 protein (25), which is functional at the permissive temperature of 32°C but defective at the restrictive temperature of 39.5°C. The resulting AdE2ts virus as well as empty Ad virus stock as a negative control were titered by plaque assays on 293 assays. Expression of E2ts protein was confirmed by Western blot analysis after infection of 293 cells at the permissive temperature (data not shown). We tested temperature-dependent E2 transcriptional activity by using an E2-dependent reporter plasmid (27) (Fig. 1A). HeLa cells were transfected with the reporter and subsequently superinfected with either AdE2ts or empty Ad with multiplicities of infection of 0.1, 1, and 10. Superinfection with empty Ad did not result in expression of the reporter at either temperature. Superinfection with AdE2ts resulted in a 75-fold induction at 32°C, compared with a minor induction at 39.5°C. In agreement with previous data (12, 17, 19, 29), the hypophosphorylation of pRB and up-regulated p107 expression were observed only in the presence of AdE2ts at 32°C and were not observed with AdE2ts at 39.5°C or in the presence of empty Ad at either temperature (data not shown). In addition, we found that the p53 protein and its transcriptional target p21CIP were induced by AdE2ts at 32°C but not at 39.5°C. Notably, both p53 and p21CIP also were heat shock sensitive in this system (see Fig. 4A).
Fig. 1.
Adenovirus-mediated expression of a temperature-sensitive E2 gene. (A) E2 transcriptional activity was determined at the permissive and restrictive temperatures by using an E2-dependent reporter plasmid. HeLa cells were transfected with the reporter plasmid and subsequently infected with 0.1 (white), 1 (gray), or 10 (black) plaque-forming units per cell. Luciferase activities are expressed as fold induction over baseline luciferase activities at the respective temperature. (B) HeLa cells were infected with AdE2ts and control empty Ad at the restrictive temperature. Some cultures were shifted to the permissive temperature for senescence induction, whereas control cultures were maintained at the restrictive temperature. The percentage of cells with the enlarged, flat-cell phenotype was determined on day 7 in three random fields relative to total cell counts. SDs are indicated. (C) Cells were infected with the AdE2ts virus and cultured at either the restrictive (39.5°C) or permissive (32°C) temperature. SAβ-gal staining was performed 3 weeks postsenescence induction.
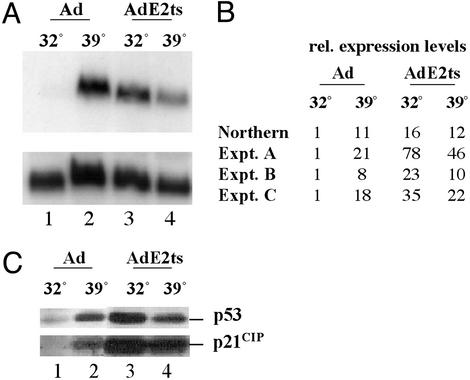
Fig. 4.
Expression levels of p21CIP.(A) Northern blot analysis with p21CIP- and GAPDH-specific probes. HeLa cells were infected with empty Ad and AdE2ts followed by temperature shift as described in the Fig. 3 legend. Lanes 1 and 2 represent samples infected with empty Ad at 32°C and 39.5°C, and lanes 3 and 4 represent samples infected with AdE2ts at 32°C and 39.5°C. Blots hybridized with p21CIP probe were stripped and reprobed with GAPDH for normalization. (B) Normalized p21CIP values from A are represented relative to the Ad sample at 32°C and are compared with the data from three independent microarray experiments. (C) HeLa cells were infected as in A, protein extracts were prepared on day 3 postsenescence induction, and equal amounts of proteins were subjected to Western blot analysis by using p53- and p21CIP-specific antibodies.
We next quantitated the early, senescence-associated flat-cell changes in response to viral infection (Fig. 1B). HeLa cells were infected with AdE2ts or the empty Ad vector at a multiplicity of infection of 10 and cultured at either 32°C or 39.5°C. Seven days postinfection, the percentage of enlarged cells was assessed relative to the total cell number in three randomly chosen fields. Senescent changes were observed only in the AdE2ts-infected cells at 32°C (Fig. 1B). The senescent phenotype after E2ts expression was maintained and was associated with positive staining of >80% of the cells for the senescence-specific SAβ-gal marker at 3 weeks postinfection (Fig. 1C).
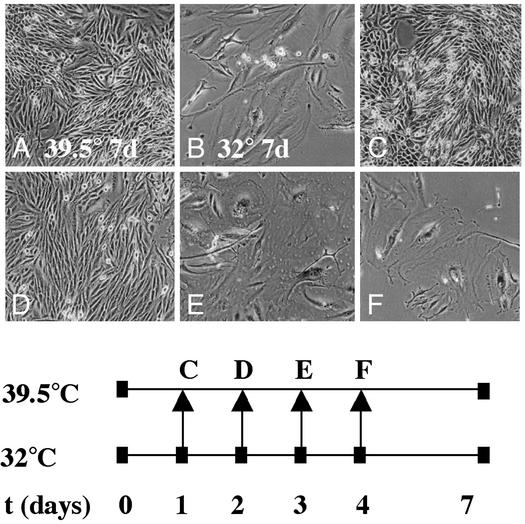
Determination of the Point of Senescence Irreversibility After E2ts Expression. We exploited the temperature-sensitive nature of the E2ts protein to determine the point of senescence irreversibility by using temperature-shift experiments (Fig. 2). HeLa cells were infected twice with 10 plaque-forming units per cell of AdE2ts at the restrictive temperature by following the strategy used previously for the simian virus 40-based PAVA viral vector system (29). The cells were split 1:5 the day after and placed at 32°C for the synchronized induction of senescence (Fig. 2; t = 0). Incubation at 32°C for 7 days yielded a senescent phenotype (28). In contrast, incubation at 39.5° did not yield a senescent phenotype (compare Fig. 2 A and B). Aliquots of infected cells were shifted on days 1, 2, 3, and 4 from 32°C to the restrictive temperature of 39.5°C to inactivate E2 expression. The cells were left at the restrictive temperature until day 7 (Fig. 2 C–F). Cells expressing functional E2 at the permissive temperature for 1 or 2 days before shift to the permissive temperature were identical to the negative control (compare Fig. 2 C and D with A). In contrast, cells expressing functional E2 for 3 or 4 days before the shift to the restrictive temperature had a senescent phenotype (compare Fig. 2 E and F with B). The observed nonsenescent and senescent phenotypes, respectively, were maintained for at least 2 additional weeks in culture after day 7 (data not shown). We therefore concluded that HeLa cells transit into the irreversible phase of the senescence program between days 2 and 3 after the expression of E2. We chose day 3 as the relevant time point for the transcriptional profiling of senescence-associated genes.
Fig. 2.
Senescence irreversibility in E2-expressing HeLa cells. HeLa cells were infected on 2 consecutive days with the AdE2ts virus at 39.5°C. The cells were split the following day and placed at 32°C for synchronized senescence induction (t = 0). Control cells remained at either 39.5°C (A) or 32°C (B) for 7 days. Aliquots were shifted from the permissive temperature of 32°C to the restrictive temperature of 39.5°C on days 1–4 for the timed inactivation of E2 (C–F). SAβ-gal staining was performed on all samples at 7 days postsenescence induction.
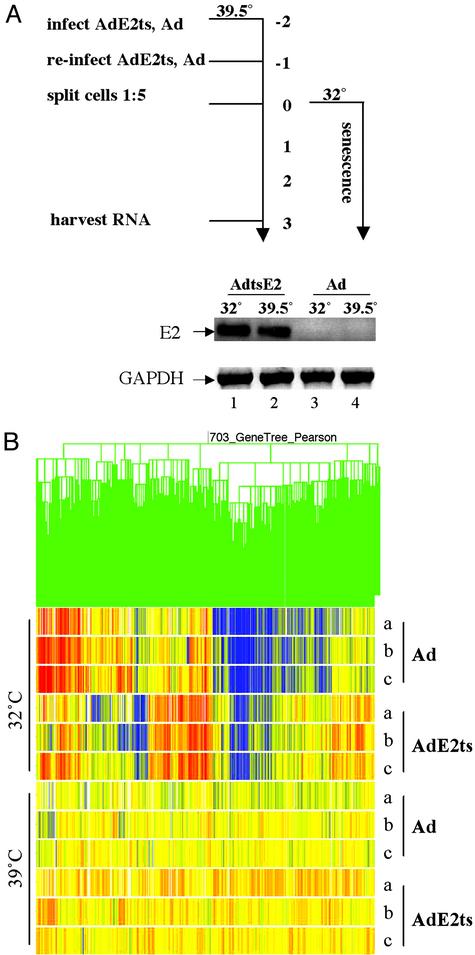
Microarray Experiments. Our experimental approach for the profiling of senescence-associated genes is depicted in Fig. 3A. HeLa cells were infected twice with a multiplicity of infection of 10 of AdE2ts at 39.5°C to allow for the accumulation of E2ts message. The day after, the cells were split 1:5 and placed at the permissive temperature of 32°C for the synchronized induction of senescence. Control cells were placed at the restrictive temperature of 39.5°C for the remainder of the experiment. Ad-infected cells were treated in the same fashion to account for any transcriptional changes that might be associated with temperature shift alone in the presence of various expressed adenoviral gene products. Each experiment therefore contained four samples, AdE2ts and Ad at both the permissive (32°C) and restrictive (39.5°C) temperatures, and was performed in triplicate. The expected senescence and control phenotypes were confirmed in each case in parallel experiments by using cell morphology and SAβ-gal staining (data not shown).
Fig. 3.
Experimental design and transcriptional profiling of senescent cells. (A) RNA samples used for microarray experiments were prepared from cells infected with AdE2ts and empty Ad at the permissive and restrictive temperatures as outlined schematically. Senescence induction was observed only with the AdE2ts sample at 32°C (lane 1). Bovine papillomavirus E2 and GAPDH messages were detected by RT-PCR, using gene-specific primers. (B) Results from clustering three independent experiments (a–c) within the four treatment groups are depicted as a gene tree (experiment tree not shown). Pearson correlation was applied to the sample measurements by using the 703 gene list derived from statistical group comparisons. Transcriptionally induced genes are indicated in red; repressed genes are indicated in blue.
Total RNA was harvested on day 3 postsenescence induction and used to generate microarray targets for Affymetrix high-density oligonucleotide arrays representing ≈10,000 full-length human genes. As shown in Fig. 3A, RT-PCR confirmed the specific expression of E2 in AdE2ts-infected cells in contrast to Ad-infected cells, whereas GAPDH was expressed in all four samples. The arrays were hybridized, scanned, and analyzed as described in Materials and Methods. To optimize our view of E2 effects at the permissive temperature, we normalized the data relative to the average of empty Ad and AdE2ts at 39°C. We then filtered in an unbiased fashion for genes that are regulated significantly in the AdE2ts relative to the Ad sample at 32°C by using a number of statistical group comparisons. Fig. 3B depicts a hierarchical gene tree with expression patterns of the resulting 703 genes, where repressed genes are represented in blue and induced genes, in red. The fully annotated dataset is available at http://genet.chmcc.org.
Up-Regulation of p21CIP mRNA. Of all genes analyzed in the microarray experiment, p21CIP displayed the highest degree of regulation. We verified the observed p21CIP expression by using Northern blot analysis in an independent temperature-shift experiment (Fig. 4A). Relative expression levels normalized to GAPDH levels (Northern) are displayed in Fig. 4B in comparison with the microarray data (Fig. 4B, experiments A–C). Levels of p21CIP message were highly induced in AdE2ts-infected compared with Ad-infected cells at the permissive temperature. Up-regulation also was observed at the restrictive temperature in both cases, consistent with the observed p21CIP protein response to heat shock (Fig. 4C). The regulation of p21CIP mRNA in three independent experiments correlated with the levels of p21CIP protein and its transcriptional activator p53 as observed in Western blot analyses (Fig. 4C). The cells were infected as for the microarray studies (Fig. 3A), and protein extracts were harvested on day 3 postsenescence induction. We found that the p53 protein and its transcriptional target p21CIP were heat shock-sensitive proteins in this system and were induced at 39.5°C in empty Ad and AdE2ts-infected cells (Fig. 3A, lanes 2 and 4 compared with lane 1). We also observed strong up-regulation of p53 and p21CIP after infection with AdE2ts at the permissive temperature (Fig. 3A, compare lanes 1 and 3). Increased levels of p21CIP protein after AdE2ts infection (Fig. 4C, lane 4) compared with Ad infection (Fig. 4C, lane 2) may be a result of experimental variability. Alternatively, leaky expression of functional tsE2 protein at the restrictive temperature (Fig. 1 A) may affect p21CIP protein levels in a p53-independent fashion.
Discussion
By limiting the number of times that a cell can divide in response to intrinsic or extrinsic signals, cellular senescence is thought to represent a natural barrier to cancer development. This notion is emphasized further by the fact that tumor suppressors such as p53 or pRB, which are inactivated frequently in human cancers in vivo, play important roles during cellular senescence in vitro (30, 31). Recent findings by Schmitt et al. (32) strongly support an antagonistic relationship between cellular senescence in vitro and carcinogenesis in vivo and positively correlate intact senescence pathways with tumor regression after chemotherapy. Despite the intriguing physiological implications of these observations, senescence remains a poorly understood phenomenon. Underlying this lack of knowledge and resulting from it is a scarcity of bona fide senescence markers. To identify potential markers and regulators of cellular senescence, we set out to determine the transcriptome of senescent cells at an early, yet irreversibly committed stage by using E2 senescence in HeLa cells as a model.
Generation of an E2-Based Inducible Senescence System. The experimental system using infection of HeLa cells with the AdE2ts virus or the empty vector control at both permissive and restrictive temperatures necessitated the analysis under four experimental conditions. A comparison of these four experimental conditions was designed to (i) account for transcriptional changes that resulted from temperature shifts alone or in the presence of various expressed Ad gene products and (ii) include genes that were temperature-sensitive as well as sensitive to E2 expression.
The regulation of p21CIP mRNA in three independent microarray experiments directly correlated with protein expression. Indeed, of all genes analyzed, p21CIP displayed the greatest induction. Levels of p21CIP message were induced between 20- and 80-fold in AdtsE2-infected cells compared with Ad-infected cells at the permissive temperature (Fig. 4B). Up-regulation also was observed in both samples at the restrictive temperature, consistent with the observed p21CIP response to heat shock. This regulated p21 expression was verified by using Northern blot analysis (Fig. 4A). We interpret the successful detection of the p21CIP senescence marker as proof of concept for the validation of this genomics approach to identify E2-responsive, senescence-associated genes.
It is important to recognize that an optimally controlled temperature-shift experiment such as the one described here does not provide a simple on–off situation. Each of the experimental steps outlined in Fig. 3 introduces variability and, consequently, differences in the signal intensities between experiments. We observed such variability for a number of genes across experiments B–D (see Fig. 4 for p21CIP as an example). Each experiment was monitored in parallel for successful senescence induction with AdE2ts at the permissive temperature compared with the controls to rule out the possibility that data variations may result from phenotypical variability. Consistent differences that we observed for specific genes in this experimental setting therefore may represent stringently, perhaps obligatorily, regulated players.
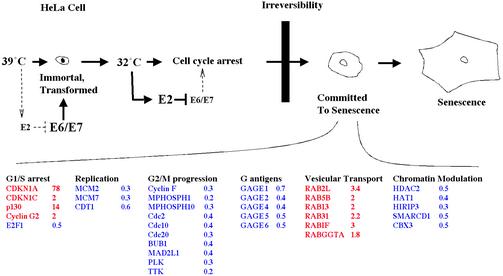
The Antagonism Between E2 Senescence and HPV Immortality. A model of E2ts senescence (Fig. 5) depicts several functional gene groups that are regulated as predicted from the cellular growth-arrest phenotype. With respect to the G1/S growth arrest that is observed in senescent cells, p21CIP (CDKN1A) up-regulation is accompanied by the induction of the cyclin/cyclin-dependent kinase inhibitor p57KIP (CDKN1C), the p130 retinoblastoma family member, and the antiproliferative cyclin G2 as well as by the repression of the S phase-promoting E2F1 gene. A number of DNA replication and G2/M phase-promoting genes also are found repressed during this time. The latter group of genes includes cyclin F, BUB1, Cdc2, 10 and 20, Polo-like kinase, and TTK protein kinase. Strikingly, several of the above proteins, most notably PLAB, BUB1, cdc20, and Polo-like kinase, recently were reported to display the opposite expression patterns in gene array studies with keratinocytes that were immortalized by the high-risk HPV31 E6 and E7 oncogenes (33). It is likely that inverse expression patterns in the immortalized vs. senescent cell system reflect the ability of E2 to repress E6/E7 expression and, thus, release downstream targets from oncogene control.
Fig. 5.
A window of gene expression changes during E2-mediated senescence induction. Expression of E2 in HPV-positive cells causes senescence via the repression of the viral E6/E7 oncogenes. Several gene groups that are regulated during senescence induction were categorized according to biological processes that they are known to regulate. Induced genes are depicted in red; repressed genes are depicted in blue. For each gene, the most significant observed expression change, of three independent experiments, is depicted as the ratio of expression signal obtained with AdE2ts to that obtained with Ad at 32°C. The data were normalized to the average of AdE2ts and Ad at 39.5°C as described in Materials and Methods. Expression changes for these genes in all three experiments are shown at http://genet.chmcc.org.
We note the coordinated repression of a group of tumor-associated antigens, GAGE 1, 2, 4, 5, and 6, by E2. GAGE genes belong to a group of tumor-specific antigens that are presented by HLA class I molecules and recognized by cytolytic T lymphocytes. Because of their tumor-specific expression, GAGE antigens are promising targets for cancer immunotherapy. Down-regulation of GAGE genes by E2 not only emphasizes a reversal of the tumorigenic phenotype through E2 senescence but also suggests the prospect for GAGE-specific immunotherapy in cervical cancer treatment.
Implications for Cellular Senescence Pathways. The molecular signals triggering cellular transit from a reversible growth arrest to the irreversible establishment of senescence currently are unknown. As depicted in Fig. 5, we identified a group of six induced members and regulators of the Ras-related RAB family of small GTPases: RAB2, 5B, 13, 31, RAB interacting factor, and the RAB geranylgeranyltransferase α-subunit. Little is known about the transcriptional regulation of RAB genes, and our results suggest the coordinated regulation of some members of the RAB family during E2 senescence. RAB GTPases are highly conserved, specialized regulators of fluid-phase and receptor-mediated endocytosis, transport between intracellular compartments, and exocytosis (for a review, see ref. 34). Coordinate transcriptional regulation of members and regulators of the RAB GTPase family in senescent cells may suggest either a global up-regulation or the specific modulation of vesicular transport during senescence. HeLa cells originally were derived from an HPV-positive adenocarcinoma of the cervix, and it is possible that the observed vesicular regulation may recapitulate aspects of the glandular differentiation process. There are a number of parallels between senescence and differentiation, including permanent cell cycle arrest, up-regulation of cyclin/ cyclin-dependent kinase inhibitors, and distinct morphological features, which have led to speculations that senescence may represent a specialized form of differentiation rather than a completely independent biological process. In this case, the induction of vesicular transport mediators after E2 expression may indicate similarities between senescence and differentiation. Such a scenario may be supported further by the up-regulated expression of the differentiation-specific cyclin G during E2 senescence. An alternative hypothesis is based on the specific microenvironment that a senescent cell provides via the secretion of molecules such as metalloproteases, inflammatory cytokines, and growth factors (35). It is conceivable that, in addition to the overexpression of relevant paracrine factors, their secretion may be influenced by RAB GTPase activities. Regardless of the implications, our data suggest the existence of regulatory crosstalk between oncogene-governed signal-transduction pathways and RAB-dependent protein trafficking.
Senescence may be linked intricately to chromatin structure and organization (36). Listed in Fig. 5 are the six chromatin regulatory genes, HDAC2, HAT1, HIRIP3, SMARCD1, and CBX3, that we identified within the transcriptome signature of senescent HeLa cells (Fig. 5). Interestingly, all of these genes are repressed during senescence. The down-regulation of HDAC2 expression may be functionally relevant, because chemical histone deacetylase inhibitors already are known to trigger senescence in human fibroblasts (37) and HPV-positive cervical cancer cells (38). However, we also found HAT1 repressed during E2 senescence, which may indicate that rather than a sufficient level of deacetylation, a critical balance between acetylated and deacetylated chromatin is required for cellular proliferation. Disturbing this balance by either excessive histone acetyltransferase or deacetylase activities then may lead to senescence or apoptosis (39) induction depending on additional internal or external parameters. This hypothesis was supported recently by the finding that not only interference with histone deacetylase activity as described above, but also interference with p300/CBP histone acetylase activities can serve as a senescence trigger in human cells (40). Repression of the SWI/ SNF-associated chromatin regulators SMARCD1, the histone-associated HIRIP3 protein, and the Drosophila HP1 human homolog CBX3 may play a similar role in the senescence phenotype. Taken together, our data support the notion that cancer cell senescence is accompanied, perhaps even actively controlled, by the decreased activity of several evolutionarily highly conserved cellular chromatin-remodeling machineries.
Acknowledgments
S.I.W. was supported by a fellowship from The Medical Foundation, Boston. This research was supported by National Cancer Institute Grant RO1 CA77385 (to P.M.H.), American Cancer Society Institutional Grant 92-026-09 (to S.I.W.), and the American Cancer Society Ohio Division Supported Research grant for the year 2002 (to S.I.W.).
Abbreviations: HPV, human papillomavirus; SAβ-gal, senescence assay–β-galactosidase.
References
- 1.Howley, P. M. & Lowy, D. R. (2001) in Fields Virology, eds. Howley, P. M. & Knipe, D. M. (Lippincott, Philadelphia), Vol. 2, pp. 2197-2229. [Google Scholar]
- 2.Munger, K. & Howley, P. M. (2002) Virus Res. 89, 213-228. [DOI] [PubMed] [Google Scholar]
- 3.Durst, M., Kleinheinz, A., Hotz, M. & Gissmann, L. (1985) J. Gen. Virol. 66, 1515-1522. [DOI] [PubMed] [Google Scholar]
- 4.Schwarz, E., Freese, U. K., Gissman, L., Mayer, W., Roggenbuck, B., Stremlau, A. & zur Hausen, H. (1985) Nature 314, 111-114. [DOI] [PubMed] [Google Scholar]
- 5.Bernard, B. A., Bailly, C., Lenoir, M. C., Darmon, M., Thierry, F. & Yaniv, M. (1989) J. Virol. 63, 4317-4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Demeret, C., Desaintes, C., Yaniv, M. & Thierry, F. (1997) J. Virol. 71, 9343-9349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DiMaio, D., Metherall, J., Neary, K. & Guralski, D. (1986) J. Virol. 57, 475-480.3003380 [Google Scholar]
- 8.Romanczuk, H., Thierry, F. & Howley, P. M. (1990) J. Virol. 64, 2849-2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tan, S.-H., Gloss, B. & Bernard, H.-U. (1992) Nucleic Acids Res. 20, 251-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thierry, F. & Howley, P. M. (1991) New Biol. 3, 90-100. [PubMed] [Google Scholar]
- 11.Thierry, F. & Yaniv, M. (1987) EMBO J. 6, 3391-3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dowhanick, J. J., McBride, A. A. & Howley, P. M. (1995) J. Virol. 69, 7791-7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goodwin, E. C., Naeger, L. K., Breiding, D. E., Androphy, E. J. & DiMaio, D. (1998) J. Virol. 72, 3925-3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nishimura, A., Ono, T., Ishimoto, A., Dowhanick, J. J., Frizzell, M. A., Howley, P. M. & Sakai, H. (2000) J. Virol. 74, 3752-3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Desaintes, C., Demeret, C., Goyat, S., Yaniv, M. & Thierry, F. (1997) EMBO J. 16, 504-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hwang, E. S., Riese, D. J., Settleman, J., Nilson, L. A., Honig, J., Flynn, S. & DiMaio, D. (1993) J. Virol. 67, 3720-3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goodwin, E. C. & DiMaio, D. (2001) Cell Growth Differ. 12, 525-534. [PubMed] [Google Scholar]
- 18.Lee, C. J., Suh, E. J., Kang, H. T., Im, J. S., Um, S. J., Park, J. S. & Hwang, E. S. (2002) Exp. Cell Res. 277, 173-182. [DOI] [PubMed] [Google Scholar]
- 19.Wells, S. I., Francis, D. A., Karpova, A. Y., Dowhanick, J. J., Benson, J. D. & Howley, P. M. (2000) EMBO J. 19, 5762-5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Francis, D. A., Schmid, S. I. & Howley, P. M. (2000) J. Virol. 74, 2679-2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Desaintes, C., Goyat, S., Garbay, S., Yaniv, M. & Thierry, F. (1999) Oncogene 18, 4538-4545. [DOI] [PubMed] [Google Scholar]
- 22.DeFilippis, R. A., Goodwin, E. C., Wu, L. & DiMaio, D. (2003) J. Virol. 77, 1551-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Campisi, J. (2000) In Vivo 14, 183-188. [PubMed] [Google Scholar]
- 24.Lloyd, A. C. (2002) Nat. Cell Biol. 4, E25-E27. [DOI] [PubMed] [Google Scholar]
- 25.DiMaio, D. & Settleman, J. (1988) EMBO J. 7, 1197-1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He, T. C., Zhou, S., da Costa, L. T., Yu, J., Kinzler, K. W. & Vogelstein, B. (1998) Proc. Natl. Acad. Sci. USA 95, 2509-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kovelman, R., Bilter, G. K., Glezer, E., Tsou, A. Y. & Barbosa, M. S. (1996) J. Virol. 70, 7549-7560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dimri, G. P., Lee, X., Basile, G., Acosta, M., Scott, G., Roskelley, C., Medrano, E. E., Linskens, M., Rubelj, I., Pereira-Smith, O., et al. (1995) Proc. Natl. Acad. Sci. USA 92, 9363-9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goodwin, E. C. & DiMaio, D. (2000) Proc. Natl. Acad. Sci. USA 97, 12513-12518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Campisi, J. (2001) Trends Cell Biol. 11, S27-S31. [DOI] [PubMed] [Google Scholar]
- 31.Serrano, M. & Blasco, M. A. (2001) Curr. Opin. Cell Biol. 13, 748-753. [DOI] [PubMed] [Google Scholar]
- 32.Schmitt, C. A., Fridman, J. S., Yang, M., Lee, S., Baranov, E., Hoffman, R. M. & Lowe, S. W. (2002) Cell 109, 335-346. [DOI] [PubMed] [Google Scholar]
- 33.Nees, M., Geoghegan, J. M., Hyman, T., Frank, S., Miller, L. & Woodworth, C. D. (2001) J. Virol. 75, 4283-4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Segev, N. (2001) Sci. STKE 2001, RE11. [DOI] [PubMed] [Google Scholar]
- 35.Krtolica, A., Parrinello, S., Lockett, S., Desprez, P.-Y. & Campisi, J. (2001) Proc. Natl. Acad. Sci. USA 98, 12072-12077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Howard, B. H. (1996) Exp. Gerontol. 31, 281-293. [DOI] [PubMed] [Google Scholar]
- 37.Ogryzko, V. V., Hirai, T. H., Russanova, V. R., Barbie, D. A. & Howard, B. H. (1996) Mol. Cell. Biol. 16, 5210-5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Terao, Y., Nishida, J., Horiuchi, S., Rong, F., Ueoka, Y., Matsuda, T., Kato, H., Furugen, Y., Yoshida, K., Kato, K., et al. (2001) Int. J. Cancer 94, 257-267. [DOI] [PubMed] [Google Scholar]
- 39.Finzer, P., Kuntzen, C., Soto, U., zur Hausen, H. & Rosl, F. (2001) Oncogene 20, 4768-4776. [DOI] [PubMed] [Google Scholar]
- 40.Bandyopadhyay, D., Okan, N. A., Bales, E., Nascimento, L., Cole, P. A. & Medrano, E. E. (2002) Cancer Res. 62, 6231-6239. [PubMed] [Google Scholar]