Abstract
The Escherichia coli BglF protein is a sugar-sensor that controls the activity of the transcriptional antiterminator BglG by reversibly phosphorylating it, depending on β-glucoside availability. BglF is a membrane-bound protein, whereas BglG is a soluble protein, and they are both present in the cell in minute amounts. How do BglF and BglG find each other to initiate signal transduction efficiently? Using bacterial two-hybrid systems and the Far-Western technique, we demonstrated unequivocally that BglG binds to BglF and to its active site-containing domain in vivo and in vitro. Measurements by surface plasmon resonance corroborated that the affinity between these proteins is high enough to enable their stable binding. To visualize the subcellular localization of BglG, we used fluorescence microscopy. In cells lacking BglF, the BglG-GFP fusion protein was evenly distributed throughout the cytoplasm. In contrast, in cells producing BglF, BglG-GFP was localized to the membrane. On addition of β-glucoside, BglG-GFP was released from the membrane, becoming evenly distributed throughout the cell. Using mutant proteins and genetic backgrounds that impede phosphorylation of the Bgl proteins, we demonstrated that BglG-BglF binding and recruitment of BglG to the membrane sensor requires phosphorylation but does not depend on the individual phosphorylation sites of the Bgl proteins. We suggest a mechanism for rapid response to environmental changes by preassembly of signaling complexes, which contain transcription regulators recruited by their cognate sensors-kinases, under nonstimulating conditions, and release of the regulators to the cytoplasm on stimulation. This mechanism might be applicable to signaling cascades in prokaryotes and eukaryotes.
What are the mechanisms that enable rapid changes in gene expression in response to external stimuli? How do membrane-bound sensors and soluble signaling proteins communicate efficiently on stimulation? These are central questions in understanding signal transduction. We asked these questions with respect to the bgl sensory system in Escherichia coli. Expression of the bgl operon is regulated by BglG, a transcriptional regulator, and BglF, a membrane-bound sugar sensor (1, 2). Transcription from the bgl promoter initiates constitutively, but in the absence of β-glucosides, most transcripts terminate prematurely at one of two ρ-independent terminators within the operon; in the presence of an inducer, BglG allows transcription through these sites by binding to the bgl transcript (3–5). BglG also recognizes and interacts with the β′ subunit of E. coli RNA polymerase (6). BglF regulates BglG activity by reversibly phosphorylating it depending on β-glucoside availability (7–9), thus modulating its dimeric state (10). Interestingly, BglF, an enzyme II of the phosphoenolpyruvate-dependent phosphotransferase system (PTS) that catalyzes transport and phosphorylation of β-glucosides, uses the same active site residue, Cys-24, to phosphorylate the sugar and BglG (11) and to dephosphorylate BglG (12). Homologues of the Bgl proteins have been identified in many bacterial species (13). Activity of these proteins as antiterminators is regulated by the PTS and for some, negative regulation by BglF-like sugar phosphotransferases has been demonstrated (14).
BglF is a membrane-bound sensor, whereas the BglG transcriptional regulator is soluble, and both are present in small amounts in uninduced cells. How do BglF and BglG find each other upon inducer-mediated stimulation of BglF? A possible mechanism, that would ensure an efficient response to environmental changes, is recruitment of BglG to the membrane by BglF and rapid release of BglG to the cytoplasm on stimulation. To test this hypothesis, we took several experimental approaches. We first asked whether BglG binds to BglF. Using the Far-Western technique and bacterial two-hybrid systems, we demonstrated that BglG interacts with BglF and with its active site-containing domain both in vitro and in vivo. Using surface plasmon resonance (SPR), we showed that the affinity of BglG for the active site containing domain of BglF is strong (KD ≈ 3 × 10-7) and supports the idea of stable binding. Using fluorescence microscopy, we showed that BglG fused to GFP is located at the cell membrane only in the presence BglF and is released to the cytoplasm after addition of β-glucosides. Neither Cys-24 of BglF, which reversibly phosphorylates BglG, nor the BglG phosphorylation site His-208 is crucial for BglF–BglG interaction. However, phosphorylation is required for the interaction, as indicated by the lack of interaction and recruitment of BglG in a Δpts strain, or when both proteins lack phosphorylation sites.
Materials and Methods
Strain Construction. A Δpts derivative of SU202 (15) was isolated by transducing SU202 to TetR by using P1 phage grown on PPA310 that harbors a deletion of the pts operon, obtained from P. Postma (University of Amsterdam). A Δpts derivative of MG1655 was isolated by transducing MG1655 to KanR using P1 phage grown on TP2811 (16).
Chemicals. N-hydroxysuccinimide, N-ethyl-N-(3-diethylaminopropyl) carbodimide, ethanolamine hydrochloride, and HBS buffer (10 mM Hepes, pH 7.4/150 mM NaCl/3.4 mM EDTA/0.005% P-20) were obtained from BIAcore AB (Uppsala).
Plasmids. All plasmids used in this study and the proteins they encode are listed in Table 3, which is published as supporting information on the PNAS web site, www.pnas.org. Construction of plasmids is provided as supporting information. Plasmids used for the LexA-based two-hybrid system were derived from pMS604 and pDP804 (15). Plasmids used for Far Western and SPR were constructed by cloning the bglF and bglG alleles in pET15b (Novagen) or in pST6#1, a derivative of MBPL–/gp21(338-445) (17), obtained from P. Poumbourios (St. Vincent's Institute of Medical Research, Victoria, Australia). Plasmids used for fluorscence microscopy: pJS185 encodes BglG-GFP; the bglF alleles were cloned in pBAD18 (18).
Two-Hybrid Analysis. SU202 strain and its Δpts derivative were cotransformed with the derivatives of pDP804 and pMS604. Cells were grown until midlogarithmic phase at 37°C in M63 medium (19) containing succinate (0.4%) as a carbon source and 0.5 mM isopropyl β-D-thiogalactoside. Interaction between the two hybrid proteins was monitored on MacConkey–maltose indicator plates and quantitated by β-galactosidase assays (19).
Affinity Chromatography. MBP-BglG (MBP, maltose-binding protein) was purified as described in ref. 11. His-tagged proteins were expressed in BL21(DE3) and purified as described (20), except that the extracts were incubated for only 1h with the Ni-NTA resin. For His-tagged BglF, disruption of cells was performed by resuspending pelleted cells in lysis buffer (30 mM Tris·HCl, pH 8/20% sucrose/10 mM EDTA), supplemented with 50 μg/ml lysozyme. After 15 min incubation at 37°C, DNase (5 μg/ml) and MgCl2 (15 mM) were added, and the extract was incubated for an additional 15 min. After centrifugation at 20,800 × g in the cold, the membrane fractions were resuspended in 2 ml of PBS buffer (80 mM Na2HPO4/20 mM NaH2PO4/100 mM NaCl) containing 0.5% SDS and 6 M urea.
Far-Western Analysis. Proteins were separated on 10% SDS-polyacrylamide gels. Gels were subjected to Far-Western analysis as described (6) or stained with Coomassie blue.
SPR. SPR measurements were preformed with a BIAcore3000 system (BIAcore, Uppsala, Sweden). All procedures were performed at 25°C. His-tagged WT BglG protein and its mutants (5 μg/ml) in 10 mM sodium acetate, pH 3.5, were immobilized on the dextran surface of CM5 sensorchips by the standard amino coupling method (21), except that 70 μl of the activators N-hydroxysuccinimide, N-ethyl-N-(3-diethylaminopropyl) carbodimide, and the blocker ethanol amine were injected at a flow rate of 10 μl/min. One flow cell with no coupled protein served as reference. Once the His-tagged BglG proteins were immobilized, serial dilutions ranging from 1.25 to 10 μM of IIBbgl or IIBbgl (C24S) were injected at a flow rate of 30 μl/min in HBS buffer containing 1 mg/ml carboxy methyl dextran to minimize nonspecific interactions. A sample injection of 90 μl was followed by buffer flow for 3 min for dissociation. Regeneration was achieved by a 20-μl pulse of 50 mM phosphoric acid. Binding constants were determined by using BIAEVALUATION software. The KD values for the interaction of BglG with IIBbgl were calculated from the apparent kinetic constants by fit to a first-order kinetic model. Values were estimated from at least three independent experiments, using different chips and protein preparations.
Fluorescence Microscopy and Photography. MG1655 and its Δpts derivative were transformed with the indicated plasmids. Cells were grown to early log phase at 30°C in either LB or M9 medium (19) containing 0.4% glycerol. Expression of the bglF alleles, cloned in pBAD18, was induced by adding 0.1% arabinose for 1 h. Expression of the BglG-GFP fusion was not induced, to keep the level of the fusion protein low. When indicated, 0.5% arbutin was added to the growing culture and BglG-GFP localization was examined at 5-min intervals, after the addition of arbutin, for 2 h.
Microscopy was performed by using an Olympus (Melville, NY) BX60 microscope with an Olympus PlanApo 100 × 1.4 n.a. oil-immersion objective. An Olympus MWIB photocube transmitting a wavelength of 460–490 nm was used to stimulate GFP fluorescence. Images were obtained by using cells that had been washed and resuspended in saline and placed on a microscope slide under a coverslip. Pictures were obtained by using a Hamamatsu Photonics (Hamamatsu City, Japan) C4742-95 digital charge-coupled device camera and an automatic light shutter (MAC2000, LudI Electronics, Hawthorne, NY). The exposure time was the same in all cases. OPENLAB 3.02 imaging software (Improvision, Lexington, MA) controlled image acquisition and control of the light shutter, as well as colorization of the greyscale images acquired.
Results
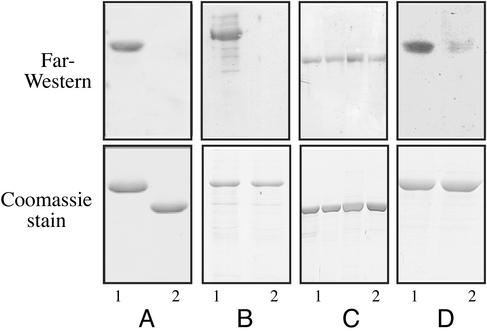
The Transcriptional Regulator BglG Binds to the BglF Sensor and to Its Active Site-Containing Domain in Vitro and in Vivo To examine the possibility that the transcriptional regulator BglG binds to the BglF sensor, we first tested whether BglG binds to IIBbgl, the active site-containing domain of BglF, in vitro, by using the Far-Western technique. Purified MBP-BglG (BglG fused to maltose-binding protein) was subjected to SDS/PAGE and blotted onto a nitrocellulose filter. When the membrane was incubated with His-tagged IIBbgl (IIBbgl fused to six histidines) and then with antibodies against the His tag, a strong signal was detected (Fig. 1A, lane 1). When MBP alone was probed with His-IIBbgl, no binding was observed (Fig. 1A, lane 2), indicating that IIBbgl interacted with BglG and not with the MBP moiety of MBP-BglG. Next, we probed filter-immobilized His-tagged BglF with either MBP-BglG or MBP and then with antibodies against MBP. An interaction between BglF and MBP-BglG was observed, but not with MBP (Fig. 1B). These results imply that the interaction of BglG with BglF and with its active site-containing domain in vitro is direct and does not require auxiliary proteins.
Fig. 1.
Far-Western analysis of the interaction between BglG and BglF or its active site-containing domain, IIBbgl. Purified proteins were fractionated by SDS/PAGE, blotted onto nitrocellulose filters, probed with secondary proteins and then with anti-His or anti-MBP antibodies (Upper), or stained with Coomassie blue (Lower). (A) MBP-BglG (lane 1) or MBP (lane 2) was probed with His-IIBbgl and then with anti-His antibodies. (B) His-BglF was probed with either MBP-BglG (lane 1) or MBP (lane 2) and then with anti-MBP antibodies. (C) MBP-BglG (lane 1) or its mutant derivatives MBP-BglG(H208R) (lane 2), MBP-BglG(H160Y) (lane 3), and MBP-BglG(D100N) (lane 4) were probed with His-IIBbgl and then with anti-His antibodies. (D) MBP-BglG (lane 1) or MBP-BglG(H208R) was probed with His-IIBbgl (C24S) and then with anti-His antibodies.
To test whether BglG binds to BglF in vivo, we used the LexA-based bacterial two-hybrid system (15). In this system, the proteins of interest are fused either to a WT LexA repressor DNA-binding domain (LexADBD) or to an altered specificity LexADBD and introduced into a strain that harbors a chromosomal copy of lacZ under the control of a LexA hybrid operator (SU202). Transcriptional repression is achieved on coexpression of both hybrid proteins, provided they bind to each other. The combination of IIBbgl fused to the WT LexADBD and BglG fused to mutant LexADBD resulted in a stable, biologically active heterodimer, as indicated by the 76% transcriptional repression (Table 1). The leucine zipper domains of Fos and Jun fused to the WT and mutant LexADBD, respectively, and chimeras between BglG and the two LexADBD served as positive controls (98% and 97% repression), and the two LexADBD served as a negative control (0% repression) (Table 1). Similar results were obtained with the adenylate cyclase-based two-hybrid system (22) (see Supporting Text, which is published as supporting information on the PNAS web site).
Table 1. Analysis of the interaction between BglG and IIBbgl and derivatives of these proteins impaired in phosphorylation by the two-hybrid LexA-based system.
| Protein fused to WT LexADBD* | Protein fused to mutant LexADBD† | Repression‡ of PlacUV5§-lacZ, % |
|---|---|---|
| — | — | 0 |
| Fos zipper | Jun zipper | 98 |
| BglG(WT) | BglG(WT) | 97 |
| IIBbgl(WT) | BglG(WT) | 76 |
| IIBbgl(WT) | BglG(H208R) | 78 |
| IIBbgl(WT) | BglG(H160Y) | 78 |
| IIBbgl(WT) | BglG(D100N) | 62 |
| IIBbgl(C24S) | BglG(WT) | 79 |
| IIBbgl(C24S) | BglG(H208R) | 52 |
| IIBbgl(C24S) | BglG(H160Y) | 75 |
| IIBbgl(C24S) | BglG(D100N) | 72 |
The experiment was performed with E. coli strain SU202, which carries a hybrid LexA operator op408/op+::lacZ fusion on its chromosome (15). The values represent the average of at least four independent measurements. Standard deviation ranged from 2% to 5% repression
Cloned in pLL3, a derivative of pMS604
Cloned in pLL1, a derivative of pDP804
Percent repression was calculated as: [1-(β-galactosidase activity with repressor/β-galactosidase activity without repressor)] × 100
A modified PlacUV5 bearing a mutation that decreases the level of expression in the presence of inducer IPTG (15)
We anticipated that fusion of the full-length BglF protein to LexADBD would result in a membrane-anchored protein, which might not bind DNA efficiently. Nevertheless, we fused BglF to the mutant LexADBD, expressed from a reduced copy number plasmid, to avoid overproduction. Coexpression of this fusion with BglG fused to WT LexADBD yielded 59% repression (Table 2). Therefore, despite the constraint caused by anchoring one of the repressor monomers to the membrane, repression was evidently achieved, corroborating BglG–BglF interaction.
Table 2. Analysis of the interaction between BglG and BglF and derivatives of these proteins impaired in phosphorylation, by the two-hybrid LexA-based system.
| Repression‡ of PlacUV5§-lacZ, %
|
|||
|---|---|---|---|
| Protein fused to mutant LexADBD* | Protein fused to WT LexADBD† | pts+ | Δpts |
| — | — | 0 | 0 |
| Jun zipper | Fos zipper | 99 | 99 |
| BglF(WT) | BglG(WT) | 59 | 12 |
| BglF(WT) | BglG(H208R) | 71 | 0 |
| BglF(WT) | BglG(H160Y) | 42 | 0 |
| BglF(WT) | BglG(D100N) | 48 | 11 |
| BglF(C24S) | BglG(WT) | 40 | 3 |
| BglF(C24S) | BglG(H208R) | 14 | 0 |
| BglF(C24S) | BglG(H160Y) | 13 | 0 |
| BglF(C24S) | BglG(D100N) | 41 | 0 |
The experiment was performed with E. coli strain SU202 or its Δpts derivative that carry a hybrid LexA operator op408/op+::lacZ fusion on their chromosome (15). The values represent the average of at least four independent measurements. Standard deviation ranged from 2% to 5% repression
Cloned in pLL1, a derivative of pDP804
Cloned in pLL3, a derivative of pMS604
Percent repression was calculated as described in Table 1
A modified PlacUV5 as described in Table 1
Taken together, the results with the two-hybrid system demonstrate that BglG interacts with the active site-containing domain of BglF, as well as with the entire BglF protein, in vivo.
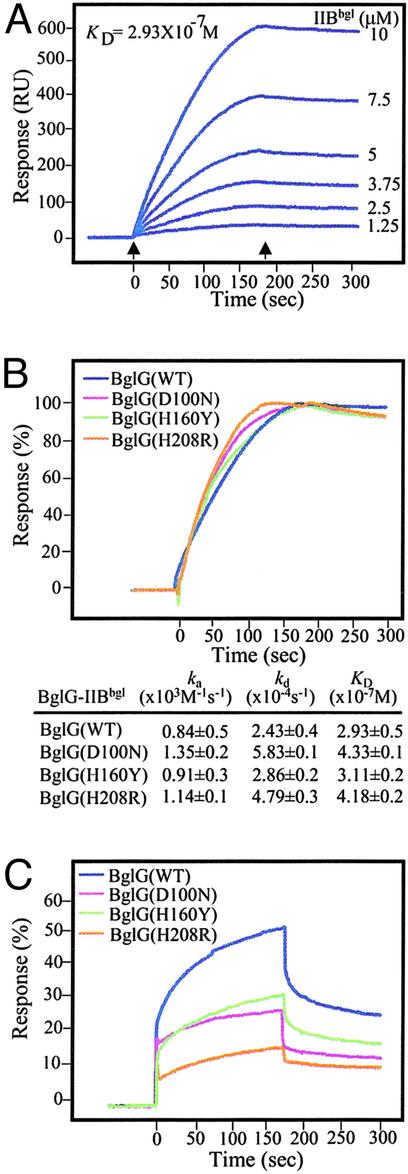
Kinetic Analysis of BglG–IIBbgl Interaction. To study the kinetics of the interaction between BglG and IIBbgl, we used SPR, which enables the measurement of rates of association (ka) and dissociation (kd). BglG was immobilized on a sensorchip, leaving one flow-cell blank, and various concentrations of IIBbgl were passed over the chip. The response from the reference surface was subtracted from the response in the channel with BglG to give a signal (resonance units) that is directly proportional to the amount of bound compound. As can be seen in Fig. 2A, BglG and IIBbgl gave binding signals in a concentration-dependent manner. The binding signal decayed very slowly after completion of the injection, indicating that the complex is highly stable. An equilibrium constant (KD) value of 2.93 × 10-7 was calculated for the BglG–IIBbgl complex. The high affinity between BglG and IIBbgl is characterized by a low dissociation rate constant (kd 2.43 × 10-4·sec-1).
Fig. 2.
SPR analysis of the interaction between BglG and BglF active site-containing domain, IIBbgl. His-BglG or its derivatives [≈1,000 resonance units (RU)] was immobilized on BIAcore sensorchips, each onto a different flow-cell. In each sensorchip, flow-cell 1 (Fc1) with no bound protein served as a reference. IIBbgl was injected over the immobilized proteins at concentrations that ranged from 1.25 to 10 μM in all cases. The figure shows the subtracted (Fcx-Fc1) sensorgrams that allowed direct visualization of specific BglG-IIBbgl binding. The association kinetics was followed for 3 min after the injection start (left arrow) and the dissociation kinetics for 3 min after the injection stop (right arrow). (A) Sensorgrams obtained when injecting IIBbgl at the indicated concentrations over immobilized BglG. (B) Sensorgrams obtained when injecting IIBbgl at 10 μM over a sensorchip with bound His-tagged BglG(WT) (blue line), BglG(D100N) (purple line), BglG(H160Y) (green line), and BglG(H208R) (orange line). The response unit differences have been normalized by using the ΔRUmax values estimated for each protein–protein complex. The KD values were calculated on the basis of measurements at various concentrations of IIBbgl.(C)Asin B but with IIBbgl(C24S) injected over the flow-cell.
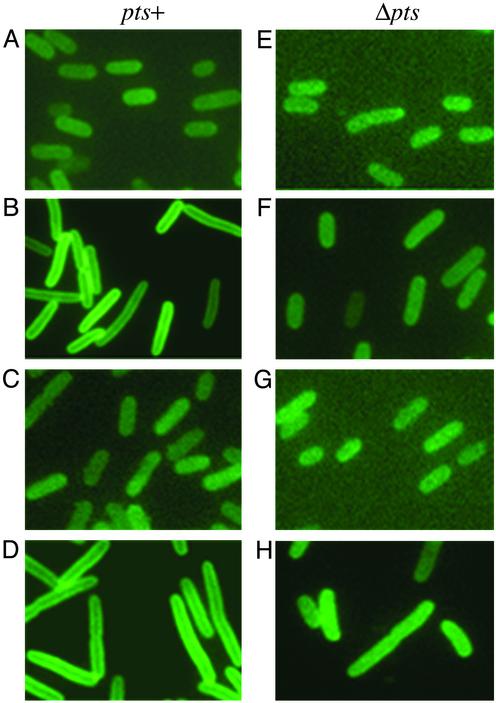
BglG Is Recruited to the Cell Membrane Only When BglF Is Expressed in the Cell and Is Released from the Membrane After Addition of the Stimulating Sugar. Does the affinity between the BglG transcriptional regulator and the BglF membrane-bound sensor lead to recruitment of BglG to the cell membrane? Previous results suggested that at least a fraction of the cellular BglG associates with the membrane (O.A.-C., unpublished data; ref. 23). Using fluorescence microscopy, we examined the subcellular localization of a BglG-GFP fusion. This fusion protein is fully functional as a transcriptional antiterminator (results not shown). As shown in Fig. 3A, BglG-GFP was evenly distributed in cells lacking BglF. In contrast, in cells producing BglF, BglG-GFP was observed mainly as a bright ring around the periphery of the cell (Fig. 3B). These results show that BglG-GFP is tethered to the cell membrane only in the presence BglF. When the soluble domain of BglF, IIBbgl, was expressed in the cells, instead of BglF, BglG-GFP was not seen associated with the membrane (Fig. 3C).
Fig. 3.
Recruitment of BglG to the membrane in cells expressing BglF. Fluorescence micrographs of E. coli MG1655 pts+ (A–D)or Δpts (E–H) cells expressing the BglG-GFP fusion and: no BglF derivative (A and E); WT BglF (B and F); IIBbgl (C and G); BglF(C24S) (D and H). Cells were grown in rich medium at 30°C. BglG-GFP was expressed from pJS185 without induction. Expression of BglF, IIBbgl and BglF(C24S), cloned in pBAD, was induced by arabinose (0.2%). cAMP (1 mM) was added twice during growth to the Δpts cells. Measurements of cell length (average of at least 100 cells): 3.492 μm(A); 6.139 μm(B); 4.12 μm(C); 6.588 μm (D); 3.618 μm(E); 5.17 μm(F); 3.36 μm(G); 5.16 μm(H).
In the experiment presented in Fig. 3, cells were grown in a rich medium. Cells overproducing BglF were longer than normal (Fig. 3, compare B with A and C), a phenomenon frequently observed in cells overproducing membrane proteins. When the experiment was repeated with cells grown in a minimal medium, BglG-GFP was also detected mainly at the periphery of BglF-producing cells (Fig. 4A). However, in this case, cell size was normal, presumably due to lower levels of BglF produced under these conditions. Hence, the subcellular localization of the BglG-GFP is not related to cell length. BglG-GFP in cells lacking BglF or producing IIBbgl that were grown in minimal medium was evenly distributed as in cells grown in rich medium (data not shown).
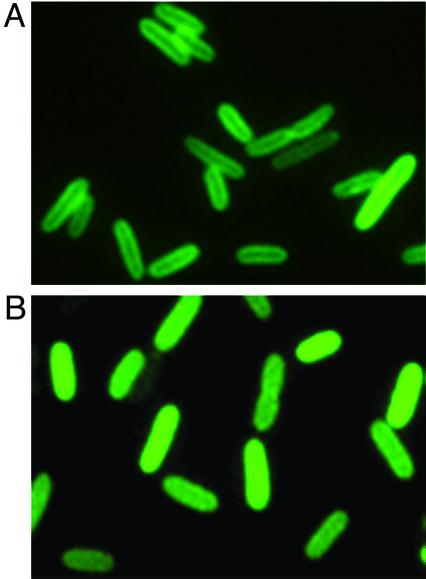
Fig. 4.
Release of BglG from the membrane after addition of the stimulating sugar. Shown are fluorescence micrographs of E. coli MG1655 cells expressing the BglG-GFP fusion and the BglF protein grown in minimal medium at 30°C. BglG-GFP was expressed from pJS185 without induction. BglF was expressed from pBAD18F, and its production was induced by arabinose (0.2%). Pictures were taken before (A) or 15 min after (B) the addition of arbutin. Measurements of cell length (average of at least 100 cells): 4.321 μm(A) and 5.354 μm(B).
Is BglG released to the cytoplasm on stimulation? To test this possibility, we grew cells in minimal medium containing glycerol as a sole carbon source and examined the subcellular localization of BglG-GFP at 5-min intervals for a period of 2 h after the addition of the β-glucoside arbutin to the medium. The fluorescent ring, observed at the cell periphery before arbutin addition (Fig. 4A), disappeared 15 min after arbutin addition, and the BglG-GFP became uniformly distributed within the cells (Fig. 4B) and remained so for the duration of the experiment. These results imply that BglG is released from the membrane to the cytoplasm shortly after addition of the stimulating sugar. To see whether BglG can relocalize to the membrane when the conditions are not in favor of β-glucoside utilization, we grew the cells in minimal medium and added arbutin for 30 min and then glucose for 30 more minutes. The fluorescent ring reappeared, indicating that BglG-GFP was retethered to the membrane (data not shown).
Taken together, the results obtained by fluorescence microscopy demonstrate that BglG is recruited to the cell membrane, provided that BglF is present in the membrane. BglG is released to the cytoplasm after stimulation of BglF with β-glucosides and stays there as long as BglF remains stimulated.
Requirements for BglF-BglG Binding and Recruitment of BglG to the Membrane. To study the requirements for the interaction between BglG and BglF, we examined the effects of mutations in BglG and BglF that impair phosphorylation on their interaction. In BglF, a mutation (C24S) in the active site prevents BglG phosphorylation and negative regulation (11). In BglG, a mutation (H208R) in the site that is phosphorylated by BglF prevents BglG regulation and phosphorylation by BglF (24). Two other mutations in BglG, D100N and H160Y, abolish negative regulation of BglG by BglF in vivo, and either severely reduce or completely abolish BglG phosphorylation by BglF in vitro, respectively (7). The three mutant BglG proteins are constitutively active in vivo.
Using the Far-Western technique, we found that the three mutant BglG proteins, each fused to MBP and immobilized on a nitrocellulose filter, gave a signal similar to that seen with WT MBP-BglG when incubated with His-IIBbgl and antibodies against the histidine tag (Fig. 1C). When a filter-immobilized WT MBP-BglG was probed with His-IIBbgl(C24S) and anti-His antibodies, a strong signal was observed (Fig. 1D, lane 1), comparable with the signal observed with His-tagged WT IIBbgl as a probe (Fig. 1A, lane 1). Hence, replacement of the phosphorylated residues on BglG and IIBbgl, as well as other residues in BglG that are required for its phosphorylation by BglF, does not seem to have an effect on BglG–IIBbgl interaction in vitro. However, when we tested the interaction between mutants of BglG and IIBbgl, both lacking their phosphorylation sites, i.e., filter immobilized MBP-BglG(H208R) probed with His-IIBbgl(C24S), a very weak signal was obtained (Fig. 1D, lane 2). It is important to mention that after purification, a fraction of the WT proteins is phosphorylated, as demonstrated by acrylamide-urea gel analysis (data not shown).
The effect of the above mutations on BglG-IIBbgl and BglG-BglF binding in vivo was examined using the LexA-based two-hybrid system. As shown in Tables 1 and 2, all four mutant proteins repressed transcription when expressed in combination with a WT partner protein, although the interaction of BglG(D100N) with IIBbgl or BglF and of BglG(H160Y) with BglF was somewhat reduced. However, the combination of IIBbgl(C24S) with BglG(H208R) reduced the interaction more profoundly (52% repression compared with 76% given by WT IIBbgl with WT BglG; Table 1). The combination of BglF(C24S) and either BglG(H208R) or BglG(H160Y) showed almost no interaction (14% and 13% repression, respectively). Hence, the results concerning the effect of the above mutations on the interaction between the studied proteins, obtained in vitro and in vivo, are in accord.
Can a mutant BglF protein that lacks the C24 phosphorylation site recruit WT BglG to the membrane? As shown in Fig. 3D, BglG-GFP was recruited to the membrane in cells expressing BglF(C24S).
Using SPR, we were able to quantify the effect of the mutations in BglG and BglF on BglG–BglF interaction. The three BglG mutant proteins were immobilized on the sensorchip, and IIBbgl, at various concentrations, was injected over these channels. A single sensorgram curve, obtained at 10 μM of IIBbgl, is presented for each channel (Fig. 2B). The binding profiles, the association and dissociation constants, and the KD values obtained for IIBbgl binding to the three BglG mutants were very similar to those obtained for IIBbgl binding to WT BglG (Fig. 2B).
A different binding profile was observed for the interaction between BglG and IIBbgl(C24S) (Fig. 2C, blue curve). The binding signal decayed rather rapidly, compared with the complex of WT IIBbgl and BglG, indicating that the BglG-IIBbgl(C24S) complex is less stable. Although kd and ka values could not be calculated accurately from the curve obtained with IIBbgl(C24S), curve simulations enabled prediction of values around 10-2·sec-1 for kd, and around 104·M-1·sec-1 for ka, compared with 10-4·sec-1 and 103·M-1·sec-1 for kd and ka of BglG-IIBbgl, respectively. These values suggest that IIBbgl(C24S) associates with and dissociates from BglG more rapidly than WT IIBbgl. Because both association and dissociation are faster, it is possible that at equilibrium the amount of IIBbgl(C24S) bound to BglG is comparable with the amount of IIBbgl. A visual comparison of the binding response obtained for BglG-IIBbgl and BglG-IIBbgl(C24S) shows a higher response level in the first case. Because the height of the plateau of the curves reflects the amount of the analyzed proteins bound to the BglG bearing channel, it is quite evident that the C24S mutation reduced the affinity toward BglG to 54% but did not abolish it (compare Fig. 2 A and B, blue curves). The binding profiles obtained for the interaction of IIBbgl(C24S) with the BglG mutants impaired in phosphorylation are similar to the profile of WT BglG with IIBbgl(C24S), i.e., fast association and dissociation. However, the affinity between IIBbgl(C24S) and the mutant BglG proteins, calculated on the basis of the height of the plateau of the curves, declined in comparison to the affinity between a pair of WT proteins or between a WT protein and a mutant protein: 33% for BglG(H160Y)-IIBbgl(C24S), 29% for BglG(D100N)-IIBbgl(C24S), and as low as 17% for BglG(H208R)-IIBbgl(C24S) (Fig. 2C, green, purple, and orange curves, respectively).
Our results imply that each individual phosphorylation site of the interacting proteins, BglG and BglF, is not crucial, by itself, for their binding. Yet, binding requires the presence of one phosphorylation site, either on BglG or on BglF, raising the possibility that the presence of a phosphoryl group on one of the interacting partners is essential for the binding.
We tested the significance of phosphorylation for BglG–BglF interaction by two experimental approaches. First, we tested the interaction between the Bgl proteins, WTs or mutants, in the LexA-based two-hybrid system in a Δpts background, in which the Bgl proteins are not phosphorylated. No interaction was observed in all cases, as indicated by the negligible or no repression (Table 2, compare values obtained in pts+ and Δpts isogenic strains).
Next, we tested the importance of phosphorylation for tethering BglG to the membrane. In a Δpts background, BglG-GFP was poorly, if at all, recruited to the membrane when coexpressed with WT BglF or with BglF(C24S), looking similar to its appearance in Δpts cells expressing IIBbgl or not expressing any BglF derivative (Fig. 3 E–H). The lack of binding between BglF and BglG, when both are nonphosphorylated, is in accord with the idea that when the sugar dephosphorylates BglF, the latter dephosphorylates BglG, releasing it to the cytoplasm to act as a transcriptional antiterminator.
Discussion
Signal transduction from ligand-activated membrane receptors to soluble proteins that function downstream in the signaling pathway usually occurs by phosphorylation, and it has been assumed that the contact is transient. How membrane-anchored receptors and soluble signaling proteins find each other rapidly on stimulation has been unclear. On the basis of theoretical calculations, it has been suggested that, to enhance signal transduction, cytoplasmic proteins should be recruited to the cell membrane by binding to membrane constituents, which need not be their cognate partners (25). It is easy to see why clusters of molecules in permanent association would be well suited to perform signaling. Solid-state interactions should be more rapid, efficient, and noise-free than systems of diffusing molecules in which at least one encounter is subject to the chaotic fluctuations of thermal energy (26). The involvement of multienzyme complexes in cellular processes, such as metabolism, macromolecular synthesis, and cell cycle progression, is established, but it is only recently that their roles in cell signaling have begun to be appreciated in both prokaryotes and eukaryotes (26, 27). Our study complements recent reports on strategies for rapid signaling that involve precomplex formation. The tumor necrosis factor receptors preassemble on the cell surface before ligand binding (28). The JAK tyrosine kinases preassemble with cytokine receptors (29). The bacterial CheA kinase preassembles with chemoreceptors through the adapter protein CheW (30). However, in all these cases, the soluble regulators are presumably recruited to the complex on activation. The results presented here demonstrate that the high affinity between the membrane-bound BglF sensor and the BglG transcription regulator leads to their preassembly at the membrane, abolishing the need to bring them together on stimulation.
The release of BglG from the membrane on stimulation is not absolutely required for its activity, although it presumably increases transcription efficiency. Bacterial DNA-binding proteins, such as ToxR in Vibrio species and CadC in E. coli, function as transcription activators despite their anchoring to the membrane (31, 32). Indeed, when BglG was artificially anchored to the membrane, it could function as a transcriptional antiterminator (23). Nevertheless, our results demonstrate that this it not the case with the native BglG. We show here that, after BglF stimulation by β-glucosides, BglG is released to the cytoplasm and stays there as long as BglF remains stimulated. Because interaction of BglF with the stimulating sugar changes the conformation of BglF (33), this change may induce not only BglG dephosphorylation but also BglF-BglG separation. The release of factors that are involved in transcription from the membrane, followed by their translocation to the transcription machinery, is achieved by various strategies, including activation-induced phosphorylation, e.g., STATs (29) and SMADs (34), and regulated transmembrane proteolysis, e.g., Notch-1, SREBP-1, APP, ErbB-4 (35) and the Bacillus subtilis σE protein (27). The stimulus-triggered release of BglG from the membrane suggests that prokaryotes and eukaryotes use similar strategies for signal transmission.
Our results shed light on the requirements for BglG–BglF interaction. Because the BglF active site, C24, phosphorylates both BglG and β-glucosides (11), we anticipated that recognition of BglG would not be mediated by this site. Indeed, as shown here, a mutation in C24 does not abolish BglF interaction with BglG, although it changes the kinetics of this interaction. This change in kinetics seems to account for the activity of BglG when expressed in the cell with BglF(C24S) (11). On BglG, the three residues that were shown to be essential for its phosphorylation by BglF (7, 24) are shown here not to be absolutely essential for the interaction with BglF, although mutating D100 and H160 somewhat reduces the interaction. However, mutating both BglF and BglG phosphorylation sites reduces (for IIBbgl) or almost abolishes (for BglF) the interaction. This suggests that the presence of a phosphate, or the negative charge that it confers, stabilizes BglF–BglG interaction. Phosphorylation of BglG in cells containing plasmid-encoded BglF(C24S) or IIBbgl(C24S) might be accomplished by HPr (14, 36) or by the chromosome-encoded BglF, shown to phosphorylate a small portion of BglG in WT E. coli strains that are bgl0 (8). The role of phosphorylation in BglF–BglG interaction was substantiated by the reduced or lack of interaction between BglF and BglG in a Δpts background, where neither protein is phosphorylated. Hence, drainage of the phosphate, required for a stable BglG–BglF complex, by the sugar is likely to contribute to the release of BglG from the membrane on BglF stimulation. The relative contribution of this drainage and of the sugar-induced conformational change in BglF (33) to the release of BglG to the cytoplasm is hard to estimate. It is important to mention that recruitment to the membrane of the BglG mutants, which cannot be phosphorylated by BglF, has not been demonstrated. We have shown only that they have a certain capacity to interact with the active site-containing domain of BglF when overproduced, indicating that each of these residues alone is not crucial for BglF–BglG interaction.
On the basis of theoretical models for molecular crowding and in vitro studies that measured phosphate flux through the glucose PTS enzymes under conditions that presumably mimic intramolecular conditions, it has been speculated that PTS enzymes form multiprotein complexes that transport PTS carbohydrates into the cell and phosphorylate them (37). Enzyme I of PTS was shown to localize to the inner surface of the cytoplasmic membrane (38). It remains to be seen whether BglG and its homologues are part of these PTS complexes.
Binding of a transcription repressor, Mlc, to the membrane-bound Enzyme II of glucose, PstG, has been reported (39, 40). However, unlike BglG and BglF, Mlc and PstG do not preassemble in the absence of stimulus. On the contrary, Mlc does not bind to the phosphorylated nonstimulated PstG. It binds to PstG only after dephosphorylation of PstG by glucose. The purpose suggested for Mlc-PstG binding is membrane sequestration of Mlc to prevent it from binding to its operator. Hence, binding of Mlc to PstG occurs in response to an environmental change and cannot play a role in increasing the efficiency or the speed of initiation of signal transduction.
Most of the information gathered on signaling pathways was obtained from biochemical studies that focused mainly on the chemical changes that the signaling proteins undergo. However, to gain better understanding of signaling processes, more information on the subcellular localization of the participating proteins, their diffusion rates, and the kinetics of their interaction is needed. Preassembly of kinases and their cognate substrates seems suitable for sensory systems that control a single process and produce one type of response, as opposed to systems that produce multiple outputs, and thus require divergent circuitry that relies on recruitment of various combinations of proteins to transient complexes. Many of the bacterial two-component sensory systems, composed of histidine kinase-response regulator pairs (41), fall into this category. Indeed, interaction of the phosphoryl transfer domain of the histidine kinase UhpB with the response regulator UhpA was suggested on the basis of indirect evidence (42). Probable recognition surfaces on several response regulators for their cognate histidine kinases have also been identified (e.g., refs. 43–45). The stability and the kinetics of these interactions remain to be studied. The preassembly of the Bgl proteins may prove to be a useful paradigm for other sensory systems that control a single process and need to respond rapidly to extracellular stimuli.
Supplementary Material
Acknowledgments
We thank Dr. Uwe Stroeher for advice on the Lex-based two-hybrid system, Liat Fux for providing plasmid pLFHG, and Dr. Jatinder Singh for providing plasmid pJS185. We thank Dr. Susana Shochat for excellent technical help with the BIAcore technology. We acknowledge Grace Leung for help with fluorescence microscopy. We appreciate helpful discussions with Galia Monderer-Rothkoff, Liat Fux, and Sharon Yagur-Kroll. We thank Dr. Abraham L. Sonenshein for critical reading of the manuscript. This research was supported by Grant 1999344 from the U.S.–Israel Binational Science Foundation (BSF) and by the Israel Science Foundation founded by the Israel Academy of Sciences and Humanities.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: PTS, phosphotransferase system; SPR, surface plasmon resonance; MBP, maltose-binding protein; LexADBD, LexA repressor DNA-binding domain.
References
- 1.Mahadevan, S., Reynolds, A. E. & Wright, A. (1987) J. Bacteriol. 169, 2570-2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schnetz, K., Toloczyki, C. & Rak, B. (1987) J. Bacteriol. 169, 2579-2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mahadevan, S. & Wright, A. (1987) Cell 50, 485-494. [DOI] [PubMed] [Google Scholar]
- 4.Schnetz, K. & Rak, B. (1988) EMBO J. 7, 3271-3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Houman, F., Diaz-Torres, M. R. & Wright, A. (1990) Cell 62, 1153-1163. [DOI] [PubMed] [Google Scholar]
- 6.Nussbaum-Shochat, A. & Amster-Choder, O. (1999) Proc. Natl. Acad. Sci. USA 96, 4339-4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amster-Choder, O., Houman, F. & Wright, A. (1989) Cell 58, 847-855. [DOI] [PubMed] [Google Scholar]
- 8.Amster-Choder, O. & Wright, A. (1990) Science 249, 540-542. [DOI] [PubMed] [Google Scholar]
- 9.Schnetz, K. & Rak, B. (1990) Proc. Natl. Acad. Sci. USA 87, 5074-5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amster-Choder, O. & Wright, A. (1992) Science 257, 1395-1398. [DOI] [PubMed] [Google Scholar]
- 11.Chen, Q., Arents, J. C., Bader, R., Postma, P. & Amster-Choder, O. (1997) EMBO J. 16, 4617-4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, Q., Postma, P. W. & Amster-Choder, O. (2000) J. Bacteriol. 182, 2033-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stulke, J., Arnaud, M., Rapoport, G. & Martin-Verstraete, I. (1998) Mol. Microbiol. 28, 865-874. [DOI] [PubMed] [Google Scholar]
- 14.van Tilbeurgh, H. & Declerck, N. (2001) Curr. Opin. Struct. Biol. 11, 685-693. [DOI] [PubMed] [Google Scholar]
- 15.Dmitrova, M., Younes-Cauet, G., Oertel-Buchheit, P., Porte, D., Shnarr, M. & Granger-Shnarr, M. (1998) Mol. Gen. Genet. 257, 205-212. [DOI] [PubMed] [Google Scholar]
- 16.Levy, S., Zeng, G.-Q. & Danchin, A. (1990) Gene 86, 27-33. [DOI] [PubMed] [Google Scholar]
- 17.Center, R. J., Kobe, B., Wilson, K. A., Teh, T., Howlett, G. J., Kemp, B. E. & Poumborios, P. (1998) Protein Sci. 7, 1612-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guzman, L.-M., Belin, D., Carson, M. J. & Beckwith, J. (1995) J. Bacteriol. 177, 4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller, J. H. (1972) Experiments in Molecular Genetics (Cold Spring Harbor Lab. Press, Plainview, NY).
- 20.Chen, Q. & Amster-Choder, O. (1999) J. Bacteriol. 181, 462-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnsson, B., Lofas, S. & Lindquist, G. (1991) Anal. Biochem. 198, 268-277. [DOI] [PubMed] [Google Scholar]
- 22.Karimova, G., Pidoux, J., Ullmann, A. & Ladant, D. (1998) Proc. Natl. Acad. Sci. USA 95, 5752-5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gorke, B. & Rak, B. (2001) J. Mol. Biol. 308, 131-145. [DOI] [PubMed] [Google Scholar]
- 24.Chen, Q., Engelberg-Kulka, H. & Amster-Choder, O. (1997) J. Biol. Chem. 272, 17263-17268. [DOI] [PubMed] [Google Scholar]
- 25.Kholodenko, B., Hoek, J. & Westerhoff, H. (2000) Trends Cell Biol. 10, 173-178. [DOI] [PubMed] [Google Scholar]
- 26.Bray, D. (1998) Annu. Rev. Biophys. Biomol. Struct. 27, 59-75. [DOI] [PubMed] [Google Scholar]
- 27.Shapiro, L. & Losick, R. (2000) Cell 100, 89-98. [DOI] [PubMed] [Google Scholar]
- 28.Locksley, R., Killeen, N. & Lenardo, M. (2001) Cell 104, 487-501. [DOI] [PubMed] [Google Scholar]
- 29.Kisseleva, T., Bhattacharya, S., Braunstein, J. & Schindler, C. W. (2002) Gene 285, 1-24. [DOI] [PubMed] [Google Scholar]
- 30.Stock, J. B., Levit, M. N. & Wolanin, P. M. (2002) Sci. STKE 132, PE25. [DOI] [PubMed] [Google Scholar]
- 31.DiRita, V. (1992) Mol. Microbiol. 6, 451-458. [DOI] [PubMed] [Google Scholar]
- 32.Dell, C., Neely, M. & Olson, E. (1994) Mol. Microbiol. 14, 7-16. [DOI] [PubMed] [Google Scholar]
- 33.Chen, Q., Nussbaum-Shochat, A. & Amster-Choder, O. (2001) J. Biol. Chem. 276, 44751-44756. [DOI] [PubMed] [Google Scholar]
- 34.Attisano, L. & Wrana, J. L. (2002) Science 296, 1646-1647. [DOI] [PubMed] [Google Scholar]
- 35.Ebinu, J. O. & Yankner, B. A. (2002) Neuron 34, 499-502. [DOI] [PubMed] [Google Scholar]
- 36.Gorke, B. & Rak, B. (1999) EMBO J. 18, 3370-3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rohwer, J., Postma, P., Kholodenko, B. & Westerhoff, H. (1998) Proc. Natl. Acad. Sci. USA 95, 10547-10552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ghosh, B., Owens, K., Pietri, R. & Peterkofsky, A. (1989) Proc. Natl. Acad. Sci. USA 86, 849-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee, S., Boos, W., Bouche, J. & Plumbridge, J. (2000) EMBO J. 19, 5353-5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tanaka, Y., Kimata, K. & Aiba, H. (2000) EMBO J. 19, 5344-5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stock, A., Robinson, V. & Goudreau, P. (2000) Annu. Rev. Biochem. 69, 183-215. [DOI] [PubMed] [Google Scholar]
- 42.Wright, J. I. & Kadner, R. (2001) J. Bacteriol. 183, 3149-3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Swanson, R., Lowry, D., Matsumura, P., McEnvoy, M., Simon, I. & Dahlquist, F. (1995) Nat. Struct. Biol. 2, 906-910. [DOI] [PubMed] [Google Scholar]
- 44.Haldimann, A., Prahalad, M., Fisher, S., Kim, S.-K., Walsh, C. & Wanner, B. (1996) Proc. Natl. Acad. Sci. USA 93, 14361-14366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tzeng, Y.-L. & Hoch, J. (1997) J. Mol. Biol. 272, 200-212. [DOI] [PubMed] [Google Scholar]
- 46.Karimova, G., Pidoux, J., Ullman, A. & Ladont, D. (1998) Proc. Natl. Sci. USA 95, 5752-5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.