Abstract
Skeletal muscle adapts to chronic physical activity by inducing mitochondrial biogenesis and switching proportions of muscle fibers from type II to type I. Several major factors involved in this process have been identified, such as the calcium/calmodulin-dependent protein kinase IV (CaMKIV), calcineurin A (CnA), and the transcriptional component peroxisome proliferator-activated receptor γ coactivator 1α (PGC-1α). Transgenic expression of PGC-1α recently has been shown to dramatically increase the content of type I muscle fibers in skeletal muscle, but the relationship between PGC-1α expression and the key components in calcium signaling is not clear. In this report, we show that the PGC-1α promoter is regulated by both CaMKIV and CnA activity. CaMKIV activates PGC-1α largely through the binding of cAMP response element-binding protein to the PGC-1α promoter. Moreover, we show that a positive feedback loop exists between PGC-1α and members of the myocyte enhancer factor 2 (MEF2) family of transcription factors. MEF2s bind to the PGC-1α promoter and activate it, predominantly when coactivated by PGC-1α. MEF2 activity is stimulated further by CnA signaling. These findings imply a unified pathway, integrating key regulators of calcium signaling with the transcriptional switch PGC-1α. Furthermore, these data suggest an autofeedback loop whereby the calcium-signaling pathway may result in a stable induction of PGC-1α, contributing to the relatively stable nature of muscle fiber-type determination.
Skeletal muscle contains myofibers that differ greatly in their oxidative capacity. Prolonged electrical stimulation or exercise training can lead to a conversion of type II (fast-twitch) muscle fibers to type I (slow-twitch) fibers (1). This conversion is characterized by dramatic changes in the expression of a large number of genes that increase mitochondrial biogenesis, oxidative capacity, and amounts of a distinct set of contractile proteins characteristic of this fiber type (2). Several key factors in this signaling cascade have been identified recently. Exercise training is accompanied by an increase in motor nerve activity that subsequently elevates intracellular calcium levels in the muscle (3, 4). Calcium and the calcium-binding protein calmodulin activate both the calcium/calmodulin-dependent protein kinase IV (CaMKIV) and the protein phosphatase calcineurin A (CnA), as well as many other factors (2). Activated CaMKIV catalyzes protein phosphorylation events that result in release of the myocyte enhancer factor (MEF)2 from an inhibitory complex including the histone deacetylases HDAC1/2 and HDAC4/5, the repressor Cabin-1, and the adapter mSin3 (5). After phosphorylation by CaMKIV, these factors are exported from the nucleus; as a consequence, the MEF2s become transcriptionally active and bind coactivator proteins including CBP/p300 or the peroxisome proliferator-activated receptor γ coactivator 1α (PGC-1α) (6, 7).
In another arm of the calcium-signaling pathway, activated CnA dephosphorylates members of the nuclear factor of activated T cell (NFAT) family, thereby stimulating a cytoplasmic– nuclear translocation of these proteins (3). Activated CnA provides a further boost to this process by dephosphorylating MEF and enhancing its transcriptional activity (8). The combined actions of MEF2s and NFATs in the nucleus increase the transcription of prototypical type I muscle fiber genes and, thus, promote fiber-type switching from type II to type I muscle (9). Important experimental evidence for this general model has come from transgenic mice that express either active CnA or CaMKIV (10, 11). In these mice, the relative amount of type I muscle fibers was increased greatly compared with wild-type animals.
PGC-1α originally was cloned as a regulator of several aspects of adaptive thermogenesis in brown adipose tissue (12) and subsequently has been found to coactivate a variety of nuclear receptors and other transcription factors (13). PGC-1α also was shown to induce mitochondrial biogenesis and greatly increase respiration when expressed in cultured fat, skeletal, or cardiac muscle cells (14, 15). We recently have discovered that transgenic expression of PGC-1α driven by a muscle-specific promoter results in a dramatic increase of type I muscle fibers (16). The PGC-1α-expressing mice showed increased expression of myofibrillar proteins characteristic of type I fibers, a higher expression of components of the mitochondrial electron transport system, and greater resistance to fatigue than wild-type mice. MEF2 proteins, especially MEF2C and MEF2D, seem to be important targets of PGC-1α in the coactivation of several type I-selective promoters. Intriguingly, several recent studies show that exercise and physical activity, as well as increased calcium concentrations, elevate PGC-1α levels (17–20), although it is not clear how this regulation occurs.
In this report, the control of PGC-1α expression by CaMKIV and CnA in muscle cells is investigated. We show that the PGC-1α promoter is subject to positive regulation by these key calcium-signaling factors. In addition, PGC-1α is demonstrated to regulate its own promoter through interactions with components of the calcium-signaling pathway, resulting in an autoregulatory loop that potentially can provide a certain stability to the expression of genes characteristic of type I muscle fibers.
Methods
Plasmids and Reagents. Various fragments of the 5′ flanking sequence of the mouse PGC-1α gene were amplified by PCR and subcloned into the pGL3basic reporter gene vector (Promega). Thus, the plasmids containing the regions between +78 and -2,533 or -6,483 with respect to the transcriptional start site are referred to as 2- and 6-kb promoters, respectively. Expression plasmids for MEF2C, the dominant-negative MEF2C KR23,24ID (dnMEF2C), MEF2D, NFATc3, constitutively active CaMKIV, and constitutively active CnA were kind gifts from Eric N. Olson (University of Texas Southwestern Medical Center, Dallas). The dominant-negative cAMP response element (CRE)-binding protein (CREB), termed ACREB, was generously provided by Charles Vinson (National Cancer Institute, National Institutes of Health, Bethesda). All other reagents were obtained from Sigma.
Site-Directed Mutagenesis. Site-directed mutagenesis was performed as described (21). Briefly, PCR amplifications were performed by using overlapping primers at the target sites, and the inserts were subcloned into new reporter gene vectors. The CRE and the MEF-binding site were termed ΔCRE and ΔMEF2, respectively.
Cell Culture, Transfection, and Reporter Gene Assays. C2C12 cells were maintained in DMEM supplemented with 10% FCS and 1 μM sodium-pyruvate in subconfluent cultures. For myotube differentiation, confluent C2C12 myoblasts were cultured in differentiation medium (DMEM with 2% horse serum and 1 μM sodium-pyruvate) for 5 days with daily changes of the medium. Cells were transfected by using Lipofectamine 2000 transfection reagent (Invitrogen), and luciferase activity was determined 48 h after transfection. Expression of reporter genes was normalized to β-galactosidase levels driven by a cotransfected pSV-β-galactosidase expression vector (Promega). Finally, these relative expressions were normalized vs. empty pGL3basic reporter gene vector expression.
Electrophoretic Mobility-Shift Assays. The sequences of the MEF2 oligonucleotide probe were derived from the mouse PGC-1α promoter and consisted of the MEF2 site and the mutated ΔMEF2 site flanked by 20 bp on each side. Radiolabeling, incubation with proteins, and gel electrophoresis were performed as described (22). PGC-1α protein at concentrations of 0.8, 2, 4, and 6 μg was added as a bacterially expressed GST-PGC-1α fusion protein encoding for amino acids 1–180 or 31–797 of PGC-1α, respectively. Antibodies against MEF2 and HNF3β (used as a nonspecific antibody) were from Santa Cruz Biotechnology.
Analysis of PGC-1α Gene Expression in Wild-Type and Transgenic PGC-1α Mice. Wild-type and transgenic mice from strain 31 (see ref. 16) that express PGC-1α in muscle were killed, and plantaris muscle was collected. Total RNA was isolated and subsequently reverse-transcribed. Primers for the ABI Prism 7700 sequence detector (Applied Biosystems) were designed for the respective target genes. Transcript levels were determined from at least three wild-type and transgenic mice and subsequently normalized to 18S rRNA levels.
Adenoviral Generation and Infections. Adenoviral constructs containing GFP, PGC-1α, MEF2C, and dnMEF2C were generated by using the Ad-Easy system as published (23). C2C12 cells were infected with adenovirus; 1 day postinfection, medium was changed to differentiation medium and cells were harvested after 5 days. Total RNA was isolated and gene expression levels were determined with real-time PCR.
Results
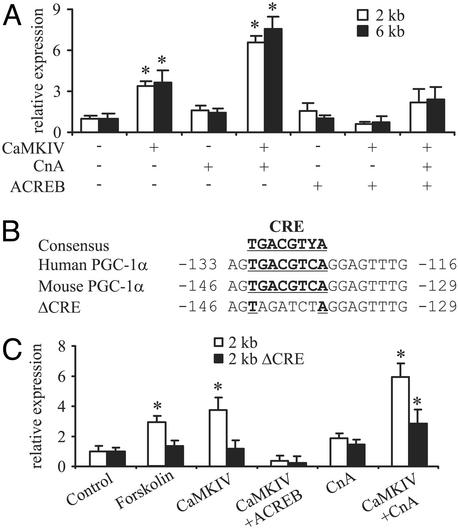
Because of the potential connections between PGC-1α gene expression and components of the calcium-signaling system, we first analyzed the PGC-1α promoter in terms of responsiveness to CaMKIV and CnA. Promoter fragments that contain either 2 or 6 kb of 5′ flanking region of mouse PGC-1α both are activated at least 3-fold by the expression of constitutively active CaMKIV (Fig. 1A). In contrast, expression of a constitutively active CnA has very little effect on reporter gene activity, consistent with the results obtained previously for PGC-1α mRNA expression in CaMKIV and CnA transgenic mice (11). However, the combination of CaMKIV and CnA has a powerful effect on the PGC-1α promoter, increasing transcription by ≈7-fold (Fig. 1 A).
Fig. 1.
Induction of PGC-1α by CaMKIV via CREB. (A) CaMKIV activation of PGC-1α is abolished by the dominant-negative ACREB. C2C12 cells were cotransfected with expression plasmids for CnA, CaMKIV, and ACREB together with reporter gene plasmids containing different fragments of the mouse PGC-1α promoter. After 48 h, cells were harvested and luciferase activity was determined. (B) Sequence of the CRE (depicted in bold and underlined) in the mouse PGC-1α promoter. Y = C or T.(C) Site-directed mutagenesis of the CRE inhibits CaMKIV-mediated activation of PGC-1α. C2C12 cells were cotransfected with expression plasmids for CnA, CaMKIV, and ACREB together with reporter gene plasmids containing 2 kb of wild-type PGC-1α promoter or a promoter with a mutation in the CRE site (ΔCRE), respectively. Cells subsequently were treated with either vehicle (0.1% DMSO) or 100 μM forskolin for 10 h and harvested 48 h after transfection before luciferase activity was determined. The values represent the average of three independent experiments, and bars represent SD. *, P < 0.5 between different treatments compared with the untreated control.
CaMKIV has been shown to phosphorylate and activate many proteins, including CREB. Because CREB has been shown to be an important activator of PGC-1α gene expression in the fasted liver (24), we used the dominant-negative ACREB protein to examine a potential role for CREB in the CaMKIV-mediated control of the PGC-1α promoter. Although ACREB alone had no effect on the PGC-1α promoter, this protein severely reduced the activation of the PGC-1α promoter by CaMKIV alone or CaMKIV in combination with CnA (Fig. 1 A).
The human PGC-1α promoter contains a CRE between -133 and -116 that is crucial for PGC-1α induction by cAMP in the liver (24). Similarly, a very conserved putative CRE can be identified in the mouse PGC-1α promoter at approximately the same distance (between -146 and -129) from the transcriptional start site (Fig. 1B). The functional role of this CRE was tested by using a mutated promoter (Fig. 1B), followed by a 10-h stimulation of cells with the cAMP-stimulating agent forskolin. As shown in Fig. 1C, mutagenesis of the CRE site abolished induction of the mouse PGC-1α promoter by forskolin, as expected. Importantly, expression of the ΔCRE PGC-1α promoter also showed a dramatically impaired response to CaMKIV alone or the combination of CaMKIV and CnA (Fig. 1C), indicating a key role for CREB in the induction of PGC-1α expression by these mediators of calcium signaling.
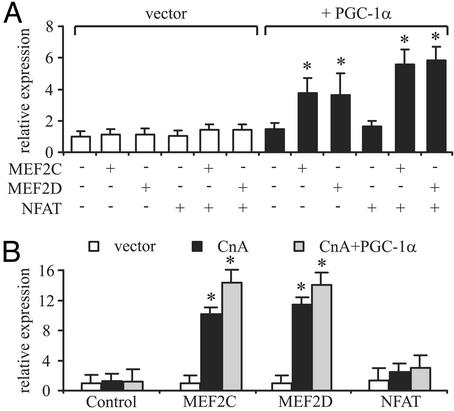
Although CREB thus seems to be an important factor in the induction of PGC-1α, the increased effect of CaMKIV in combination with CnA suggests that factors in addition to CREB are likely to be involved in the transcription of the PGC-1α gene in muscle. Because MEF2 and NFAT transcription factors are known targets of CaMKIV and CnA in muscle fiber-type determination, we tested the role of these factors in the control of the PGC-1α promoter. MEF2C, MEF2D, or NFATc3 alone did not have a significant effect on the function of the 2-kb PGC-1α promoter (Fig. 2A). However, because MEF2 proteins are known to be coactivated by PGC-1α (7), we also tested these factors in cotransfection experiments. As shown in Fig. 2 A, a coactivation by PGC-1α of both MEF2C and MEF2D, but not NFAT, was observed on the PGC-1α promoter. These data suggest that PGC-1α might participate in the activation of its own promoter and that the MEF2 proteins may function both in the regulation of PGC-1α expression and as downstream targets of PGC-1α coactivation of other muscle-selective genes.
Fig. 2.
PGC-1α coactivates MEF2s on the PGC-1α promoter. (A) MEF2C and MEF2D activate the mouse PGC-1α promoter. C2C12 cells were cotransfected with expression plasmids for MEF2C, MEF2D, NFAT, and PGC-1α together with reporter gene plasmids containing 2 kb of the mouse PGC-1α promoter. After 48 h, cells were harvested and luciferase activity was determined. (B) MEF2 activity is increased by CnA. C2C12 cells were cotransfected with expression plasmids for MEF2C, MEF2D, NFAT, CnA, and PGC-1α together with reporter gene plasmids containing 6 kb of the mouse PGC-1α promoter. After 48 h, cells were harvested and luciferase activity was determined. The values represent the average of three independent experiments, and bars represent SD. *, P < 0.5 between different treatments compared with the untreated control.
The transcriptional activities of both MEF2 and NFAT are known to be increased by CaMKIV- and CnA-mediated changes in phosphorylation status (3, 6). As shown in Fig. 2B, CnA is able to substantially increase the activity of MEF2C and MEF2D on the PGC-1α promoter. The strongest effect was observed when MEF2C or MEF2D was expressed with both CnA and PGC-1α (Fig. 2B). No effect was found by the coexpression of any of these proteins with NFAT. In contrast to the data for CnA, the effect of CaMKIV on this promoter was not changed by the addition of MEF2s or PGC-1α (data not shown). Together, these data strongly suggest that the major effect of CaMKIV on PGC-1α expression is via CREB acting on the PGC-1α promoter, whereas CnA is able to increase further the activity of MEF2s to stimulate transcription driven by the PGC-1α promoter.
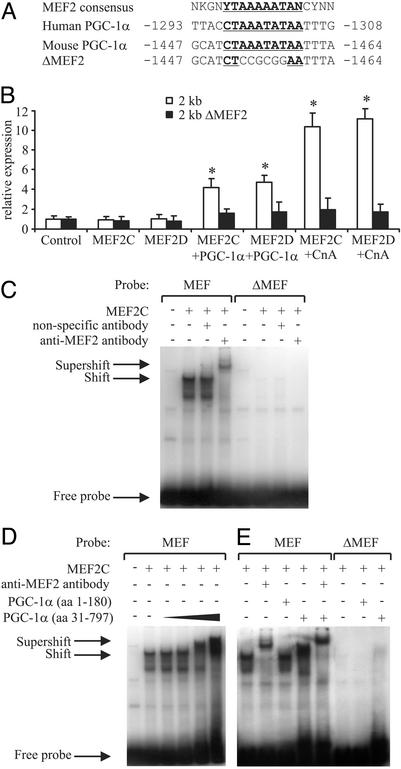
Computer-aided sequence analysis of the mouse PGC-1α 5′ flanking region revealed a potential MEF2-binding site between -1,464 and -1,447 (Fig. 3A) and a possible NFAT-binding site between -1,547 and -1,536 (25). Similar configurations of adjacent MEF2- and NFAT-binding sites previously have been described in several muscle fiber type I-specific promoters (9). We next examined whether mutations of this MEF2 site affects MEF2 activity on the PGC-1α promoter. The mutated 2-kb promoter (called ΔMEF2; see Fig. 3A) is no longer able to mediate MEF2C or MEF2D induction alone, when activated by CnA or coactivated by PGC-1α, suggesting that this site is responsible for the MEF2 action (Fig. 3B). Moreover, electrophoretic mobility-shift assays revealed binding of MEF2C to this MEF-binding site but not the mutated ΔMEF site in the PGC-1α promoter (Fig. 3C). The identity of the MEF2C–DNA complex was confirmed by using an anti-MEF2 antibody that leads to a supershift of the DNA–protein complex.
Fig. 3.
MEF2C and MEF2D activate the PGC-1α promoter via a conserved binding site. (A) Identification of putative MEF2-binding site (depicted in bold and underlined) in the human and mouse PGC-1α promoter. N = A, T, C, or G; K = G or T; Y = C or T. (B) MEF2C and MEF2D activate an MEF-binding site in the PGC-1α promoter. C2C12 cells were cotransfected with expression plasmids for MEF2C, MEF2D, CnA, and PGC-1α together with reporter gene plasmids containing 2 kb of wild-type mouse PGC-1α promoter or 2 kb of the promoter with a mutation in the MEF2-binding site (ΔMEF2). After 48 h, cells were harvested and luciferase activity was determined. (C) MEF2C binds to the MEF-binding site in the PGC-1α promoter. Electrophoretic mobility-shift assays were performed by using in vitro transcribed/translated MEF2C and radiolabeled probe encoding the MEF-binding site from the PGC-1α promoter or the mutated binding site, ΔMEF. Antibodies against HNF3β and MEF2 were used to test for the specificity of the protein–DNA interactions. (D and E) PGC-1α interacts with MEF2C on the MEF-binding site. Increasing amounts of PGC-1α protein including amino acids 31–797 (0.8, 2, 4, and 6 μg, respectively) were added to the reactions, resulting in a larger protein–DNA complex and thus a supershift (D). As a control, 6 μg of PGC-1α protein including amino acids 1–180 that lacks the MEF2–interaction domain was not able to interact with MEF2C (E). The protein–DNA complex of PGC-1α, MEF2C, and the MEF-binding site could be supershifted further when adding anti-MEF2 antibody. The values represent the average of three independent experiments, and bars represent SD. *, P < 0.5 between different treatments compared with the untreated control.
To confirm that the coactivation of MEF2C by PGC-1α observed in reporter gene assays (Fig. 3B) was linked to direct binding of these two proteins, we tested whether PGC-1α directly interacts with MEF2C on this MEF-binding site. Increasing amounts of PGC-1α protein (amino acids 31–797) decreased the mobility of the complex containing MEF2C bound to the MEF-binding site, as visualized by a supershift in electrophoretic mobility-shift assays (Fig. 3D). As a control, PGC-1α protein that lacks the MEF2C-interaction domain (amino acids 1–180; see ref. 7) was not able to bind to MEF2C. Inclusion of an MEF2-specific antibody was able to supershift further the protein–DNA complex containing MEF2C, PGC-1α, and the MEF2-binding site (Fig. 3E). Neither a shift nor a supershift could be obtained when using the mutated ΔMEF site. These results indicate that MEF2s bind to the PGC-1α promoter and that PGC-1α coactivates MEF2 proteins on the PGC-1α promoter by a direct protein–protein interaction.
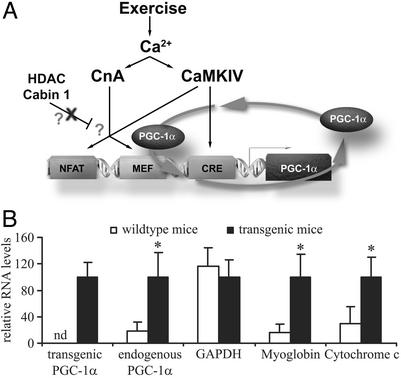
The ability of PGC-1α protein to stimulate the PGC-1α promoter via coactivation of the MEF2 proteins implies a potential autoregulatory loop. As shown in the model illustrated in Fig. 4A, exercise or increased motor neuron activity would result in the activation of CaMKIV and CnA. This, in turn, would lead to activation of the CREB and MEF2 proteins, which then would bind to and potentially activate the PGC-1α promoter. Of course, MEF2 bound to the PGC-1α promoter is also a target for PGC-1α coactivation, resulting in a stable, feed-forward regulatory loop. A key feature of this model is that PGC-1α should be a regulator of its own expression in skeletal muscle. To critically test this hypothesis, we examined whether ectopic PGC-1α expressed in the muscle of transgenic mice activates expression of the endogenous PGC-1α gene. Real-time PCR primers were designed for the PGC-1α 3′ untranslated region that allows for determination of the levels of ectopically expressed and endogenous PGC-1α. As shown in Fig. 4B and as expected, ectopic PGC-1α is detected only in the muscle of transgenic mice. Strikingly, a 7-fold elevation of endogenous PGC-1α mRNA was seen in the transgenic animals as compared with wild-type mice (Fig. 4B). A robust increase in the transcript levels of cytochrome c and myoglobin also was observed in the RNA isolated from skeletal muscle of the transgenic mice, whereas GAPDH transcript levels remained unchanged (Fig. 4B). These results indicate a regulation of PGC-1α transcription by PGC-1α protein in an apparent autoregulatory loop.
Fig. 4.
A model for the autoregulatory loop regulating the PGC-1α promoter in muscle fiber-type determination. (A) Exercise and subsequently elevated intracellular calcium levels result in an activation of both CaMKIV and CnA in skeletal muscle. Activated CaMKIV can phosphorylate CREB, which then increases transcription of PGC-1α via a conserved CREB-binding site in the proximal promoter. Moreover, CaMKIV and CnA activate the transcriptional activity of MEF2s in part by promoting the dissociation of inhibitory HDACs and Cabin1. MEF2s, potentially in combination with NFAT, bind to at least one MEF2-binding site in the PGC-1α flanking region and increase transcriptional activity. Newly synthesized PGC-1α protein can coactivate MEF2s and thus positively regulate its own transcription. PGC-1α also might compete with the inhibitory HDACs and Cabin 1 for binding to MEF2s. Together, this positive feedback loop may ensure a stable transcription of PGC-1α, leading to muscle fiber-type I determination. (B) Endogenous PGC-1α expression is increased in transgenic mice expressing ectopic PGC-1α in skeletal muscle. Total RNA from wild-type and transgenic skeletal muscle was analyzed for the expression of transgenic and endogenous PGC-1α, GAPDH, myoglobin, and cytochrome c by using real-time PCR. Relative mRNA expression levels were normalized to 18S rRNA levels. nd, not detectable. The values represent the average of three independent experiments, and bars represent SD. *, P < 0.5 between different treatments compared with the untreated control.
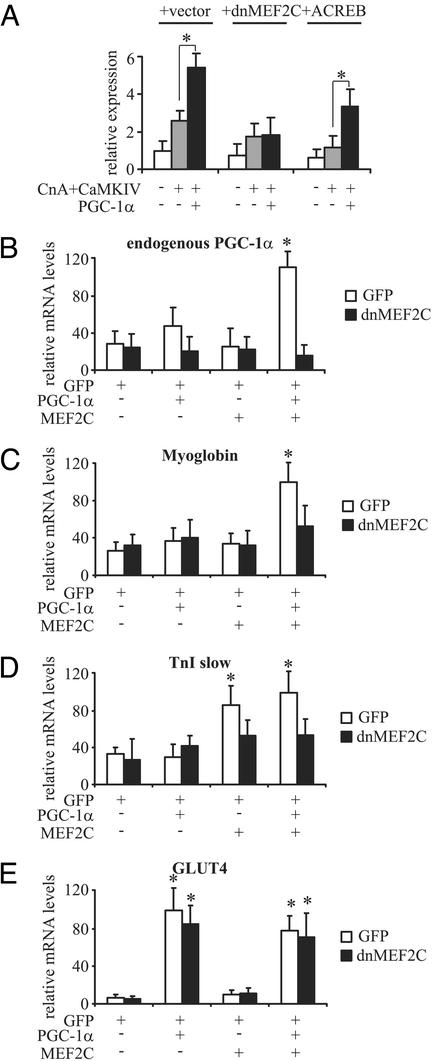
As a second critical test of our proposed model, we examined whether inhibiting MEF2 and/or CREB activity in transfections can decrease the transcription of the PGC-1α promoter stimulated by CaMKIV, CnA, and PGC-1α. To do this, we used a previously characterized dominant-negative allele of MEF2C in addition to ACREB (26). As shown in Fig. 5A, dnMEF2C reduces the effects of the combination of CnA and CaMKIV, and completely eliminates the effect of PGC-1α on the PGC-1α promoter. In contrast, the dominant-negative allele of CREB (ACREB) reduces the activation by CnA and CaMKIV but does not affect the relative fold activation by PGC-1α.
Fig. 5.
PGC-1α autoregulation can be inhibited by dnMEF2C. (A) dnMEF2C completely blocks the activation of the PGC-1α promoter by PGC-1α. C2C12 cells were cotransfected with expression plasmids for CaMKIV, CnA, ACREB, dnMEF2C, and PGC-1α together with a reporter gene plasmid containing 2 kb of the mouse PGC-1α promoter. After 48 h, cells were harvested and luciferase activity was determined. (B–E) dnMEF2C adenovirus blocks induction of PGC-1α mRNA. C2C12 myoblasts were infected with adenovirus containing GFP, PGC-1α, MEF2C, dnMEF2C, or combinations thereof. After 5 days of differentiation into myotubes, cells were harvested and mRNA levels of endogenous PGC-1α (B), myoglobin (C), troponin I slow (TnI slow, D), and the glucose transporter GLUT4 (D) were determined. Relative expression levels were normalized to 18S rRNA levels. nd, not detectable. The values represent the average of at least three independent experiments, and bars represent SD. *, P < 0.5 between different treatments compared with the untreated control unless otherwise indicated.
The effects of MEF2C, PGC-1α, and dnMEF2C on the expression of endogenous PGC-1α mRNA and on other fiber type-I specific genes also were examined. C2C12 cells were infected with adenoviruses encoding GFP, PGC-1α, MEF2C, and dnMEF2C in various combinations. Subsequently, gene expression levels were determined by using real-time PCR. Levels of endogenous PGC-1α mRNA were elevated only slightly in cells infected with PGC-1α alone, and no induction was observed when MEF2C alone was expressed (Fig. 5B). In contrast, a large induction of endogenous PGC-1α mRNA (4-fold) was seen in cells that received a combination of PGC-1α and MEF2C. The induction of endogenous PGC-1α mRNA could be blocked completely by coexpression of dnMEF2C. In all cases, similar amounts of the virally expressed PGC-1α and MEF2C mRNAs were observed (data not shown). Expression of myoglobin in C2C12 myotubes also required infection of both PGC-1α and MEF2C (Fig. 5C). Interestingly, induction of two other genes that are highly expressed in type I muscle fibers, troponin I slow and the glucose transporter GLUT4, is not dependent on the combined expression of PGC-1α and MEF2C. Troponin I slow is induced after infection with virally encoded MEF2C, and the addition of PGC-1α has no further effect (Fig. 5D). In contrast, GLUT4 transcript levels can be elevated by PGC-1α alone, independent of MEF2C infection, consistent with previous reports (7) (Fig. 5E). These data indicate that there is an autoregulatory loop whereby PGC-1α expression depends on the action of MEF2 and PGC-1α proteins on the PGC-1α promoter. Moreover, adenoviral infection of C2C12 myotubes with PGC-1α and MEF2C results in an induction of a number of muscle fiber type I-specific genes, further underlining the important role of these two proteins in triggering and maintaining a fiber type I typical gene expression.
Discussion
In this report, we describe mechanisms by which PGC-1α gene expression is regulated by calcium-signaling components in muscle cells. CaMKIV and CnA, two key enzymes in this pathway, play distinct but overlapping roles in increasing expression from the PGC-1α promoter. CaMKIV likely induces PGC-1α by activating CREB, which, in turn, binds to a conserved CRE in the PGC-1α promoter. Activated CnA acts additively or synergistically with both CaMKIV and PGC-1α. Because CnA is a known activator of MEF2 proteins, and most or all of these effects are lost when the binding site for MEF2 on the PGC-1α promoter is mutated, it is highly likely that a major portion of the effects of CnA on PGC-1α gene transcription is through the MEF2 proteins. Although expression of NFAT itself has little effect on the PGC-1α promoter, a small increase in activity is observed when NFAT is coexpressed with either MEF2s or with activated CnA. Thus, it is possible that NFAT may exert a positive effect on PGC-1α expression by interacting functionally with MEF2C or MEF2D on the PGC-1α promoter, an effect also observed on the promoters of type I myofibrillar proteins (9).
One central aspect of muscle fiber-type determination is that it represents a fairly stable, “differentiation-like” response. Muscle fiber types ordinarily can be altered only through prolonged physical activity or chronic inactivity. In this way, it is very different from classic gene induction events, where transcriptional stimulation is reversed rapidly after hormone withdrawal. The differentiation-like behavior of muscle fiber-type switching implies that there are likely to be stable regulatory loops developed, and our data here provide a potentially new insight.
Indeed, a key finding here is that MEF2C and MEF2D can increase transcription from the PGC-1α promoter, and this activity is enhanced significantly by the presence of PGC-1α itself. These data imply a positive autoregulatory loop that may help to sustain high PGC-1α levels in these cells and, thus, promote mitochondrial biogenesis and a stable expression of typical muscle fiber-type I genes. A very recent study also has shown that MEF2s bind to the PGC-1α promoter and that HDAC5 can inhibit MEF2 activity on this promoter (27). Thus, it is also possible that PGC-1α binding to MEF2 might prevent binding of the MEF2-repressing HDACs and Cabin1 proteins, an additional mechanism that warrants testing. Also highly relevant here are observations by Kelly and coworkers (15) that forced expression of PGC-1α in heart induces numerous genes involved in mitochondrial biogenesis and respiration; these studies also show elevated levels of endogenous PGC-1α, suggesting an autoregulatory loop for PGC-1α control in cardiac muscle. The regulatory mechanisms in the heart have not been explored, but because MEF2 proteins also are expressed in heart, they could be similar to the mechanisms shown in this study.
It is very likely that other mechanisms and signaling cascades not explored here also may contribute to PGC-1α expression in skeletal muscle. For example, prolonged exercise leads to release of catecholamines that can activate β-adrenergic receptors in muscle (28). Activation of these receptors increases intracellular cAMP levels and potentially could activate CREB function on the PGC-1α promoter in muscle as it does in liver (24). In addition, the increased energy demand and the resulting activation of energy-sensing signaling pathways could contribute to PGC-1α expression. The 5′ AMP-activated protein kinase is activated during exercise by low ATP and high AMP levels (29), and chemical activators of 5′ AMP-activated protein kinase have been observed to modestly increase both CaMKIV and PGC-1α mRNA levels in muscle (17, 30). Because PGC-1α protein activity and stability are modified by protein phosphorylation events, these and other protein kinases and phosphatases might regulate PGC-1α posttranscriptionally, in addition to their effect on transcription (31). The exact role of 5′ AMP-activated protein kinase and other factors involved in energy sensing must be elucidated in future studies. A role for PGC-1β in modulating the expression of PGC-1α is also possible, although PGC-1β is not elevated significantly in type I-enriched muscles (32). The PGC-1β gene also does not seem to be regulated by CREB, in that it is not activated by cAMP signaling in hepatocytes (J.L., unpublished observation).
An improved knowledge of the molecular mechanisms controlling muscle fiber types eventually could lead to new therapies for patients for whom this is a component of their pathological condition. Prominent changes in the proportion of type I and type II muscle fibers are found in a variety of diseases such as obstructive pulmonary disease, paraplegia, or age-related muscle wasting. Novel therapeutic approaches, especially those based on known molecular mechanisms, could radically alter the quality of life for many of these patients.
Acknowledgments
We thank the people mentioned in Methods for their generous gifts of plasmids. This work was supported by National Institutes of Health Grant DK54477 (to B.M.S.). C.H. is the recipient of a fellowship from the “Schweizerische Stiftung für Medizinisch-Biologische Stipendien” and the Swiss National Science Foundation. J.L. is supported by a postdoctoral fellowship from the American Heart Association.
Abbreviations: CaMKIV, calcium/calmodulin-dependent protein kinase IV; CnA, calcineurin A; MEF, myocyte enhancer factor; PGC-1α, peroxisome proliferator-activated receptor γ coactivator 1α; dnMEF2C, dominant-negative MEF2C; NFAT, nuclear factor of activated T cells; CRE, cAMP response element; CREB, CRE-binding protein; HDAC, histone deacetylase.
References
- 1.Booth, F. W. & Thomason, D. B. (1991) Physiol. Rev. 71, 541-585. [DOI] [PubMed] [Google Scholar]
- 2.Berchtold, M. W., Brinkmeier, H. & Muntener, M. (2000) Physiol. Rev. 80, 1215-1265. [DOI] [PubMed] [Google Scholar]
- 3.Olson, E. N. & Williams, R. S. (2000) Cell 101, 689-692. [DOI] [PubMed] [Google Scholar]
- 4.Hood, D. A. (2001) J. Appl. Physiol. 90, 1137-1157. [DOI] [PubMed] [Google Scholar]
- 5.Corcoran, E. E. & Means, A. R. (2001) J. Biol. Chem. 276, 2975-2978. [DOI] [PubMed] [Google Scholar]
- 6.McKinsey, T. A., Zhang, C. L. & Olson, E. N. (2002) Trends Biochem. Sci. 27, 40-47. [DOI] [PubMed] [Google Scholar]
- 7.Michael, L. F., Wu, Z., Cheatham, R. B., Puigserver, P., Adelmant, G., Lehman, J. J., Kelly, D. P. & Spiegelman, B. M. (2001) Proc. Natl. Acad. Sci. USA 98, 3820-3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu, H., Rothermel, B., Kanatous, S., Rosenberg, P., Naya, F. J., Shelton, J. M., Hutcheson, K. A., DiMaio, J. M., Olson, E. N., Bassel-Duby, R., et al. (2001) EMBO J. 20, 6414-6423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chin, E. R., Olson, E. N., Richardson, J. A., Yang, Q., Humphries, C., Shelton, J. M., Wu, H., Zhu, W., Bassel-Duby, R. & Williams, R. S. (1998) Genes Dev. 12, 2499-2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naya, F. J., Mercer, B., Shelton, J., Richardson, J. A., Williams, R. S. & Olson, E. N. (2000) J. Biol. Chem. 275, 4545-4548. [DOI] [PubMed] [Google Scholar]
- 11.Wu, H., Kanatous, S. B., Thurmond, F. A., Gallardo, T., Isotani, E., Bassel-Duby, R. & Williams, R. S. (2002) Science 296, 349-352. [DOI] [PubMed] [Google Scholar]
- 12.Puigserver, P., Wu, Z., Park, C. W., Graves, R., Wright, M. & Spiegelman, B. M. (1998) Cell 92, 829-839. [DOI] [PubMed] [Google Scholar]
- 13.Puigserver, P. & Spiegelman, B. M. (2003) Endocr. Rev. 24, 78-90. [DOI] [PubMed] [Google Scholar]
- 14.Wu, Z., Puigserver, P., Andersson, U., Zhang, C., Adelmant, G., Mootha, V., Troy, A., Cinti, S., Lowell, B., Scarpulla, R. C., et al. (1999) Cell 98, 115-124. [DOI] [PubMed] [Google Scholar]
- 15.Lehman, J. J., Barger, P. M., Kovacs, A., Saffitz, J. E., Medeiros, D. M. & Kelly, D. P. (2000) J. Clin. Invest. 106, 847-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin, J., Wu, H., Tarr, P. T., Zhang, C. Y., Wu, Z., Boss, O., Michael, L. F., Puigserver, P., Isotani, E., Olson, E. N., et al. (2002) Nature 418, 797-801. [DOI] [PubMed] [Google Scholar]
- 17.Goto, M., Terada, S., Kato, M., Katoh, M., Yokozeki, T., Tabata, I. & Shimokawa, T. (2000) Biochem. Biophys. Res. Commun. 274, 350-354. [DOI] [PubMed] [Google Scholar]
- 18.Baar, K., Wende, A. R., Jones, T. E., Marison, M., Nolte, L. A., Chen, M., Kelly, D. P. & Holloszy, J. O. (2002) FASEB J. 16, 1879-1886. [DOI] [PubMed] [Google Scholar]
- 19.Pilegaard, H., Saltin, B. & Neufer, P. D. (2003) J. Physiol. (London) 546, 851-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ojuka, E. O., Jones, T. E., Han, D. H., Chen, M. & Holloszy, J. O. (2003) FASEB J. 17, 675-681. [DOI] [PubMed] [Google Scholar]
- 21.Handschin, C. & Meyer, U. A. (2000) J. Biol. Chem. 275, 13362-13369. [DOI] [PubMed] [Google Scholar]
- 22.Rhee, J., Inoue, Y., Yoon, J. C., Puigserver, P., Fan, M., Gonzalez, F. J. & Spiegelman, B. M. (2003) Proc. Natl. Acad. Sci. USA 100, 4012-4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He, T. C., Zhou, S., da Costa, L. T., Yu, J., Kinzler, K. W. & Vogelstein, B. (1998) Proc. Natl. Acad. Sci. USA 95, 2509-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herzig, S., Long, F., Jhala, U. S., Hedrick, S., Quinn, R., Bauer, A., Rudolph, D., Schutz, G., Yoon, C., Puigserver, P., et al. (2001) Nature 413, 179-183. [DOI] [PubMed] [Google Scholar]
- 25.Quandt, K., Frech, K., Karas, H., Wingender, E. & Werner, T. (1995) Nucleic Acids Res. 23, 4878-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Molkentin, J. D., Black, B. L., Martin, J. F. & Olson, E. N. (1996) Mol. Cell. Biol. 16, 2627-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Czubryt, M. P., McAnally, J., Fishman, G. I. & Olson, E. N. (2003) Proc. Natl. Acad. Sci. USA 100, 1711-1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sakamoto, K. & Goodyear, L. J. (2002) J. Appl. Physiol. 93, 369-383. [DOI] [PubMed] [Google Scholar]
- 29.Hardie, D. G. & Hawley, S. A. (2001) BioEssays 23, 1112-1119. [DOI] [PubMed] [Google Scholar]
- 30.Zong, H., Ren, J. M., Young, L. H., Pypaert, M., Mu, J., Birnbaum, M. J. & Shulman, G. I. (2002) Proc. Natl. Acad. Sci. USA 99, 15983-15987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Puigserver, P., Rhee, J., Lin, J., Wu, Z., Yoon, J. C., Zhang, C. Y., Krauss, S., Mootha, V. K., Lowell, B. B. & Spiegelman, B. M. (2001) Mol. Cell 8, 971-982. [DOI] [PubMed] [Google Scholar]
- 32.Lin, J., Puigserver, P., Donovan, J., Tarr, P. & Spiegelman, B. M. (2002) J. Biol. Chem. 277, 1645-1648. [DOI] [PubMed] [Google Scholar]