Abstract
We have examined corticogenesis in mouse embryos lacking DNA topoisomerase IIβ (IIβ) in the brain or in all tissues. The absence of IIβ, a type II DNA topoisomerase normally expressed in postmitotic cells in the developing cortex, severely affects cerebral stratification: no subplate is discernible, and neurons born at later stages of corticogenesis fail to migrate to the superficial layers. This abnormal pattern of neuron positioning in the cerebral cortex is reminiscent of that observed in mouse mutants defective in the reelin-signaling pathway. Significantly, the level of reelin in the neocortex is much reduced when IIβ is absent. These results implicate a role of IIβ in brain development. The enzyme may be required in implementing particular genetic programs in postmitotic cells, such as reelin expression in Cajal-Retzius cells, perhaps through its action on nucleoprotein structure of particular chromosomal regions.
Mammalian DNA topoisomerase IIβ (IIβ) is a type II DNA topoisomerase that catalyzes the transport of one DNA double-helix through another (1–3). Unlike the IIα isozyme, which is specifically expressed in proliferating cells and most likely performs the essential function of resolving intertwined chromosome pairs during mitosis (4–6), IIβ is apparently dispensable in cell growth (reviewed in ref. 7). Targeted disruption of the murine TOP2β gene showed; however, that IIβ has a critical role in neural and neuromuscular development (8). Whereas top2β+/Δ mice are phenotypically TOP2β+ and indistinguishable from their TOP2β+/+ littermates, top2βΔ/Δ nullizygotes die of a breathing impairment at birth, most likely owing to neural and neuromuscular defects (8).
In mammals, IIα and IIβ are largely expressed in proliferating cells and nonproliferating cells, respectively (7). In a detailed examination of the expression of the two forms during postnatal development of rat cerebellum, a sharp transition from IIα to IIβ expression was observed in cell populations that had undergone the final division and committed to differentiate into Purkinje or granule cells (9). The amount of DNA-bound IIβ becomes undetectable, however, when cells reach terminal differentiation (10).
To further test the role of IIβ in neural development, we have examined cerebral corticogenesis in embryos of mouse lines lacking IIβ in all tissues or specifically in certain brain structures including the telencephalon. We report here that IIβ has a remarkable role in neurogenesis. The induction of IIβ expression, after the final division of cells committed to a neuronal fate, appears necessary for the developmentally regulated expression of certain specific genes in these cells. The role of IIβ is unlikely to be limited to neuronal development and may also be important in the genetic programming of other postmitotic cells.
Materials and Methods
Construction of the Targeting Vector. The left and right arms of TOP2β-derived sequences, for targeted insertion of DNA sequences between them into the mouse genome, were obtained by PCR. The right arm was amplified and subcloned as two separate fragments, with a 34-nt loxP sequence incorporated in one of the PCR primers, so as to introduce a loxP site downstream of the exons targeted for deletion by Cre-mediated recombination (11). Restriction sites were sometimes added to the ends of various subcloned fragments, during PCR and/or subcloning, to facilitate their subsequent joining. A self-excision neomycin-resistance (neoR) cassette, in which a phosphoglycerate kinase (PGK) promoter-driven neomycin-resistance marker and an angiotensin-converting enzyme (ACE) promoter-driven yeast FLP recombinase gene were placed in between a loxP and frt recombination site on one side and a second frt site on the other side, was constructed by multiple steps of PCR-based cloning. The PGK-neoR region was amplified from pPGKNeo/TK2 (8), the ACE-FLP region from pACN (12) and pFLPe (13), and the frt sites from pPRT2 (14). The design of this cassette followed that reported in ref. 12, except that the expression of FLP recombinase rather than Cre recombinase was used to delete sequences between frt rather than loxP sites. The various subcloned TOP2β fragments and neoR cassette were sequentially cloned into pPGKNeo/TK2 to give the final targeting vector pYLL22.
Gene Targeting. Strain 129SvEv TC1 embryonic stem cells (kindly provided by P. Leader and C. Deng, Harvard Medical School, Boston) were transfected with 50 μg of ClaI-cut pYLL22 by electroporation and screened for resistance to 200 μg/ml G418 and 2 μM ganciclovir (15). DNA was isolated from each drug-resistant colony and subsequently analyzed by Southern blotting and PCR. In the PCR assays, the presence of the upstream loxP site was detected as a 320-bp product by using the primer pair PR3 (5′-ATATGGTACAGCAACAAAGCATTTGACATA-3′) and PR4 (5′-GGATATAACTTCGTATAGCATACATTATAC-3′), the downstream loxP site as a 490-bp fragment by using the primer pair PR5 (5′-TAGTGCTGTTGTAGATAGGATCCTATTAAG-3′) and PR6 (5′-GCTCATAACTTCGTATAGCATACATTATAC-3′), and the WT TOP2β allele as a 620-bp fragment by using the primer pair PR1 (5′-ACACAAATGTGAACAAATTTGCTATGAC-3′) and PR3.
Generation of Mouse Top2β Mutant Lines. Two positive clones identified in both blot hybridization and PCR assays were individually injected into C57BL/6 or BALB/c blastocysts to obtain chimeric animals (performed at the Center for Animal Resource and Comparative Medicine, Brigham and Women's Hospital, Boston). Germ-line males were identified and subsequently crossed with 129SvEv females (Taconic Farms). F1 pups were genotyped by PCR for the presence of the loxP sites, which is indicative of the incorporation of a “floxed” mutation into a TOP2β allele. The occurrence of FLP-mediated excision of the DNA segment between the frt sites was assessed by the presence of a 610-bp product in a PCR by using the primer pair PR2 (5′-TCATTGGGAGGCCAGAGCATC-3′) and PR3. Heterozygous top2β mice bearing a floxed top2β allele (with or without the neoR cassette) were bred to give floxed top2β homozygotes.
Two mouse lines expressing the Cre recombinase were used to delete the DNA segment flanked by loxP sites in a floxed top2β allele. The transgenic mouse strain 129/SvJae-TgN(PRM-Cre)58Og, which expresses Cre from the Protamine 1 promoter in male germ cells, was purchased from The Jackson Laboratory. The Foxg1-Cre mice, in a predominantly 129SvJ genetic background, were kindly provided by S. K. McConnell (Stanford University, Stanford, CA). In this knock-in strain, expression of Cre occurs specifically in several brain structures during embryogenesis (16). Crossing of these lines with mice containing floxed top2β alleles, or one top2β deletion and one floxed top2β allele, was used to produce the top2βΔ/Δ allele in all tissues or specifically in the brain (see Results). Pups with a top2β deletion were identified by the presence of a 450-bp product in a PCR by using the primer pair PR3 and PR7 (5′-GAATTGTTTGCTGTGGATGCATGTA-3′).
Tissue Collection. Cohabitation of mating pairs was started late in the afternoon, and noontime of the day of vaginal plug detection was counted as embryonic day 0.5 (E0.5). Embryos were removed from killed dams at various stages of gestation and immediately placed in cold PBS. Brains of E16.5, E17.5, and E18.5 embryos and whole heads of E14.5 embryos were dissected and further processed for embedding in paraffin or OCT compound (17).
Histochemical and Immunohistochemical Analyses. Tissues embedded in paraffin blocks were sectioned to 6-μm slices for mounting on SuperFrost Plus slides (Fisher). Staining of the sections with hematoxylin and eosin or Cresyl violet was carried out according to ref. 18. For immunohistochemical examination, cryosections of OCT-embedded brains (14–20 μm) were used (17). The sources and dilutions of the Abs used were as follows. Mouse α-NeuN (Chemicon, 1:100), mouse α-Map2 (Sigma clone HM-2, 1:100), rabbit α-IIβ 779 (kindly provided by F. Boege, University of Wurzburg, Wurzburg, Germany, 1:200), rabbit α-calretinin (Chemicon, 1:1,000), mouse α-reelin CR50 (kindly provided by K. Nakajima, Jikei University School of Medicine, Tokyo, and M. Ogawa, RIKEN Brain Science Institute, Wako, Japan, 1:500), and mouse α-βIII tubulin (Chemicon, 1:200). Cy2- or Cy3-conjugated secondary Abs (Jackson ImmunoResearch) were used to visualize the primary Ab staining patterns in a fluorescence microscope.
BrdUrd Birthdating. BrdUrd labeling of Cajal-Retzius and subplate neurons was done according to ref. 19 and of cortical neurons of middle layers according to ref. 20.
Blot Hybridization of Reelin Message RELN. Freshly dissected whole brains, protected in RNAlater solution (Ambion), were used in total RNA preparations by using Trizol (GIBCO/BRL). Electrophoresis of 7.3 μg of RNA in agarose gel containing formaldehyde (17) was carried out for blot hybridization of RELN mRNA, using a PCR-generated radiolabeled probe covering nucleotides 9011–10552 of RELN cDNA (GeneBank accession no. 6755311). Mouse GAPDH mRNA was also probed to provide a loading normalization standard.
Results
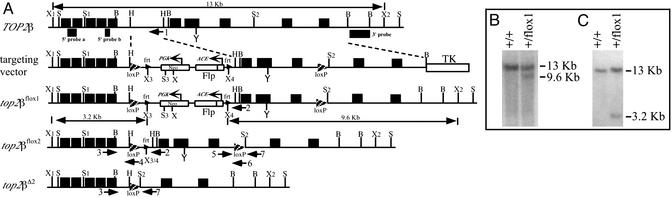
Generation of Murine Top2β Mutants by Cre Recombinase-Mediated Deletion. We have constructed mouse lines for Cre-mediated deletion within the TOP2β gene in all tissues or specifically in several brain structures including the telencephalon and olfactory bulb. The relevant region of the targeting vector used in these constructions is depicted in Fig. 1A (targeting vector). The vector was designed for the deletion of three neighboring exons that encode a IIβ segment containing the active-site tyrosyl residue (marked as Y in TOP2β). Homologous recombination between the targeting vector and the chromosomal TOP2β locus of mouse embryonic stem cells yields a mutant top2β allele, designated top2βflox1, in which the three exons are flanked by a 34-bp loxP site in the downstream intron, and a second loxP site and a neoR cassette in the upstream intron (Fig. 1 A, top2βflox1). Cre-mediated site-specific recombination would then replace the region bounded by the pair of loxP sites by a single 34-bp loxP sequence (Fig. 1 A, top2βΔ2). This top2β deletion is designated top2βΔ2 to distinguish it from a previously constructed deletion (8), which will hereafter be designated top2βΔ1.
Fig. 1.
Construction of mouse lines bearing mutations in the TOP2β gene. (A) Maps of relevant regions of TOP2β, the targeting vector, the top2βflox1, top2βflox2, and top2βΔ2 allele. The filled boxes represent exons, small and large arrowheads indicate the frt sites and the loxP sites, and larger dark arrows labeled 1–7 mark the approximate locations of PCR primers PR1–PR7, respectively. Open bars represent the transcription units neoR, Flp, and TK. Small dark bars under the line representing genomic DNA indicate the probes used in blot hybridization. X, S, B, and H mark the restriction sites of XbaI, SacI, BglII, and HindIII, respectively. Enumerated sites X1, X2, etc. designate individual sites of the particular restriction enzyme. Y marks the active-site tyrosyl residue of IIβ. (B and C) Southern hybridization of a XbaI digest of DNA from an embryonic stem cell clone. Radiolabeled 3′ probe was used in B, and 5′ probes a and b were used in C.
To avoid potential complications from the presence of an actively transcribed neoR marker in top2βflox1, the neoR cassette was constructed based on the design previously used for the deletion of specific DNA sequences during spermatogenesis (12). The testes-specific expression of the yeast recombinase gene FLP was used to first excise the neoR marker between two frt sites to give the allele top2βflox2 (Fig. 1 A, top2βflox2), which, similar to top2βflox1, could subsequently be converted to top2βΔ2 by Cre-mediated recombination.
Mouse top2β+/flox1 embryonic stem cells, identified by Southern blotting (Fig. 1 B and C) and PCR assays, were used to obtain male germ-line chimeras. These were subsequently mated with 129SvEv females to produce top2β+/flox1 or top2β+/flox2 F1s, the latter being derived from top2βflox1 through expression of the Flp recombinase during spermatogenesis. All F2 and F3 progenies, including those with only top2βflox1 or top2βflox2 alleles and no WT TOP2β, were found to be phenotypically TOP2β+. Thus the presence of loxP sites in the pair of introns flanking the three exons targeted for deletion, or the presence of a neoR marker in addition to the loxP sites, has minimal effect on TOP2β function. For brevity, the notation top2βflox will be used to represent either one of the phenotypically identical alleles top2βflox1 and top2βflox2.
To obtain top2βΔ2/Δ2 embryos or embryos specifically lacking IIβ in the brain, top2βflox mice were crossed with two different lines expressing Cre recombinase. In one, the recombinase is expressed from a testes-specific Protamine 1 promoter in a transgene, and deletion within top2βflox occurs during spermatogenesis (21). Male progenies from crosses with this line were genotyped, and those bearing both the Cre and top2βflox loci were again mated with female 129SvEv mice to generate top2β+/Δ2 pups. In the latter step, site-specific deletion of top2βflox1 to top2βΔ2 was found to be nearly quantitative, and the top2βflox2 to top2βΔ2 conversion was also highly efficient. Among the top2β+/Δ2 offspring, those without the Cre locus were back-crossed once with 129SvEv mice, and the top2β+/Δ2 progenies were then intercrossed to produce top2βΔ2/Δ2 embryos. In the other Cre-expressing line Foxg1-Cre (16), the coding sequence of the recombinase had been targeted to the Foxg1 locus, a gene specifically expressed in embryonic telencephalon and several other brain structures including olfactory bulb and hippocampus (16). This foxg1+/cre line was crossed with top2β+/Δ2, and progenies that were foxg1+/cre top2β+/Δ2 were further crossed with top2βflox/flox. Among the genotypes of the offspring, those that are foxg1+/cre top2βΔ2/flox were expected to have greatly reduced level of IIβ in brain structures that express Cre from the Foxg1 promoter (16), owing to the robust conversion of top2βΔ2/flox to top2βΔ2/Δ2.
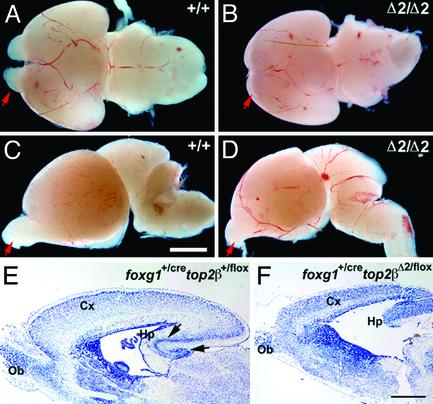
Defective Embryonic Brain Development in the Absence of IIβ. The top2βΔ2/Δ2 embryos lacking IIβ in all tissues show defects that were previously reported for the top2βΔ1/Δ1 embryos (8). Both top2βΔ1/Δ1 and top2βΔ2/Δ2 newborns have a smaller body size than their TOP2β+ (TOP2β+/+ or top2β+/Δ2) littermates and an abnormal curvature of their vertebral column; they die of respiratory failure shortly after birth. Sagittal sections of E14.5 top2βΔ2/Δ2 and TOP2β+ littermates revealed that various parts of the brain, including the olfactory bulb, cerebral cortex, hippocampus, diencephalon, midbrain, pons, and medulla, were morphologically similar in the two. At later stages of embryogenesis, however, differences between top2βΔ2/Δ2 and TOP2β+ brains are readily discernible. Comparing to the brains of the TOP2β+/+ littermates, the brains of the top2βΔ2/Δ2 nullizygotes at E16.5 and E18.5 were visibly smaller (Fig. 2, compare A and B; C and D). The olfactory bulb, a prominent protrusion at E16.5 in TOP2β+ brains, is barely identifiable in the top2βΔ2/Δ2 nullizygotes (Fig. 2 A and B); it becomes recognizable in the top2βΔ2/Δ2 brain at E18.5 but remains poorly developed (Fig. 2 C and D).
Fig. 2.
Abnormal brain development in embryos lacking IIβ in all tissues or only in the brain. (A and B) Ventral view of brains from E16.5 TOP2β+/+ and top2βΔ2/Δ2 littermates. (C and D) Lateral view of brains from E18.5 TOP2β+/+ and top2βΔ2/Δ2 littermates. Red arrows mark the olfactory bulb. (E and F) Cresyl violet staining of sagittal sections of cortex (Cx), olfactory bulb (Ob), and hippocampus (Hp) of a postnatal day 0 (P0) foxg1+/cre top2β+/flox pup (E) and a P0 foxg1+/cre top2βΔ2/flox pup (F). Black arrows in E mark the pyramidal and granule cell layers in hippocampus. (Scale bars = 1mm in A–D and 500 μm in E and F.)
Embryos bearing foxg1+/cre top2βΔ2/flox, which are expected to show IIβ deficiency in brain structures that express Cre from the Foxg1 promoter, were also found to suffer perinatal death. The foxg1+/cre top2βΔ2/flox embryos differ from the top2βΔ2/Δ2 embryos, however, in that their body size and appearance are similar to the TOP2β+ newborns. Furthermore, although the foxg1+/cre top2βΔ2/flox embryos lack the vigor of the TOP2β+ newborns, they show some body movements upon tactile stimulation. Nevertheless, they die within 30 min after birth, and the collapsed lung alveoli of the dead pups suggest a respiratory failure similar to neonates lacking IIβ in all tissues. At birth, the brains of the foxg1+/cre top2βΔ2/flox embryos and their foxg1+/cre top2β+/flox, foxg1+/+ top2βΔ2/flox, or foxg1+/+ top2β+/flox littermates show clear anatomical differences. The brain of foxg1+/cre top2βΔ2/flox embryos shows a much thinner cortex, a less developed olfactory bulb, and ill-defined pyramidal and granule cell layers in the hippocampus (Fig. 2 E and F).
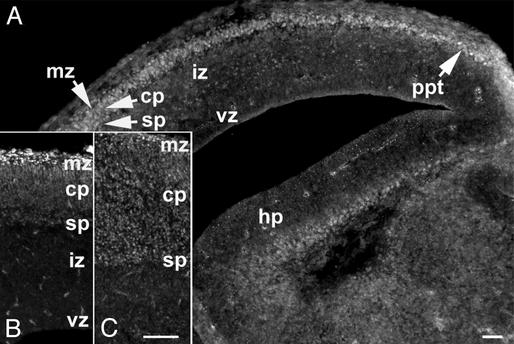
Expression of IIβ in the Neocortex. The results described above provide strong evidence that IIβ is required in normal embryonic brain development. To learn more about its role, we first examined its expression pattern in the neocortex by the use of a IIβ-specific Ab. IIβ immunoreactivity was undetectable in brain sections of top2βΔ2/Δ2 embryos, confirming the high specificity of the Ab preparation. In TOP2β+ embryos, the presence of IIβ in the neocortex becomes apparent at E14.5. The distribution of IIβ-positive cells largely coincides with the graded lateral to medial neocortical development. In the dorsal-medial region, IIβ is expressed in a thin layer of cells located immediately under the pial surface corresponding to the preplate (Fig. 3A). The layer of cells expressing IIβ becomes progressively thicker in the more lateral regions, corresponding to the marginal zone and the subplate that are derived from the preplate, as well as the cortical plate sandwiched in between (Fig. 3A). At E16.5, IIβ-positive cells greatly increase in number as the cortical plate thickens (Fig. 3B); immunoreactivity was found in strata spanning the marginal zone, the cortical plate and the subplate, but only sparsely in the ventricular zone or the intermediate zone (Fig. 3B). E18.5 brains show a IIβ pattern similar to that observed in the E16.5 embryos (Fig. 3C). These results indicate that IIβ expression is minimal in the proliferating neuronal precursors in the ventricular zone and in neurons that have just exited the cell cycle and migrated into the intermediate zone. Expression of IIβ is much elevated, however, in the postmitotic neurons that have moved out of the intermediate zone.
Fig. 3.
Expression of IIβ during corticogenesis. (A) A coronal section of E14.5 top2β+/Δ2 telencephalon immunostained with α-IIβ Ab 779. (B and C) Sagittal sections of the TOP2β+/+ neocortex stained with 779; E16.5 (B) and E18.5 (C). vz, ventricular zone; iz, intermediate zone; sp, subplate; cp, cortical plate; ppt, preplate; hp, hippocampus. (Scale bars = 100 μm.)
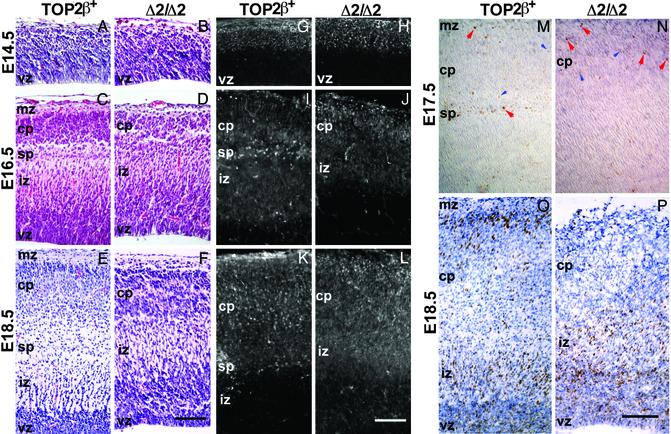
Defective Lamination During Cerebral Cortical Development in the Absence of IIβ. We next examined the layered structure of the cerebral cortex of top2βΔ2/Δ2 embryos at various stages of gestation. At E14.5, the earliest stage examined, the histological appearance of the cerebral wall of the top2βΔ2/Δ2 and their TOP2β+ littermates is not significantly different (Fig. 4 A and B). Staining with Abs specific to βIII tubulin, one of the earliest markers of postmitotic cells of a neuronal lineage in the neocortex, also showed a similar pattern in the top2βΔ2/Δ2 and TOP2β+ littermates (Fig. 4 G and H). At E16.5 and E18.5, however, histological differences in the neocortex of top2βΔ2/Δ2 and TOP2β+ littermates are readily discernible. Whereas both show a layered structure, in the top2βΔ2/Δ2 nullizygotes the cortical plate appears disorganized, the subplate is absent, and the ventricular zone is thicker than that of the TOP2β+ embryos (Fig. 4 C–F). The subplate neurons stained by the NeuN-specific Abs form a distinct boundary between the intermediate zone and the prominently stained cortical plate in the TOP2β+ embryos (Fig. 4 I and K); in the top2βΔ2/Δ2 mutant, such a boundary is absent (Fig. 4 J and L). A similar difference was observed by staining for Map2 (result not shown).
Fig. 4.
Abnormal lamination in the cortex of top2βΔ2/Δ2 embryos. (A–F) Hematoxylin-eosin-stained sagittal sections of the E14.5 (A and B), E16.5 (C and D), and E18.5 (E and F) neocortex. Genotypes of the embryos are marked on the photographs. (G and H) α-βIII tubulin-stained E14.5 top2β+/Δ2 (G) and top2βΔ2/Δ2 cortex (H). (I–L) α-NeuN stained E16.5 (I and J) and E18.5 (K and L) cortex. Genotypes of the embryos are marked on the photographs. (M–P) BrdUrd birthdating. (M and N) BrdUrd (50 μg/g of body weight) was injected on E10.5 and E11.5, and the brains were analyzed on E17.5. (C and D) BrdUrd was injected on E15.5, and the brains were analyzed on E18.5. Sagittal sections of the neocortex were first stained for BrdUrd-positive cells (brown color) and then counterstained with hematoxylin. A few examples of BrdUrd-positive cells are marked by arrows (red, strongly stained; blue, less strongly stained). vz, ventricular zone: iz, intermediate zone; sp, subplate; cp, cortical plate; mz, marginal zone. (Scale bars = 100 μm.)
To test whether the lamination defects in the top2βΔ2/Δ2 neocortex reflect an abnormality in neuronal migration or positioning, cell birthdating by BrdUrd labeling was carried out. In one experiment, BrdUrd was successively injected into a pregnant dam to mark postmitotic cells born at E10.5 and E11.5, and embryos were subsequently removed on E17.5 and genotyped and examined for the locations of these neurons. As shown in Fig. 4M, in the TOP2β+ E17.5 embryos a small number of labeled neurons, representing those that had undergone their last division around the times of BrdUrd injection, were found in the marginal zone under the pial surface, and in the subplate region under the cortical plate. This distribution is expected from the known temporal order of corticogenesis, in which the final division of the precursors of Cajal-Retzius cells and the subplate neurons mainly occur around E10.5 and E11.5, respectively (19). In the top2βΔ2/Δ2 littermates, a small number of BrdUrd-labeled cells were detected in the region under the pial surface but not in regions deep in the cerebral cortex (Fig. 4N). When BrdUrd was injected at E15.5 and the cerebral cortex subsequently examined at E18.5, a large number of BrdUrd-labeled neurons were seen in both TOP2β+ and top2βΔ2/Δ2 littermates (Fig. 4 O and P), but there is a striking difference in their distribution patterns. In the TOP2β+ neocortex, pronounced labeling of the superficial layer of the cortical plate under the marginal zone is observed. BrdUrd-labeled cells are also observed in the ventricular and intermediate zones and the layers in between (Fig. 4O). In contrast, in the top2βΔ2/Δ2 littermates the majority of BrdUrd-labeled cells are found in the intermediate and ventricular zones; some labeled cells are found to scatter across the entire cortical plate, but there is no concentration of labeled cells in the superficial layers of the neocortex (Fig. 4P). Thus in contrast to the “inside-out” lamination in the normal cortex, in which the later-born neurons migrate through layers of the earlier born neurons to locate in the superficial layers (22), the ordered outward movement of the later-born neurons is defective in the top2βΔ2/Δ2 neocortex.
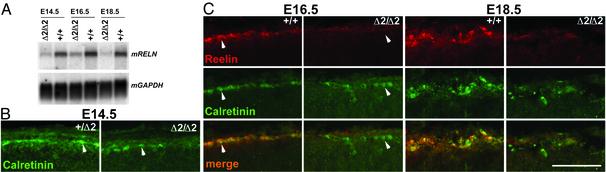
Reduced Expression of reelin in the Mouse Neocortex Lacking IIβ. The abnormal cerebral stratification in top2βΔ2/Δ2 embryos resembles that in mutant mice defective in reelin signaling. In the reeler mice lacking reelin, e.g., separation of the marginal zone and the subplate during neocorticogenesis does not occur, and neurons born at later embryonic stages are located in the deeper rather than the more superficial layers of the neocortex (ref. 23 and references therein). We thus examined the expression of reelin in the cerebral cortex of the top2βΔ2/Δ2 embryos. Reelin is normally expressed and secreted by Cajal-Retzius cells in the marginal zone of the neocortex (23). Blot hybridization for RELN mRNA indicates that in the brains of E14.5, E16.5, and E18.5 top2βΔ2/Δ2 embryos its level is much reduced relative to that in the TOP2β+ controls at the same stage of gestation (Fig. 5A). To examine more specifically the expression of reelin in the Cajal-Retzius cells, histocytochemical examination of brain sections of top2βΔ2/Δ2 and TOP2β+ littermates was carried out by using appropriate Abs. Cajal-Retzius cells in the outermost layer of E14.5 mouse neocortex are identifiable as calretinin-positive cells, in both groups (Fig. 5B). At E16.5 and E18.5, however, there appears to be a sparsity of these cells in the nullizygotes. Significantly, expression of reelin by the calretinin-positive cells is evident only in the TOP2β+ controls and not in the top2βΔ2/Δ2 embryos (Fig. 5C).
Fig. 5.
Reduction in RELN expression in top2βΔ2/Δ2 embryos. (A) Analysis of RELN message by blot hybridization. (B) Immunostaining of the E14.5 top2β+/Δ2 and top2βΔ2/Δ2 neocortex with α-calretinin Ab. Arrows point to examples of the calretinin-positive cells. (C) Double-immunostaining of E16.5 and E18.5 TOP2β+/+ and top2βΔ2/Δ2 cortex with an α-reelin Ab CR50 (red) and an α-calretinin Ab (green). Merged images are shown at the bottom. Arrows shown in the E16.5 images point to examples of calretinin-positive cells in TOP2β+/+ embryos that express reelin and calretinin-positive cells in top2βΔ2/Δ2 that do not express reelin. (Scale bar = 100 μm, B and C.)
Discussion
Our results show that targeted deletion of the mouse TOP2β gene, in all tissues or specifically in the brain, leads to defective brain development and perinatal death. What might be the function of IIβ in brain development? The onset of robust IIβ expression in postmitotic neurons during prenatal corticogenesis (this work) is strikingly similar to that observed in the postnatal neuronal development of the cerebellar cortex (9). This common pattern of IIβ induction in cells of different neuronal lineages undergoing differentiation at different developmental stages raises the possibility of a general role of the enzyme in differentiating cells. In the top2βΔ/Δ nullizygotes, however, cell differentiation and organ development appear to be largely normal (this work and ref. 8). Thus any role that IIβ might assume in differentiation must be of a more specific nature.
During embryonic development of the cerebral cortex, the laminar locations and phenotypes of cortical neurons are determined mainly during the course of their last cell division (24), before the elevation of IIβ expression in these cells. Thus IIβ is unlikely to have a direct role in laminar fate determination of neurons. As the postmitotic neurons further differentiate to become progressively more specialized, however, different nuclear events are presumably necessary to modulate the expression of various genes. The absence of IIβ may directly or indirectly affect the expression of a subset of such genes, particularly RELN, and thus result in a cortical lamination pattern resembling that seen in mutants of the reelin-signaling pathway. However, the effects of IIβ inactivation apparently go much beyond a reduction in RELN expression. Whereas mice lacking reelin are viable, perinatal death of mice embryos occurs even when IIβ inactivation is limited to the telencephalon and a few other embryonic brain structures.
How might the expression of a particular collection of genes in postmitotic cells be affected by the absence of IIβ? Although IIβ has been shown to associate with intracellular DNA undergoing transcription (25, 26), it seems unlikely that the enzyme is specifically required for relieving template supercoiling during transcription (27). In yeast, inactivation of DNA topoisomerase II has rather minor effects on transcription, which is significantly reduced only when both DNA topoisomerases I and II are inactivated (28).
It is plausible that IIβ may affect genetic programming in postmitotic cells through a role in the folding and organization of chromatin. Different postmitotic cells might have distinct nuclear architectures and characteristic higher-order chromosomal organizations (29, 30), and there appears to be a close correlation between chromatin structure and gene activation (31–33). During cellular differentiation, the changes in gene expression patterns may require local or more global alterations of chromatin structures, such as condensation or decondensation of specific chromosomal regions to different degrees of compaction, and thus may require the participation of a type II DNA topoisomerase. The possibility of a structural role of a type II DNA topoisomerase in the scaffolding of chromosomal loops has also been discussed (34).
Another possibility is that IIβ, most likely in association with other proteins, may specifically affect the regulation of particular genes. Type II DNA topoisomerases of various organisms were reported to physically interact with a diverse collection of proteins (35–50). Several of these proteins are of particular interest. The Drosophila barren protein, in association with the polyhomeotic protein of the polycomb group, has been suggested to regulate the chromatin structure and expression of the bithorax gene complex (50). The localization of the Drosophila “supercoiling factor” to natural and ecdysteroid-induced polytene puffs is suggestive of its involvement in the formation of active chromatin (39). Equally significant is the association of DNA topoisomerase II with components of chromatin remodeling complexes (45–47).
In conclusion, our studies of the top2β mouse lines clearly demonstrate that IIβ plays an important role in the development of neurons. The enzyme appears to affect the expression of particular genes in postmitotic neurons and probably in other postmitotic cells as well. Whereas IIα appears to have assumed the central role in chromosome segregation, IIβ has apparently evolved into a unique role in genetic programming in postmitotic cells.
Acknowledgments
We are most grateful to those who generously shared reagents with us, especially S. M. Dymecki, P. Leader, C. Deng, F. Boege, K. Nakajima, M. Ogawa, and S. K. McConnell. We thank K. Kwan and L. Cai for technical advice, L. Du for blastocyst injections, and A. Greenwood and O. Martinez for help with histology and immunohistochemical analysis. This work was supported by National Institutes of Health Grant GM24544.
Abbreviations: IIβ, DNA topoisomerase IIβ;En, embryonic day n; neoR, neomycin resistance.
References
- 1.Wang, J. C. (1996) Annu. Rev. Biochem. 65, 635-692. [DOI] [PubMed] [Google Scholar]
- 2.Wang, J. C. (2002) Nat. Rev. Mol. Cell Biol. 3, 430-440. [DOI] [PubMed] [Google Scholar]
- 3.Champoux, J. J. (2001) Annu. Rev. Biochem. 70, 369-413. [DOI] [PubMed] [Google Scholar]
- 4.DiNardo, S., Voelkel, K. & Sternglanz, R. (1984) Proc. Natl. Acad. Sci. USA 81, 2616-2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holm, C., Goto, T., Wang, J. C. & Botstein, D. (1985) Cell 41, 553-563. [DOI] [PubMed] [Google Scholar]
- 6.Uemura, T., Ohkura, H., Adachi, Y., Morino, K., Shiozaki, K. & Yanagida, M. (1987) Cell 50, 917-925. [DOI] [PubMed] [Google Scholar]
- 7.Austin, C. A. & Marsh, K. L. (1998) BioEssays 20, 215-226. [DOI] [PubMed] [Google Scholar]
- 8.Yang, X., Li, W., Prescott, E. D., Burden, S. J. & Wang, J. C. (2000) Science 287, 131-134. [DOI] [PubMed] [Google Scholar]
- 9.Tsutsui, K., Hosoya, O., Sano, K. & Tokunaga, A. (2001) J. Comp. Neurol. 431, 228-239. [DOI] [PubMed] [Google Scholar]
- 10.Tsutsui, K., Sano, K., Kikuchi, A. & Tokunaga, A. (2001) J. Biol. Chem. 276, 5769-5778. [DOI] [PubMed] [Google Scholar]
- 11.Sauer, B. & Henderson, N. (1988) Proc. Natl. Acad. Sci. USA 85, 5166-5170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bunting, M., Bernstein, K. E., Greer, J. M., Capecchi, M. R. & Thomas, K. R. (1999) Genes Dev. 13, 1524-1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodriguez, C. I., Buchholz, F., Galloway, J., Sequerra, R., Kasper, J., Ayala, R., Stewart, A. F. & Dymecki, S. M. (2000) Nat. Genet. 25, 139-140. [DOI] [PubMed] [Google Scholar]
- 14.Dymecki, S. M. (1996) Gene 171, 197-201. [DOI] [PubMed] [Google Scholar]
- 15.Hogan, B., Beddington, R., Constantini, F. & Lacy, E. (1994) Manipulating the Mouse Embryo: A Laboratory Annual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 16.Hebert, J. M. & McConnell, S. K. (2000) Dev. Biol. 222, 296-306. [DOI] [PubMed] [Google Scholar]
- 17.Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A. & Struhl, K. (1996) Current Protocols in Molecular Biology (Wiley, New York).
- 18.Presnell, J. K. & Schreibman, M. P. (1997) Animal Tissue Techniques (The Johns Hopkins Univ. Press, Baltimore).
- 19.Magdaleno, S., Keshvara, L. & Curran, T. (2002) Neuron 33, 573-586. [DOI] [PubMed] [Google Scholar]
- 20.Caviness, V. S., Jr. (1982) Brain Res. 256, 293-302. [DOI] [PubMed] [Google Scholar]
- 21.O'Gorman, S., Dagenais, N. A., Qian, M. & Marchuk, Y. (1997) Proc. Natl. Acad. Sci. USA 94, 14602-14607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Angevine, J. B., Jr., & Sidman, R. L. (1961) Nature 192, 766-768. [DOI] [PubMed] [Google Scholar]
- 23.Rice, D. S. & Curran, T. (2001) Annu. Rev. Neurosci. 24, 1005-1039. [DOI] [PubMed] [Google Scholar]
- 24.McConnell, S. K. & Kaznowski, C. E. (1991) Science 254, 282-285. [DOI] [PubMed] [Google Scholar]
- 25.Govoni, M., Neri, S., Labella, T., Sylvester, J. E., Novello, F. & Pession, A. (1995) Biochem. Biophys. Res. Commun. 213, 282-288. [DOI] [PubMed] [Google Scholar]
- 26.Mao, Y., Desai, S. D., Ting, C. Y., Hwang, J. & Liu, L. F. (2001) J. Biol. Chem. 276, 40652-40658. [DOI] [PubMed] [Google Scholar]
- 27.Liu, L. F. & Wang, J. C. (1987) Proc. Natl. Acad. Sci. USA 84, 7024-7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yanagida, M. & Sternglanz, R. (1990) in DNA Topology and its Biological Effects, eds. Cozzarelli, N. R. & Wang, J. C. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 299-320.
- 29.Cremer, T., Kreth, G., Koester, H., Fink, R. H., Heintzmann, R., Cremer, M., Solovei, I., Zink, D. & Cremer, C. (2000) Crit. Rev. Eukaryotic Gene Expression 10, 179-212. [PubMed] [Google Scholar]
- 30.Martou, G. & De Boni, U. (2000) Exp. Cell Res. 256, 131-139. [DOI] [PubMed] [Google Scholar]
- 31.Tsukamoto, T., Hashiguchi, N., Janicki, S. M., Tumbar, T., Belmont, A. S. & Spector, D. L. (2000) Nat. Cell Biol. 2, 871-878. [DOI] [PubMed] [Google Scholar]
- 32.Muller, C. & Leutz, A. (2001) Curr. Opin. Genet. Dev. 11, 167-174. [DOI] [PubMed] [Google Scholar]
- 33.Mahy, N. L., Perry, P. E. & Bickmore, W. A. (2002) J. Cell Biol. 159, 753-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Warburton, P. E. & Earnshaw, W. C. (1997) BioEssays 19, 97-99. [DOI] [PubMed] [Google Scholar]
- 35.Wang, X., Watt, P. M., Borts, R. H., Louis, E. J. & Hickson, I. D. (1999) Mol. Gen. Genet. 261, 831-840. [DOI] [PubMed] [Google Scholar]
- 36.Bhat, M. A., Philp, A. V., Glover, D. M. & Bellen, H. J. (1996) Cell 87, 1103-1114. [DOI] [PubMed] [Google Scholar]
- 37.Hirano, T., Kobayashi, R. & Hirano, M. (1997) Cell 89, 511-521. [DOI] [PubMed] [Google Scholar]
- 38.Nakano, H., Yamazaki, T., Miyatake, S., Nozaki, N., Kikuchi, A. & Saito, T. (1996) J. Biol. Chem. 271, 6483-6489. [DOI] [PubMed] [Google Scholar]
- 39.Kobayashi, M., Aita, N., Hayashi, S., Okada, K., Ohta, T. & Hirose, S. (1998) Mol. Cell. Biol. 18, 6737-6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bhat, U. G., Raychaudhuri, P. & Beck, W. T. (1999) Proc. Natl. Acad. Sci. USA 96, 7859-7864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mao, Y., Desai, S. D. & Liu, L. F. (2000) J. Biol. Chem. 275, 26066-26073. [DOI] [PubMed] [Google Scholar]
- 42.Cowell, I. G., Okorokov, A. L., Cutts, S. A., Padget, K., Bell, M., Milner, J. & Austin, C. A. (2000) Exp. Cell Res. 255, 86-94. [DOI] [PubMed] [Google Scholar]
- 43.Kurz, E. U., Leader, K. B., Kroll, D. J., Clark, M. & Gieseler, F. (2000) J. Biol. Chem. 275, 13948-13954. [DOI] [PubMed] [Google Scholar]
- 44.Durrieu, F., Samejima, K., Fortune, J. M., Kandels-Lewis, S., Osheroff, N. & Earnshaw, W. C. (2000) Curr. Biol. 10, 923-926. [DOI] [PubMed] [Google Scholar]
- 45.Tsai, S. C., Valkov, N., Yang, W. M., Gump, J., Sullivan, D. & Seto, E. (2000) Nat. Genet. 26, 349-353. [DOI] [PubMed] [Google Scholar]
- 46.Johnson, C. A., Padget, K., Austin, C. A. & Turner, B. M. (2001) J. Biol. Chem. 276, 4539-4542. [DOI] [PubMed] [Google Scholar]
- 47.LeRoy, G., Loyola, A., Lane, W. S. & Reinberg, D. (2000) J. Biol. Chem. 275, 14787-14790. [DOI] [PubMed] [Google Scholar]
- 48.Park, G. H., Lee, Y. T. & Bae, Y. S. (2001) Mol. Cell 11, 82-88. [PubMed] [Google Scholar]
- 49.Escargueil, A. E., Plisov, S. Y., Skladanowski, A., Borgne, A., Meijer, L., Gorbsky, G. J. & Larsen, A. K. (2001) FASEB J. 15, 2288-2290. [DOI] [PubMed] [Google Scholar]
- 50.Lupo, R., Breiling, A., Bianchi, M. E. & Orlando, V. (2001) Mol. Cell 7, 127-136. [DOI] [PubMed] [Google Scholar]