Abstract
Virtually all smooth muscle genes analyzed to date contain two or more essential binding sites for serum response factor (SRF) in their control regions. Because SRF is expressed in a wide range of cell types, it alone cannot account for smooth muscle-specific gene expression. We show that myocardin, a cardiac muscle- and smooth muscle-specific transcriptional coactivator of SRF, can activate smooth muscle gene expression in a variety of nonmuscle cell types via its association with SRF. Homodimerization of myocardin is required for maximal transcriptional activity and provides a mechanism for cooperative activation of smooth muscle genes by SRF–myocardin complexes bound to different SRF binding sites. These findings identify myocardin as a master regulator of smooth muscle gene expression and explain how SRF conveys smooth muscle specificity to its target genes.
Development of cardiac muscle, skeletal muscle, and smooth muscle cells is accompanied by transcriptional activation of overlapping but distinct sets of muscle-specific genes. Differentiation of skeletal muscle cells is controlled by members of the MyoD family of basic helix–loop–helix transcription factors, which have the remarkable ability to activate skeletal muscle gene expression when expressed in nonmuscle cell types (reviewed in refs. 1 and 2). No single factor has been found to be sufficient to activate the cardiac muscle or smooth muscle gene programs. Whether skeletal muscle is unique with respect to its induction by a single transcription factor or whether as-yet-unidentified master regulators govern cardiac muscle and smooth muscle development remains to be determined.
Smooth muscle genes share the property of being regulated by serum response factor (SRF), a ubiquitous MADS (MCM1, Agamous, Deficiens, SRF) box transcription factor that binds as a homodimer to the DNA consensus sequence CC(A/T)6GG, known as a CArG box (3, 4). Virtually every smooth muscle gene analyzed to date contains at least two CArG boxes in its control region, which act cooperatively to govern smooth muscle-specific transcription (5–11). Blockade of SRF activity with a dominant negative SRF mutant has also been shown to prevent expression of smooth muscle genes in proepicardial explants (12). However, the mechanism for SRF-dependent activation of smooth muscle genes has not been fully resolved and is complicated by the fact that SRF is not smooth muscle-specific.
Recently, we discovered an SRF transcriptional coactivator called myocardin that is expressed specifically in smooth and cardiac muscle cell lineages (13, 14). Myocardin belongs to the SAP domain family of transcription factors (15) and activates smooth and cardiac muscle reporter genes by interacting with SRF (13, 14). Dominant negative myocardin mutants that compete with the wild-type myocardin protein for interaction with SRF block cardiac gene expression in injected Xenopus embryos (13), suggesting an essential early role for myocardin in heart development.
Here we show that myocardin is sufficient to activate the program of smooth muscle differentiation. The promyogenic activity of myocardin requires association with SRF and is augmented by homodimerization, which provides a molecular basis for the cooperativity among CArG boxes that is required for smooth muscle gene activation.
Methods
Cell Culture and Transfection. 10T1/2 cells were maintained at low density (≈30% confluence) in DMEM with 10% FBS. Transfections were conducted with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Two days after transfection, cells were shifted to differentiation medium (DMEM with 2% horse serum). Five days later, further analyses, including immunocytochemistry, Western blot, and RT-PCR, were performed. Generally, 0.5 μg of plasmid was used for each well in a 12-well plate.
To obtain cardiac fibroblasts, neonatal rat hearts were digested as described (16), and the fibroblast fraction was purified by differential plating for 2 h on tissue culture plastic. Adherent fibroblasts from this plating were passaged twice, plated at low density (5 × 104 cells per cm2), and grown to subconfluence in 10% FBS in DMEM. These cultures were washed extensively to remove serum and infected with adenoviruses encoding lacZ or residues 128–935 of myocardin in serum-free medium at a multiplicity of infection of 100 for 2 h at 37°C. Cells were cultured for 14–21 days, fixed in 4% formaldehyde in PBS for 30 min, and stained for smooth muscle (SM)-α-actin as described below.
The PAC1 (17) and A10 (18) smooth muscle cell lines were maintained in DMEM with 10% FBS and were infected with adenovirus encoding lacZ or a myocardin dominant negative mutant lacking the transcription activation domain (TAD) (13).
COS cell transfections and luciferase assays were performed as described (13). Unless otherwise indicated, 100 ng of reporter plasmid and 100 ng of each activator plasmid were used. The total amount of DNA per well was kept constant by adding the corresponding amount of expression vector without a cDNA insert.
Myocardin and SRF expression vectors have been described (13, 14). Myocardin mutants were generated through PCR-based mutagenesis by using the QuikChange kit from Stratagene. All mutations were confirmed by DNA sequencing. The myocardin mutant NLS-Δbasic contains an SV40 nuclear localization sequence (NLS) at the amino terminus and an internal deletion of the basic region. The SV40 NLS is necessary for this mutant protein to become localized to the nucleus. MyoD was expressed by using the pcDNA3.1 expression plasmid (Invitrogen). The SM22-luciferase reporter contained the 1434-bp promoter (6, 13, 14). The 3xc-fos-SRE-luciferase reporter was constructed by linking three tandem copies of the c-fos serum response element to a thymidine kinase promoter-driven luciferase reporter. CMV-lacZ was included as an internal control for variations in transfection efficiency.
Immunocytochemistry and Western Blot. Immunostaining was performed as described (19). Myogenic conversion assays were performed as described (20), except that rabbit anti-SM-myosin heavy chain (MHC) antibody was used. MF20 antisarcomeric myosin antibody was collected from the supernatant of cultured hybridoma cells from the Developmental Studies Hybridoma Bank (University of Iowa, Iowa City). Mouse anti-skeletal myosin (MY32), mouse anti-SM-α-actin (1A4), mouse anti-SM calponin, mouse anti-h caldesmon, and mouse anti-SM myosin light chain kinase (K36) were purchased from Sigma. Rabbit anti-SM22 antibodies were provided by Drs. Michael Parmacek (University of Pennsylvania, Philadelphia) and Lucia Schuger (Wayne State University, Detroit). Rabbit anti-SM-MHC was purchased from Biomedical Technologies (Stoughton, MA). Mouse anti-SM-γ-actin was purchased from ICN. All primary antibodies were used at 1:200 dilutions, except for anti-SM-γ-actin, which was used at 1:1,000 dilution to avoid crossreaction with SM-α-actin. FITC or Texas red-conjugated anti-mouse or anti-rabbit secondary antibody (Vector Laboratories) was used at a 1:200 dilution.
Western blots on cell extracts were performed as described (19) by using the antibodies described above in addition to an anti-α-tubulin antibody (Sigma). As secondary antibodies, horseradish peroxidase-conjugated donkey antibodies were used.
Gel Mobility Shift Assays. Gel mobility shift assays were performed as described (13). In vitro translated proteins and 32P-labeled probe corresponding to the c-fos CArG box were used.
RT-PCR. Total RNA was isolated with TRIzol reagent (Invitrogen). After treatment with DNaseI, 1 μg of RNA was used as a template for reverse transcription with random hexamer primers. Sequences of PCR primers are available upon request. All PCR products span intron regions of the genes. RT-PCRs were performed under conditions of linearity with respect to input RNA.
Results
Activation of Smooth Muscle Gene Expression by Myocardin. Because myocardin is expressed specifically in cardiac muscle and smooth muscle cells and potently transactivates cardiac muscle and smooth muscle reporter genes (13, 14), we tested whether it was sufficient to activate endogenous muscle genes in transfected 10T1/2 fibroblasts. In parallel, we transfected cells with MyoD (21) to compare the potential myogenic activity of the two proteins.
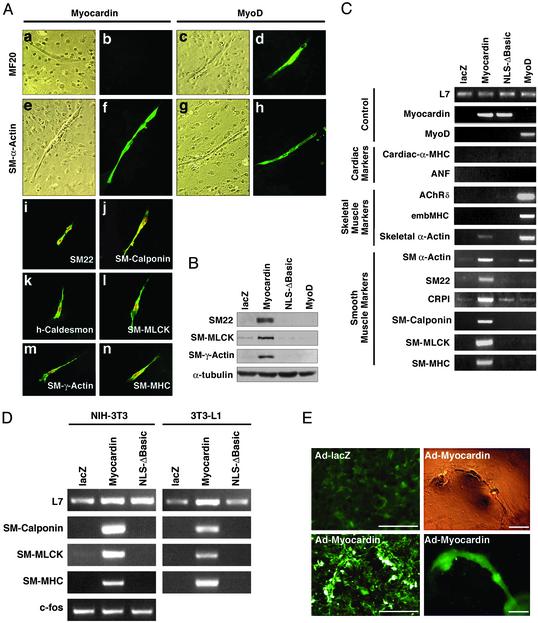
Cells expressing myocardin adopted an elongated morphology reminiscent of differentiated myocytes (Fig. 1A). MyoD-transfected cells stained with MF20 antibody, which recognizes cardiac and skeletal muscle myosin, but myocardin did not induce these muscle markers. Instead, cells transfected with myocardin stained intensely for expression of α-SM actin. This actin isoform is also expressed in cardiac myocytes, as well as in skeletal muscle cells, as demonstrated by its up-regulation by MyoD (Fig. 1 A). Myocardin also induced expression of SM22, SM-calponin, h-caldesmon, SM myosin light chain kinase, SM γ-actin, and SM-MHC proteins, which are markers of smooth muscle (Fig. 1 A). Myocardin-induced expression of smooth muscle proteins was further confirmed by Western blot analysis (Fig. 1B). The activation of smooth muscle gene expression by myocardin depended on reducing the concentration of FBS in the medium from 10% to 2% horse serum, suggesting that cell proliferation was unfavorable to its promyogenic activity.
Fig. 1.
Activation of smooth muscle cell differentiation by myocardin. (A) 10T1/2 cells were transfected with myocardin (a, b, e, f, and i–n) or MyoD (c, d, g, and h) expression vectors and muscle markers were assayed by immunostaining. Bright and dark field images are shown in a, c, e, and g and b, d, f, and h, respectively. FLAG-tagged myocardin was also detected within the nuclei of transfected cells in i–n. (B) 10T1/2 cells were transfected with expression vectors encoding lacZ (as a negative control), myocardin, myocardin-NLSΔbasic, and MyoD. Protein extracts were assayed by Western blot by using antibodies against SM22, SM-γ-actin, and SM myosin light chain kinase. α-Tubulin was detected as a loading control. (C and D) 10T1/2 cells (C) and NIH 3T3 or 3T3-L1 cells (D) were transfected with the expression vectors indicated above each lane. RNA was isolated and muscle gene expression was assayed by RT-PCR. L7 was measured as a loading control. (E) Primary rat cardiac fibroblasts were infected with adenoviruses encoding lacZ or myocardin and stained for SM-α-actin expression. Only background staining was seen with Ad-lacZ, whereas intensely stained cells were seen with Ad-myocardin. The organization of these cells can also be seen at low magnifications (Right). (Upper Right) A phase–contrast image. (Bars, 200 μm.)
We also measured the expression of smooth, cardiac, and skeletal muscle markers by RT-PCR (Fig. 1C). Myocardin specifically activated expression of smooth muscle markers in 10T1/2 cells, whereas MyoD up-regulated expression of skeletal muscle markers such as the acetylcholine receptor δ subunit and embryonic MHC. Neither myocardin nor MyoD activated expression of the cardiac markers cardiac α-MHC or atrial natriuretic factor (Fig. 1C).
Myocardin also induced the expression of smooth muscle genes in NIH 3T3 cells and 3T3-L1 cells (Fig. 1D). In contrast, HeLa cells and COS cells were refractory to the full promyogenic activity of myocardin (data not shown).
Because c-fos is also controlled by a CArG box, referred to as the serum response element, we assayed its expression in myocardin-transfected NIH 3T3 cells. Consistent with previous studies showing that myocardin cannot activate a reporter gene linked to the c-fos promoter (13), there was no change in c-fos expression in transfected cells (Fig. 1D).
We also expressed myocardin in rat primary cardiac fibroblasts. As shown in Fig. 1E, these cells showed robust expression of α-SM actin when infected with an adenovirus encoding myocardin, whereas cells infected with a lacZ adenovirus were negative for this and other smooth muscle markers (data not shown). Intriguingly, cardiac fibroblasts that expressed ectopic myocardin became organized into three-dimensional structures resembling primitive vessels (Fig. 1E). We never observed these sorts of structures in 10T1/2 cells expressing myocardin. The potential significance of this morphological response remains to be investigated.
Structure–Function Studies. The conserved N-terminal domain of myocardin was dispensable, whereas the TAD was required for generation of the smooth muscle phenotype, as shown by the lack of myogenic activity of C-terminal deletion mutants lacking this region (mutants 128–713 and 128–513; Fig. 2A). Replacement of the myocardin TAD with that of the viral coactivator VP16 restored full myogenic activity, indicating that the TAD plays a general role in transcriptional activation but does not confer target gene specificity to the process. Deletion of the basic region, which is required for SRF interaction (13), also abolished myogenic activity of myocardin (mutant NLS-Δbasic; Figs. 1B and 2 A), and mutation of the SAP domain reduced myogenic activity (Fig. 2 A).
Fig. 2.
Analysis of myocardin mutants. (A) Domains of myocardin required for smooth muscle gene expression. 10T1/2 cells were transfected with expression vectors encoding the indicated myocardin constructs. Values are expressed as the number of SM-MHC-positive cells with each mutant relative to the number in cultures transfected with the wild-type myocardin expression plasmid, which was assigned a value of 100. (B) Activation of smooth muscle gene expression by myocardin and MRTF-A. 10T1/2 cells were transfected with expression vectors encoding myocardin, MRTF-A, or MRTF-B and scored for SM-MHC-positive cells as in A.(C) Inhibition of smooth muscle gene expression by dominant negative myocardin. PAC1 and A10 cells were infected with adenovirus encoding lacZ or the myocardin dominant negative mutant 128–513. Smooth muscle gene expression was assayed by RT-PCR. GAPDH was measured as a loading control.
Myocardin shares high homology with two myocardin-related transcription factors (MRTFs) A and B (14). MRTF-A was as effective as myocardin in activating smooth muscle gene expression in 10T1/2 cells, whereas MRTF-B was inactive in this assay (Fig. 2B).
To determine whether myocardin was necessary for smooth muscle differentiation, we infected the PAC1 and A10 smooth muscle cell lines, which express myocardin, with an adenovirus encoding residues 128–513 of myocardin, which functions as a dominant negative mutant (13, 14). The dominant negative myocardin mutant specifically suppressed the expression of smooth muscle genes by at least 5-fold in both cell lines (Fig. 2C). We conclude that myocardin is sufficient for smooth muscle differentiation and that a dominant negative myocardin mutant can perturb the smooth muscle program.
Dimerization Is Required for Maximal Activity of Myocardin. Smooth muscle genes generally require pairs of CArG boxes for transcriptional activation (6–11), whereas the c-fos promoter, which is activated by proliferative signals, contains only a single CArG box. Although myocardin can form a ternary complex with SRF on a single CArG box, it preferentially activates muscle genes that contain multiple CArG boxes (13). Thus, there must be a mechanism that enables myocardin to discriminate between target genes based on the number of SRF binding sites.
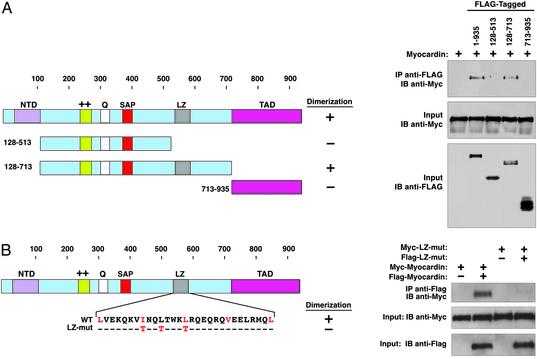
To determine whether the selective responsiveness of multiple CArG boxes to myocardin might reflect a requirement for cooperative interactions of myocardin, we tested its ability to self-associate. Indeed, immunoprecipitation experiments showed that myocardin homodimerizes (Fig. 3A). Deletion mapping localized the dimerization domain to residues 513–713, which contain a coiled-coil motif resembling a leucine zipper (Fig. 3B). Replacement of aliphatic residues at position 7 within each heptad repeat in the putative leucine zipper with threonines, which disrupts homodimerization of other leucine zipper proteins (22), prevented myocardin dimerization (Fig. 3B) but did not alter its ability to form a ternary complex with SRF bound to the CArG box (Fig. 4A). The wild-type protein and leucine zipper mutant yielded SRF ternary complexes with identical mobilities, suggesting that myocardin binds SRF as a monomer and that dimerization likely occurs between myocardin molecules bound to different CArG boxes. Despite its ability to associate with SRF, the leucine zipper mutant was compromised in its ability to activate CArG box-dependent promoters and endogenous smooth muscle genes in 10T1/2 cells (Fig. 4 B–D), suggesting that dimerization is essential for maximal activity of myocardin.
Fig. 3.
Homooligomerization of myocardin mediated by the coiled-coil domain. (A) Schematic of myocardin and myocardin deletion mutants. Deletion mutants with FLAG tags and Myc-tagged myocardin were expressed in transfected COS cells. Cell extracts were immunoprecipitated (IP) with anti-FLAG antibody and analyzed by immunoblot (IB) with anti-Myc antibody. Blots are shown (Right). LZ, leucine zipper. (B) The sequence of the leucine zipper region of myocardin and the amino acid changes in the leucine zipper mutant (LZ-mut) are shown. Wild-type and LZ-mutant myocardin were expressed in COS cells, and protein–protein interactions were detected by immunoprecipitation followed by immunoblotting, as indicated.
Fig. 4.
Functional analysis of myocardin dimerization mutant. (A) Gel mobility shift assays were performed with in vitro translated SRF, myocardin (WT), and the myocardin LZ-mutant and a radiolabeled probe corresponding to the c-fos serum response element. Only the region of the gel containing shifted probe is shown. (B) COS cells were transiently transfected with expression vectors for myocardin and the myocardin LZ-mutant and luciferase reporters linked to the SM22 promoter or three copies of the c-fos CArG box, and luciferase activity was measured. (C) 10T1/2 cells were transfected with expression vectors for myocardin or the myocardin LZ-mutant, and SM-MHC-positive cells were scored as in Fig. 2 A.(D) 10T1/2 cells were transfected with expression vectors encoding myocardin or the LZ-mutant. RNA was isolated and transcripts were assayed by RT-PCR.
Discussion
Smooth muscle cells are essential for the contractility, structure, and functions of the cardiovascular, pulmonary, digestive, and genitourinary systems and express a unique array of muscle-specific genes. Yet the molecular mechanisms that govern smooth muscle gene expression are poorly understood. In this study, we demonstrate that myocardin is sufficient to activate smooth muscle genes in nonmuscle cells.
Myocardin Is a Master Regulator of SM Differentiation. The dominant smooth muscle myogenic activity of myocardin is reminiscent of the activity of MyoD, which can convert nonmuscle cells to skeletal muscle (1, 2, 21). Given that most, if not all, smooth muscle genes contain essential CArG boxes in their control regions (5–11), it is likely that myocardin activates these genes directly through an obligate interaction with SRF. Consistent with this conclusion, mutations in the SRF binding region of myocardin abolish its myogenic activity, and myocardin is unable to activate SRF-dependent reporter genes in SRF-deficient embryonic stem cells (13, 14).
The embryonic expression pattern of myocardin is consistent with its involvement in smooth muscle gene activation (13, 14). Intriguingly, MRTF-A was also an effective activator of smooth muscle gene expression, but its embryonic expression pattern is not restricted to smooth and cardiac muscle cells (14). It is unclear why smooth muscle genes are not expressed in many of the cell types that express MRTF-A in vivo. Perhaps its level of expression in these tissues is insufficient to activate the smooth muscle program, or perhaps other factors or signals affect its transcriptional activity. MRTF-B shares high homology with myocardin and MRTF-A in the SRF binding region, but it was ineffective in activating smooth muscle gene expression. Similarly, MRTF-B is a weak SRF coactivator (14).
Myocardin and MRTF-A are the only transcription factors shown to have the potential to activate the smooth muscle differentiation program. If cofactors in addition to SRF are required for this activity, they must be expressed in 10T1/2 cells, or myocardin and MRTF-A must induce their expression. Overexpression of the combination of SRF, GATA6, and LIM-domain proteins of the CRP family has been shown to stimulate smooth muscle gene expression, apparently by enhancing SRF DNA binding activity (23). However, none of these factors shows myogenic activity alone. Two AT-rich DNA binding factors, known as Mrf2α and Mrf2β, have also been reported to activate expression of early smooth muscle lineage markers characteristic of myofibroblasts (24), but not the complete smooth muscle program as observed with myocardin. Whether these factors act downstream or in parallel to myocardin remains to be determined.
A recent study reported that overexpression of myocardin in skeletal and smooth muscle cells could elevate the expression of α-SM actin and calponin transcripts, as detected by RT-PCR (25). Because skeletal muscle cells express some smooth muscle genes at low levels, it was unclear from those findings whether myocardin could induce the complete smooth muscle phenotype in a nonmuscle cell. As this work was being completed, two other studies also reported the ability of myocardin to activate smooth muscle gene expression in mouse embryonic stem cells and 10T1/2 cells (26, 27).
The suppression of smooth muscle gene expression in the PAC1 and A10 cell lines by a dominant negative myocardin mutant suggests that myocardin is necessary for smooth muscle differentiation. Although we favor the interpretation that this mutant acts by preventing the formation of transcriptionally active SRF–myocardin complexes, it is also possible that it acts by another mechanism.
Preferential Activation Through Multiple CArG Boxes by Myocardin. The number and positions of SRF binding sites and cofactor associations confer specificity to the expression patterns of SRF target genes. Transcriptional activation of smooth muscle genes typically requires at least two CArG boxes that are often located at a distance (5–11). In contrast, the c-fos gene is controlled by one CArG box close to the transcription initiation site (28). Because of the inability of myocardin to effectively activate transcription through a single CArG box (13), such genes would be expected to be exempt from the activity of myocardin.
Our findings demonstrate that the leucine zipper-like structure in myocardin mediates homodimerization and is required for efficient activation of smooth muscle genes. We believe dimerization occurs between myocardin proteins bound to SRF homodimers on different CArG boxes, which would provide an explanation for the dependence of smooth muscle genes on multiple CArG boxes. We favor the possibility that dimerization exposes the TAD of myocardin, which is otherwise cryptic (13), or promotes the assembly of a transcriptional complex on muscle target genes by juxtaposing distant CArG boxes in the vicinity of the transcriptional machinery (Fig. 5). Selective interactions of myocardin with other promoter-specific cofactors are likely to provide further specificity to target gene activation.
Fig. 5.
A model for the regulation of smooth muscle genes by SRF. Myocardin preferentially activates smooth muscle genes controlled by pairs of CArG boxes. The c-fos promoter contains a single CArG box and is not efficiently activated by myocardin. Dimerization of myocardin through the leucine zipper (LZ) may expose the TAD with resulting activation of muscle gene expression. Other promoter-specific factors (X) cooperate with SRF and myocardin.
Roles for Myocardin in Smooth Versus Cardiac Gene Expression. It is notable that CArG boxes also control expression of many cardiac and skeletal muscle genes that cannot be activated by myocardin in transfected fibroblasts (29). Nevertheless, a dominant negative myocardin mutant can suppress cardiac gene expression in Xenopus embryos (13), suggesting that myocardin is required for cardiac gene expression during embryogenesis, despite its inability to activate cardiac gene expression in 10T1/2 cells. Perhaps myocardin cooperates with other factors to control cardiac gene expression in vivo, whereas in transfected fibroblasts it is only able to activate smooth muscle genes because of the absence of essential cardiac cofactors or the presence of negative regulators of cardiac genes that myocardin cannot override.
Further understanding of the roles of myocardin and MRTFs in smooth and cardiac muscle development will be provided by the analysis of loss-of-function phenotypes in mice. It will also be of particular interest to explore the potential involvement of these transcriptional coactivators in smooth and cardiac muscle disease.
Acknowledgments
We thank Drs. A. Sharrocks, M. Parmacek, L. Schuger, and K. Kamm for reagents, A. Tizenor for graphics, J. Page for editorial assistance, L. Sutherland for technical assistance, R. Gerard for adenoviruses, and R. Bassel-Duby, H. Yanagisawa, and A. Nordheim for helpful discussions. This work was supported by grants from the National Institutes of Health, the McGowan Foundation, the Donald W. Reynolds Foundation, and the Robert A. Welch Foundation (to E.N.O.), and from the Muscular Dystrophy Association to (D.-Z.W.), and by a postdoctoral fellowship from the National Institutes of Health (to G.C.T.P.).
Abbreviations: SRF, serum response factor; SM, smooth muscle; NLS, nuclear localization sequence; MHC, myosin heavy chain; TAD, transcription activation domain; MRTF, myocardin-related transcription factor.
References
- 1.Olson, E. N. (1990) Genes Dev. 4, 1454-1461. [DOI] [PubMed] [Google Scholar]
- 2.Hauschka, S. D. (1994) in Myology, eds. Engel, A. G. & Franzini-Armstrong, C. (McGraw–Hill, New York), 2nd Ed., pp. 3-73.
- 3.Norman, C., Runswick, M. & Pollock, R. (1988) Cell 55, 989-1003. [DOI] [PubMed] [Google Scholar]
- 4.Treisman, R. (1995) EMBO J. 14, 4905-4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Owens, G. K. (1995) Physiol. Rev. 75, 487-517. [DOI] [PubMed] [Google Scholar]
- 6.Li, L., Liu, Z., Mercer, B., Overbeek, P. & Olson, E. N. (1997) Dev. Biol. 187, 311-321. [DOI] [PubMed] [Google Scholar]
- 7.Kim, S., Ip, H. S., Lu, M. M., Clendenin, C. & Parmacek, M. S. (1997) Mol. Cell. Biol. 17, 2266-2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mack, C. P & Owens, G. K. (1999) Circ. Res. 84, 852-861. [DOI] [PubMed] [Google Scholar]
- 9.Miano, J. M., Carlson, M. J., Spencer, J. A. & Misra, R. P. (2000) J. Biol. Chem. 275, 9814-9822. [DOI] [PubMed] [Google Scholar]
- 10.Manabe, I. & Owens, G. K. (2001) J. Clin. Invest. 107, 823-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lilly, B., Olson, E. N. & Beckerle, M. C. (2001) Dev. Biol. 240, 531-547. [DOI] [PubMed] [Google Scholar]
- 12.Landerholm, T. E., Dong, X-R., Lu, J., Belaguli, N. S., Schwartz, R. J. & Majesky, M. W. (1999) Development 126, 2053-2062. [DOI] [PubMed] [Google Scholar]
- 13.Wang, D-Z., Chang, P., Wang, Z., Sutherland, L., Small, E., Krieg, P. A. & Olson, E. N. (2001) Cell 105, 851-862. [DOI] [PubMed] [Google Scholar]
- 14.Wang, D-Z., Li, S., Hockemeyer, D., Sutherland, L., Wang, Z., Schratt, G., Richardson, J. A., Nordheim, A. & Olson, E. N. (2002) Proc. Natl. Acad. Sci. USA 99, 14855-14860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Avarvind, L. & Koonin, E. V. (2000) Trends Biochem. Sci. 25, 112-114. [DOI] [PubMed] [Google Scholar]
- 16.Molkentin, J. D., Lu, J. R., Antos, C. L., Markham, B., Richardson, J., Robbins, J., Grant, S. R. & Olson, E. N. (1998) Cell 93, 215-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rothman, A., Kulik, T. J., Taubman, M. B., Berk, B. C., Smith, C. W. & Nadal-Ginard, B. (1992) Circulation 86, 1977-1986. [DOI] [PubMed] [Google Scholar]
- 18.Kimes, B. W. & Brandt, B. L. (1976) Exp. Cell Res. 98, 349-366. [DOI] [PubMed] [Google Scholar]
- 19.McKinsey, T. A., Zhang, C. L., Lu, J. & Olson, E. N. (2000) Nature 408, 106-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu, J., McKinsey, T. A., Zhang, C. L., Lu, J. & Olson, E. N. (2000) Mol. Cell 6, 233-244. [DOI] [PubMed] [Google Scholar]
- 21.Davis, R. L., Weintraub, H. & Lassar, A. B. (1987) Cell 51, 987-1000. [DOI] [PubMed] [Google Scholar]
- 22.Deminoff, S. J. & Santangelo, G. M. (2001) Genetics 158, 133-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang, D. F., Belaguli, N. S., Iyer, D., Roberts, W. B., Wu, S. P., Dong, X. R., Marx, J. G., Moore, M. S., Beckerle, M. C., Majesky, M. W. & Schwartz, R. J. (2003) Dev. Cell 4, 107-118. [DOI] [PubMed] [Google Scholar]
- 24.Watanabe, M., Layne, M. D., Hsieh, C.-M., Maemura, K., Gray, S., Lee, M.-E. & Jain, M. K. (2002) Circ. Res. 91, 382-389. [DOI] [PubMed] [Google Scholar]
- 25.Chen, J., Kitchen, C. M., Streb, J. W. & Miano, J. M. (2002) J. Mol. Cell. Cardiol. 34, 1345-1356. [DOI] [PubMed] [Google Scholar]
- 26.Du, K. L., Ip, H. S., Li, J., Chen, M., Dandre, F., Yu, W., Lu, M. M., Owens, G. K. & Parmacek, M. S. (2003) Mol. Cell. Biol. 23, 2425-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoshida, T., Sinha, S., Dandre, F., Wamhoff, B. R., Hoofnagle, N. H., Kremer, B. E., Wang, D. Z., Olson, E. N. & Owens, G. K. (2003) Circ. Res. 92, 856-864. [DOI] [PubMed] [Google Scholar]
- 28.Hipskind, R. A., Baccarini, M. & Nordheim, A. (1994) Mol. Cell. Biol. 14, 6219-6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reecy, J. M., Belaguli, N. S. & Schwartz, R. J. (1999) in Heart Development, eds. Harvey, R. P. & Rosenthal, N. (Academic, New York), pp. 273-287.