Abstract
Sympatric speciation through intraspecific social parasitism has been proposed for the evolution of Hymenopteran workerless parasites. Such inquilines exploit related host taxa to produce their own sexual offspring. The relatedness of inquilines to their hosts has been generalized in Emery's rule, suggesting that social parasites are close or the closest relatives to their host species. If the closest relative of each parasite is its host, then multiple independent origins of the parasite species are implied even within a single genus, probably through sympatric speciation. To test the plausibility of sympatric speciation in inquilines, we conducted a mitochondrial DNA phylogenetic analysis in three inquiline–host pairs of Myrmica ant species. We show that congeneric inquilines have originated independently several times. We also show that two of the inqulines are more closely related to their hosts than to any other species. Our results suggest sympatric speciation of Myrmica inquilines. Sympatric speciation is probably facilitated by the social biology and ecology of Myrmica, with polygyny as a prerequisite for the evolution of intraspecific parasitism.
Within the framework of the New Evolutionary Synthesis, developed in the 1930s and early 1940s, speciation proceeds through accumulation of genetic differences during extensive periods in allopatry, resulting ultimately in genetic incompatibility and reproductive isolation between populations (1, 2). The Synthesis was built on the foundations of population genetics which described the evolutionary processes within populations and the restrictions gene flow imposes on divergent evolution. Gene flow seemed such a fundamental obstacle to divergence in sympatry that the goal of speciation studies was “to search for genetic reasons that necessitate geographic isolation during speciation” (2).
The allopatric speciation dogma contradicts, however, with recent evidence for rapid evolution of reproductive isolation as a result of divergent selection in sympatry (3, 4). Consequently, during the past decade, speciation studies have shifted from focusing on geography, back to Darwin's (5) emphasis on the role of ecology and selection in species formation (4, 6–10); correspondingly, the plausibility of sympatric speciation has been accepted (3, 4, 9, 11, 12). Although ecological speciation need not be restricted to sympatry, it seems now likely that most sympatric speciation is ecological, mediated by competition for habitat or resources, or by host shifts and host race formation (7, 8, 13–15).
Owing to the multifarious nature of speciation, verbal models and generalizations about data are superior to mathematical theory (16). In recent mathematical models of sympatric speciation through natural or sexual selection (17–20), however, the combination of reality and generality strongly supports the interpretation that diverse sympatric sets of related taxa and ecological forms have resulted from divergence in sympatry; for example, closely related lake fish species (21–22), sympatric host races of herbivorous insects (4, 14, 15, 23, 24), and snail ecomorphs (25). Sympatric speciation is further supported by laboratory studies concluding that it is likely via pleiotropy or physical linkage disequilibrium where there is strong divergent selection relative to gene flow (26), a conclusion corroborated by quantitative trait locus mapping (15). Thus, theory, experiments, and field studies all suggest that a recurrent theme in sympatric speciation is divergent selection that leads to assortative mating through direct mate choice, or through choice of mating habitat or host plant. Also, during early phases of divergence in sympatry, factors acting against hybrids are ecological rather than genetic (15).
So far, the ecological conditions thought to facilitate sympatric speciation include sympatric potential for habitat and host shifts (4). An alternative scenario applies to the case of intraspecific parasitism in social Hymenoptera (27–32). Societies of many ant species accept surplus queens into the nest. The advantage of such polygyny is ecological (29). Its disadvantage springs from the possibility that some of the surplus queens may produce almost exclusively sexual offspring raised by workers of nestmate queens, thus acting as intraspecific parasites (29).
It has been suggested that completely or nearly workerless social parasites, inquilines, arose from intraspecific parasites, which are likely cases of sympatric speciation (32). The suggestion is based on the observation that all inquiline ants are either closely related to their hosts (loose form of Emery's rule), or are the closest relatives to their hosts (strict form of Emery's rule) (33). Although the evidence for the loose form of Emery's rule is convincing (34), phylogeny-based studies on inquiline ants, bumblebees, and wasps have rejected the strict form of Emery's rule (35–38); consequently, the studied inquilines cannot support sympatric speciation.
The validity of the strict form of Emery's rule and sympatric speciation has, however, been suggested repeatedly for Myrmica inquilines (28, 29). Here we test the strict form of Emery's rule in Myrmica ants, and examine phylogenetic support for the evolution of inquilines in sympatry. In phylogenetic terms, we test the following three alternative hypotheses.
Hypothesis 1
The inquilines are polyphyletic and obey Emery's rule. Because the inquilines and their hosts in this study all belong to the genus Myrmica, the loose form of Emery's rule maintains by definition. Adherence to the strict form of Emery's rule, i.e., each inquiline in this study is the closest relative of its host, would support the interpretation that each inquiline originated as an intraspecific social parasite; such evolution of inquilinism would imply multiple sympatric speciation. Under the alternative, allopatric model, a new prospective inquiline species should first colonize the range of its present closest relative and host species, then evolve social parasitism within its new range, whereas its free-living immediate ancestor would go extinct (32). Even more complicated alternative allopatric scenarios can easily be derived.
Hypothesis 2
The inquilines are polyphyletic, but are located essentially randomly within the phylogeny. Such polyphyly would suggest that each inquiline has arisen independently without any correlation either to its host or to the other inquilines; consequently, the strict form of Emery's rule would be rejected, and the mode of speciation would remain open.
Hypothesis 3
The inquilines group monophyletically. Only if their positions in the phylogeny are within the clade including all their known hosts, would the loose form of Emery's rule (but not the strict form) maintain. Only if the inquiline clade would be the closest relative to one of the host species would a single sympatric speciation event through intraspecific parasitism be supported; radiation and speciation of the common ancestor of the present inquilines through host shifts could then explain the evolution of the present inquilines. A modification of this hypothesis states that the inquilines form two clades; the interpretation of such a pattern would depend on the positions of the inquilines in the phylogeny relative to their hosts, following the logic of the first two hypotheses. Similarly, in case of parapatry, ad hoc interpretation of the results would be necessary, depending on the pattern of paraphyly.
Materials and Methods
Myrmica and Its Social Parasites. The genus Myrmica (Myrmicinae) includes ≈110 described species in the Holarctic region (39); of these, 14 (≈13%) are inquilines (40, 41). Myrmica ants are suitable candidates of sympatric speciation owing to their social biology and ecology: most species are polygynous, and they develop locally high nest densities, which are necessary for the maintenance of inquiline populations (29, 42–44).
Inquilines tend to be rare, but if the nest density of a suitable host is high, some inquiline species may be locally abundant (42, 45). Inquilines do not usually produce worker offspring (28, 41, 46), which are supplied by the host. The inquilines, thus, depend on their host for all colony tasks and cannot live without them.
Taxa Sampled. Two of the inquiline species included in the analyses, Myrmica hirsuta and Myrmica microrubra, are host specific (28, 41), whereas Myrmica karavajevi uses as host Myrmica scabrinodis (the host in this study), Myrmica rugulosa, and Myrmica sabuleti (40). We collected three inquiline–host pairs in Finland, a second sample of hirsuta–sabuleti in Germany, and obtained another microrubra–rubra from England. Each inquiline and its host came from the same nest. The four additional species of the genus Myrmica (all from unparasitized nests) were Myrmica ruginodis, M. rugulosa, Myrmica hellenica, and Myrmica sulcinodis; of these, the first two species are known to host inquilines. As outgroup species we used Manica rubida (Myrmicinae), and Camponotus herculeanus (Formicinae). To analyze the inquiline–host relationships, our taxon sampling is comprehensive: it is unlikely that the Myrmica species not included in the study would alter the interpretations of the results (47). Specimen localities are available in Table 5, which is published as supporting information on the PNAS web site, www.pnas.org.
Molecular Methods. Mitochondrial DNA was extracted from ethanol-preserved individual ants by using the Nucleo Spin Tissue kit (Macherey–Nagel). Parts of the cytochrome oxidase one (COI), cytochrome oxidase two (COII), and cytochrome b (Cyt b) genes were amplified and sequenced by using universal and ant-specific primers (Table 1). Final concentrations of reagents in a 20-μl PCR were 0.75× Taq buffer/0.09 mM dNTP/3.1 mM MgCl2/0.5 μM of each primer/0.5 units of Taq polymerase. DNA was amplified through 30 cycles of 30 s at 94°C, 45 s at 46–49°C, and 2 min at 72°C. The PCR products were purified with GFX PCR DNA and Gel Band Purification kit (Amersham Pharmacia). In the sequencing reaction, BigDye (Original) Terminator Cycle Sequencing kit (Applied Biosystems) was used. The cycle sequencing reactions were purified with Centri-sep spin columns (Princeton Separations, Adelphia, NJ), and both strands were sequenced on an automated DNA sequencer ABI 377. The sequences were compiled, edited, and aligned in SEQUENCHER, version 3.0 (Gene Codes, Ann Arbor, MI).
Table 1. Oligonucleotide primers used for the three mitochondrial genes.
| Primer name | Primer sequence 5′-3′ | Ref. |
|---|---|---|
| COI | ||
| LCO1490 | GGTCAACAAATCATAAAGATATTGG | 48 |
| HCO2198 | TAAACTTCAGGGTGACCAAAAAATCA | 48 |
| C1-J-1751 | GGATCACCTGATATAGCATTCCC | 49 |
| C1-J-2183 | CAACATTTATTTTGATTTTTTGG | 49 |
| TL2-N-3014T | CCAATGCACTAATCTGCCATATTA | 49 |
| COII | ||
| C2-N-3389 | TCATAAGTTCARTATCATTG | 49 |
| Cyt b | ||
| CB1 | TATGTACTACCATGAGGACAAATATC | 50 |
| CB2 | ATTACACCTCCTAATTTATTAGGAAT | 50 |
| CB5 | GAAAATCCCCCTCAGATTCA* | 50 |
| CB7 | CTCCCAACTCCTATTAATATTTCTT | 51 |
| tRs | TATTTCTTTATTATGTTTTCAAAAC | 51 |
Original primer modified for Myrmica by R.S
Substitution Model. To find the best-fit substitution model for the data, we used MODELTEST version 3.06 (52). The program compares nested models of DNA substitution in a hierarchical hypothesis testing framework. The program does not, however, include the site-specific rates model (53), often used for proteincoding genes. To calculate the log-likelihood score and parameter estimates for the general-time-reversible model with site-specific substitution rates, with one rate category for each codon position, we used PAUP 4.0b10 (54). To compare the site-specific rates model (alternative model) and the best model produced by MODELTEST (null model), we could not use the χ2 approximation, because the site-specific rates model is not nested with the models in MODELTEST. Thus, to compare the models statistically, we used SEQ-GEN version 1.2.4 (55) to produce by Monte Carlo simulations 100 data sets the size of the original data, with model parameters estimated from the original data according to the null model. We then calculated likelihood scores of the 100 simulated data sets under both the null and the alternative model, and the difference δ between each pair of scores, thus obtaining the null distribution of δ (56). If the value of δ from the original data fell within the five largest values of the null distribution, then the null model was rejected in favor of the alternative model at the 5% level.
Phylogenetic Analyses. We applied Bayesian phylogenetic inference using mrbayes version 3.0b3 (57, 58). mrbayes implements a Metropolis-coupled Markov chain Monte Carlo algorithm that runs several chains simultaneously; the method ensures that trees are sampled in proportion to their probability of occurrence under the model of evolution. We used the best-fit substitution model to carry out two separate runs with four Markov chains, each starting from a random tree. The Markov chains were run for one million generations, sampling every 100th generation for a total of 10,000 trees each run. We excluded the first 1,000 trees (10%) that were sampled before the chains converged on a stable value. Of the remaining 9,000 trees, we obtained in PAUP a majority-rule consensus tree and the posterior probability for each clade of the tree.
We applied Bayesian inference to study congruence among the three gene partitions (59). For each partition and for the three partitions combined, we estimated the 0.95 posterior intervals of tree topologies, while assuming for each partition the best-fit model for the combined data. We then determined whether the topology for the combined data with the highest posterior probability lay within the 0.95 posterior interval of a partition. If the combined-data topology was within the 0.95 interval of a partition, we assumed that the topology could have produced the gene partition; otherwise, we interpreted the topology to be incongruent with the data.
We tested the alternative hypotheses of inquiline evolution [monophyletic vs. polyphyletic (or paraphyletic) origin of inquilines, the latter including the alternatives that inquilines are closely related to their hosts vs. randomly located in the topology] by considering the trees sampled by the Markov chains. The proportion of trees that were consistent with monophyly of all 9,000 trees sampled was an approximation of the posterior probability of monophyly. We used the same logic to find the posterior probabilities of paraphyly and polyphyly; the hypothesis of random location was evaluated on the basis of the probabilities of mono-, para-, and polyphyly.
We also conducted an equally weighted cladistic parsimony analysis, by using the branch-and-bound search in PAUP, with default settings, to find the most parsimonious tree. Support for the clades came from 1,000 bootstrap replicates with the branch-and-bound search. We excluded the parsimony-uninformative characters from these analyses.
Results
After excluding the noncoding region between COI and COII, the sequence alignment of the three genes for the 16 taxa consisted of 2,772 base pairs (Table 2). GenBank accession numbers of the sequences are available in Table 5.
Table 2. Base numbers for COI, COII, and Cyt b genes, and for the combined data.
| COI | COII | Cyt b | Combined data | |
|---|---|---|---|---|
| No. of sites | 1,446 | 297 | 1,029 | 2,772 |
| Variable sites | 512 | 123 | 446 | 1,081 |
| MP sites* | 330 | 55 | 279 | 664 |
MP sites refer to informative sites in cladistic parsimony analysis
Best-Fit Substitution Model. The best-fit model found by MODEL-TEST was the general time reversible model with γ-distributed substitution rates and a proportion of invariable sites (GTR+Γ+I; –lnL0 = 1,2127.39). The site-specific rates model, with one rate category for each codon position over the three genes, had a higher likelihood score (GTR+SS3; -lnL1 = 1,1763.00) than GTR+Γ+I. The likelihood-ratio statistic for these models was δ = (lnL1 - lnL0) = 364.39. Because this value was larger than any of the 100 δ values within the simulated null distribution, we rejected GTR+Γ+I in favor of GTR+ SS3 (P < 0.01).
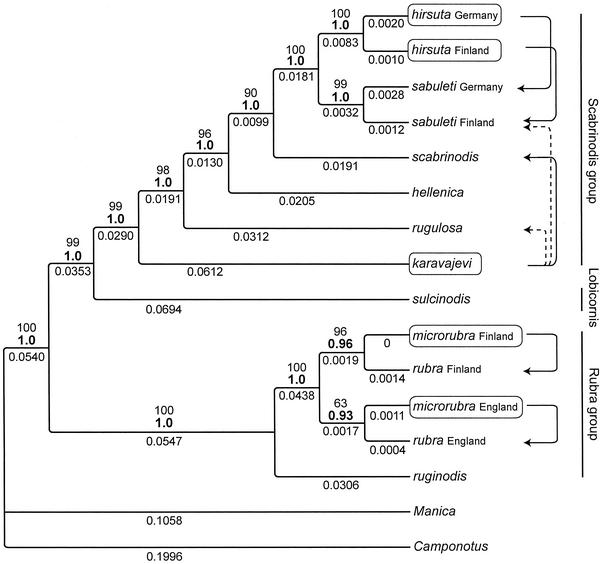
Parasite–Host Relationships. The majority-rule consensus tree of 9,000 Bayesian trees, based on the GTR+SS3 model, shows that the inquilines of Myrmica are polyphyletic (Fig. 1). The probability of this topology (Hypothesis 1 in the introduction) is 0.945, and the 0.95 posterior interval includes only this tree (see Table 3 for parameter estimates for the combined data). The nodes of the tree are very well supported; all but two nodes have a posterior probability of 1.0, and the remaining nodes have posterior probabilities of 0.98 and 0.97 (Fig. 1). The replicate run resulted in an identical topology and probabilities to that of the above one, except for the remaining two nodes (0.96 and 0.98). Cladistic parsimony analysis produced a single most parsimonious tree with an identical topology to that of the Bayesian consensus tree (tree length = 1,551 steps, retention index = 0.75).
Fig. 1.
Majority-rule consensus tree of 9,000 Bayesian trees. Bold numbers above branches represent the posterior probability that the clade is correct, given the model of evolution, and numbers above the posterior probabilities are bootstrap proportions; average branch length estimates from the Bayesian analysis are given below the branches (tree length = 1.206). The single most parsimonious tree topology is identical to that of the Bayesian consensus tree. Parasites are given in frames with solid arrow lines pointing to their hosts in this study; other documented karavajevi hosts, rugulosa and sabuleti (43), are shown with dashed lines. The three species groups shown with vertical lines are defined on the basis of worker morphology, excluding the inquilines (47).
Table 3. Mean parameter estimates and their 0.95 posterior intervals of the combined data using the GTR+SS3 model.
| Parameter | Mean (interval) |
|---|---|
| Tree length | 1.206 (1.126-1.296) |
| πA | 0.336 (0.321-0.351) |
| πC | 0.146 (0.137-0.155) |
| πG | 0.090 (0.080-0.010) |
| πT | 0.428 (0.414-0.442) |
| RAC | 4.449 (2.659-6.950) |
| RAG | 10.713 (6.741-16.965) |
| RAT | 4.144 (2.605-6.387) |
| RCG | 3.039 (1.393-5.749) |
| RCT | 27.890 (17.769-42.491) |
| Rate for 1. position | 0.421 (0.375-0.467) |
| Rate for 2. position | 0.121 (0.101-0.143) |
| Rate for 3. position | 2.458 (2.404-2.510) |
π refers to base frequency, and R refers to substitution rate
Hypothesis 3, assuming a single origin of the inquilines was significantly worse-fitting than the optimal polyphyletic solution; the five parasites did not occur as a monophyletic group in any of the 9,000 trees, thus giving a probability of <1/9,000 for their common ancestry. Hypothesis 2, suggesting random location of the inquilines in the topology, could also be rejected on the basis of the strong support for Hypothesis 1 (above).
The three inquilines differed in their phylogenetic positions and DNA sequence divergences (Fig. 1). M. karavajevi was at the base of the clade containing all its known hosts, having the longest branch of the three parasite taxa (Fig. 1). M. hirsuta was the sister group of its M. sabuleti host, but the two geographic replicates of both the parasite and the host were monophyletic (P = 1.0 for both groups). The two M. microrubra differed genetically very little from their hosts, and M. microrubra grouped with M. rubra in both geographical areas, thus revealing polyphyly of the inquiline and its host (Fig. 1). The posterior probability for polyphyly was P > 0.97, and for paraphyly, P < 0.03. The two M. microrubra never formed a monophyletic group in any of the 9,000 Bayesian trees, thus giving a probability <1/9,000 for their monophyly.
Congruence Among Data Partitions. In the above analyses, we have combined all three genes. However, when the genes were analyzed separately, the tree topology based on the combined data were, satisfyingly, within the 0.95 posterior interval for the COI and the Cyt b genes, but not for the COII gene (Table 4). The majority-rule consensus tree based on 9,000 trees of COII was incompletely resolved owing to short sequence length (Fig. 2, which is published as supporting information on the PNAS web site), but despite the apparent incongruence, its topology agreed, for the most part, with the topology of the combined data (Fig. 1).
Table 4. Posterior probability (Pr) of the Bayesian topology based on combined data, estimated from different gene partitions.
| COI | COII | Cyt b | Combined data | |
|---|---|---|---|---|
| Pr | 0.109 | - | 0.685 | 0.945 |
| Cumulative Pr | 0.752 | - | 0.685 | 0.945 |
| No. of trees in 0.95 posterior interval | 9 | 4,306 | 4 | 1 |
Cumulative Pr is the sum of the Pr of that topology and all topologies with a greater Pr
Discussion
Before discussing the significance of our results, the well known risk of using mtDNA data in reconstructing phylogenies ought to be evaluated. This is especially relevant, because our phylogeny based on the nuclear gene 28S, although consistent with the mtDNA tree, resolved only two species groups as given in Fig. 1 (unpublished data). The mtDNA is prone to introgression, and hybridization among species could thus confound the phylogeny (60). Although interspecific hybridization is documented in many European ants on the basis of intricate morphological analyses, there is no evidence of hybridization among Myrmica species (61). This finding, together with our very well supported mtDNA phylogeny, which is consistent with the Myrmica species groups distinguished on the basis of morphology (47) (Fig. 1), suggests that introgression is not a likely source of bias in this study.
Our three inquilines form a unique sequence of parasite–host relationships, which probably reflects the evolutionary ages of their parasitic behavior. This is indicated by their phylogenetic positions and DNA sequence divergences (Fig. 1). The location of M. karavajevi at the base of the clade containing all its known hosts, fits the loose form of Emery's rule. The two other inquiline–host pairs obey the strict form of Emery's rule, but differ in details. M. hirsuta is a full-blown sister species of M. sabuleti, and is monophyletic. M. microrubra, on the other hand, is polyphyletic.
Polyphyly is expected during early phases of speciation (62), and M. microrubra presumably is an incipient species. In this case, the two divergent populations would still be undergoing lineage sorting, the mtDNA not yet having coalesced. Alternatively, M. microrubra may evolve in parallel (see refs. 3 and 8) in different regions. In lack of more detailed molecular, behavioral and ecological data it is not possible to distinguish between the two alternatives. Finally, M. microrubra may be an intraspecific parasite of M. rubra, and the polyphyletic mtDNA pattern would testify of ongoing gene flow between the parasite and its host. Further divergence and evolution of reproductive isolation could take place, if fortuitous pleiotropic or linkage-disequilibrium correlations developed between parasitic behavior and changes in time or place of mating, leading to positive assortative mating of M. microrubra females and males, as seems the case in pea aphid host races (15).
The relative ages of the inquilines are also reflected in their number of host species, morphology and reduction of the workforce. M. karavajevi uses several host species, is workerless, morphologically very differentiated, and has traditionally been classified as Sifolinia or Symbiomyrma (40). M. hirsuta is host-specific, easily distinguishable from its host, and occasionally produces a few workers. M. microrubra is host-specific, produces a strongly reduced number of workers (63), and is separable from M. rubra by subtle morphometric differences, mainly smaller size (41).
Our results reject Hypothesis 2 and Hypothesis 3 as formulated at the end of the Introduction, but agree with Hypothesis 1. Adherence to the strict form of Emery's rule by M. hirsuta and M. microrubra strongly supports sympatric speciation of the inquilines, which is also consistent with the “karavajevi” ancestor. The present M. karavajevi obeys the loose form of Emery's rule. Its multiple hosts, belonging to the same clade as the inquiline, and four other karavajevi-like species parasitizing other Myrmica species (40), may be a result of host shifts of “karavajevi” ancestor, resulting in host expansion and later host specialization and speciation. This mode of radiation has been documented in various parasitic taxa, such as mistletoes (64), red algae (65), and gall wasps (66), and seems plausible also in bumblebee (35) and paper wasp inquilines (37). Alternatively, “karavajevi” may have increased its host array concurrently with speciation events in its original host lineage. It has also been suggested that, given that the host species have been properly identified, the present M. karavajevi may be a complex of host-specific sister species (40).
Accounting for the phylogenetic relationships of Myrmica inquilines, which support the independent origins of the inquilines and obey the strict form of Emery's rule in two of the inquilines, through allopatric speciation (see Hypothesis 1) would involve complicating factors such as (i) after divergence in geographical isolation, recolonization of former range, (ii) evolution of parasitism to use its seemingly closest relative as host, and (iii) extinction of its free-living immediate ancestor, i.e., its real closest relative (32). On the other hand, the model of evolution of inquilinism in sympatry builds on known features of the polygyny syndrome. Its key feature is polygyny, the presence of multiple queens in the nest (27, 29, 31, 32), which could start a process leading to cheating where a female produces mainly sexual offspring at the expense of other queens producing the workforce, and miniaturization of the sexual offspring by the cheating queens. These changes would automatically shift the timing of mating between the parasite and the host. Incipient reproductive isolation could be strengthened by changes in the mating site of the parasite. In the following, we will discuss the process in more detail.
Polygyny is advantageous in specific ecological conditions, where suitable, patchily distributed but relatively extensive and stable habitat is open for colonization (67). There polygynous societies are able to grow rapidly to a dense multinest system that fills the free space. This polygyne tactic is, in contrast to dispersal by flight and claustral colony foundation by single queens, most economically realized by producing numerous small, cooperating queens.
The ecologically well studied M. ruginodis is a good example of a species using the above alternative social tactics. Polygynous colonies containing more than one functional, small queen (microgyne) specialize on stable habitats, e.g., heather moorland. Monogynous colonies usually have a large queen (macrogyne) and use shorter-lived habitats (68). However, both queen types can be polygynous, and a substantial proportion of colonies contain a mixture of small and large queens (69).
In certain conditions, alternative adaptations may facilitate strong diversifying selection and speciation: alternative social behavior is here a special case (70). In social insects, acceptance of extra queens into the colony makes the system vulnerable to a parasitic tactic in which some queens use the existing workforce to produce mainly sexual offspring (29). It may not be a mere coincidence that the genus Myrmica has an exceptionally high number of inquiline species (40) and that most, if not all, of its well studied species are known to have polygynous societies (29, 42–44).
The ant tribe Formicoxenini (Myrmicinae) is comparable to Myrmica in its relatively high proportion of polygyny and inquiline species (as implied in refs. 30 and 31). Basic similarities in the social biology and ecology between many Formicoxenini (e.g., Leptothorax) and Myrmica species suggest evolution of inquilinism through intraspecific parasitism also in this tribe. In contrast, functionally monogynous species of ants, wasps, and bumblebees, used to test the strict Emery's rule phylogenetically, have rejected the rule (refs. 35–38; gyny assessed from refs. 36, 42, 71, and 72). Consequently, in these cases, evolution of inquilinism through intraspecific parasitism is rejected. The low proportion of inquilines in monogynous taxa also suggests that monogynous species resist inquilines better than do polygynous species.
Normally the presence of a fertile queen stimulates workers to favor smaller worker-biased larvae, while inhibiting the growth of larger larvae due to develop into gynes (here, young unfertilized, alate queens) (29). Notably, this “queen effect” is not species-specific in Myrmica (73). A further step toward the evolution of true social parasites is miniaturization of the polygyne queen (27, 29, 74); of our inquilines, the queens of M. microrubra and M. hirsuta are, on an average, only slightly larger than their host workers, and the queen of M. karavajevi is substantially smaller than its host workers.
Miniaturization of the intraspecific parasite may help its gyne offspring to escape the queen effect. For example, M. microrubra completely suppresses production of sexuals by its host queens, raises only a few workers, but a 37-fold number of gynes relative to nonparasitized M. rubra (63). M. microrubra lacks the queen effect on its own offspring (41), and small gyne larvae of the inquiline may develop as fast as the host workers do in the presence of the host queen, and faster than M. rubra gynes in nonparasitized nests. This could cause earlier maturation of the parasites, and through earlier mating facilitate evolution of reproductive isolation between the parasite and its host (32). Instead of mating at swarming sites outside the nest, and dispersing by flight to found a new colony alone as macrogynes do, microgynes may mate in or close to the nest, and return to the nest or a neighboring nest, where they are accepted as extra queens (27, 29, 31, 32).
There is a fair probability that M. microrubra would mate in the nest with males of its kin rather than with males of M. rubra. This follows from differences in the production of males in the two species (29). By necessity, the practically workerless queen of M. microrubra has to produce both female and male offspring. In contrast, M. rubra female offspring are produced by the queen(s), but male offspring (mostly) by workers; however, workers will produce males only in queenless nests. Further, because the queen effect is not species-specific, M. microrubra prevents production of sexual offspring of its host female; consequently, given that M. microrubra sexuals mate in the nest, interbreeding of M. microrubra and M. rubra would be efficiently inhibited. Indeed, intranidal mating seems to predominate in two groups of ants: social parasites and highly polygynous tramp species (75).
Empirical studies, as well as theory emphasizing the active role of natural selection in sympatric speciation, suggest that sympatric speciation is likely to occur faster and involve fewer loci than does allopatric speciation (4); actually, sympatric speciation could produce punctuated patterns of evolution, socially parasitic species serving as especially convincing examples (70). The recent finding that a major gene can switch the colony organization of the fire ant Solenopsis invicta from monogyny to polygyny (76) demonstrates how a single genetic change, in a suitable ecological context, can start a strong runaway process leading to social parasitism. Given polygyny and the possibility of cheating, selection should strongly favor miniaturization of the polygyne queens, changed mating behavior, and fidelity to the birth habitat. However, details of the evolution of reproductive isolation between the incipient inquiline and its host are still unknown, and one of the most crucial future fields of study of sympatric speciation of social parasites.
Supplementary Material
Acknowledgments
We thank Tiina Marjomaa and Nina Siitonen for the laboratory work, Graham Elmes for the English microrubra–rubra samples, and Wojciech Czechowski for Manica rubida. Comments by Allen Herre, John Huelsenbeck, Fredrick Ronquist, two anonymous referees, and the Population Biology Discussion Group of the University of Copenhagen helped to improve the text. R.S. was funded by the Academy of Finland.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: COI, cytochrome oxidase one; COII, cytochrome oxidase two; Cyt b, cytochrome b.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. AY280591–AY280606 (COI and COII) and AY280575–AY280590 (Cyt b)].
See commentary on page 6896.
References
- 1.Mayr, E. (1942) Systematics and the Origin of Species (Columbia Univ. Press, New York).
- 2.Mayr, E. (1963) Animal Species and Evolution (Harvard Univ. Press, Cambridge, MA).
- 3.Johannesson, K. (2001) Trends Ecol. Evol. 16, 148-153. [DOI] [PubMed] [Google Scholar]
- 4.Via, S. (2001) Trends Ecol. Evol. 16, 381-390. [DOI] [PubMed] [Google Scholar]
- 5.Darwin, C. (1859) On the Origin of Species by Means of Natural Selection (Murray, London).
- 6.Smith, T. B., Wayne, R. K., Girman, D. J. & Bruford, M. W. (1997) Science 276, 1855-1857. [Google Scholar]
- 7.Orr, M. R. & Smith, T. B. (1998) Trends Ecol. Evol. 13, 502-506. [DOI] [PubMed] [Google Scholar]
- 8.Schluter, D. (1998) in Endless Forms: Species and Speciation, eds. Howard, D. J. & Berlocher, S. H. (Oxford Univ. Press, Oxford), pp. 114-129.
- 9.Schluter, D. (2000) The Ecology of Adaptive Radiation (Oxford Univ. Press, Oxford).
- 10.Schneider, C. J., Smith, T. B., Larison, B. & Moritz, C. (1999) Proc. Natl. Acad. Sci. USA 96, 13869-13873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tauber, C. A. & Tauber, M. J. (1989) in Speciation and its Consequences, eds. Otte, D. & Endler, J. A. (Sinauer, Sunderland, MA), pp. 307-344.
- 12.Bush, G. L. (1994) Trends Ecol. Evol. 9, 285-288. [DOI] [PubMed] [Google Scholar]
- 13.Berlocher, S. H. (1998) in Endless Forms: Species and Speciation, eds. Howard, D. J. & Berlocher, S. H. (Oxford Univ. Press, Oxford), pp. 99-113.
- 14.Feder, J. L. (1998) in Endless Forms: Species and Speciation, eds. Howard, D. J. & Berlocher, S. H. (Oxford Univ. Press, Oxford), pp. 130-144.
- 15.Via, S. & Hawthorne, D. J. (2002) Am. Nat. 159, S76-S88. [DOI] [PubMed] [Google Scholar]
- 16.Turelli, M., Barton, N. H. & Coyne, J. A. (2001) Trends Ecol. Evol. 16, 330-343. [DOI] [PubMed] [Google Scholar]
- 17.Dieckmann, U. & Doebeli, M. (1999) Nature 400, 354-357. [DOI] [PubMed] [Google Scholar]
- 18.Kondrashov, A. S. & Kondrashov, F. A. (1999) Nature 400, 351-354. [DOI] [PubMed] [Google Scholar]
- 19.Gavrilets, S. & Waxman, D. (2002) Proc. Natl. Acad. Sci. USA 99, 10533-10538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Higashi, M., Takimoto, G. & Yamamura, N. (1999) Nature 402, 523-526. [DOI] [PubMed] [Google Scholar]
- 21.Schliewen, U. K., Tautz, D. & Pääbo, S. (1994) Nature 368, 629-632. [DOI] [PubMed] [Google Scholar]
- 22.Albertson, R. C., Markert, J. A., Danley, P. D. & Kocher, T. D. (1999) Proc. Natl. Acad. Sci. USA 96, 5107-5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bush, G. L. (1969) Evolution (Lawrence, Kans.) 23, 237-251. [Google Scholar]
- 24.Menken, S. B. J. & P. Roessingh, P. (1998) in Endless Forms: Species and Speciation, eds. Howard, D. J. & Berlocher, S. H. (Oxford Univ. Press, Oxford), pp. 154-156.
- 25.Johannesson, K., Rolán-Alvarez, E. & Ekendahl, A. (1995) Evolution (Lawrence, Kans.) 49, 1180-1190. [DOI] [PubMed] [Google Scholar]
- 26.Rice, W. R. & Hostert, E. E. (1993) Evolution (Lawrence, Kans.) 47, 1637-1653. [DOI] [PubMed] [Google Scholar]
- 27.Elmes, G. W. (1976) Insectes Soc. 23, 3-21. [Google Scholar]
- 28.Elmes, G. W. (1978) Syst. Entomol. 3, 131-145. [Google Scholar]
- 29.Elmes, G. W. (1991) Actes Coll. Ins. Soc. 7, 17-34. [Google Scholar]
- 30.Buschinger, A. (1970) Biol. Zentralblatt 88, 273-299. [Google Scholar]
- 31.Buschinger, A. (1990) Z. Zool. Syst. Evol.-Forsch. 28, 241-260. [Google Scholar]
- 32.Bourke, A. F. G. & Franks, N. R. (1991) Biol. J. Linn. Soc. 43, 157-178. [Google Scholar]
- 33.Ward, P. S. (1989) in The Genetics of Social Evolution, eds. Breed, M. D. & Page, R. D. (Westview, Boulder, CO), pp. 261-270.
- 34.Wilson, E. O. (1971) Insect Societies (Harvard Univ. Press, Cambridge, MA).
- 35.Pedersen, B. V. (1996) Mol. Phylogenet. Evol. 5, 289-297. [DOI] [PubMed] [Google Scholar]
- 36.Ward, P. S. (1996) Syst. Entomol. 21, 253-263. [Google Scholar]
- 37.Carpenter, J. M. (1997) Mém. Mus. Natn. Hist. Nat. 173, 135-161. [Google Scholar]
- 38.Sanetra, M. & Buschinger, A. (2000) Eur. J. Entomol. 97, 95-117. [Google Scholar]
- 39.Bolton, B. (1995) A New General Catalogue of the Ants of the World (Harvard Univ. Press, Cambridge, MA).
- 40.Bolton, B. (1988) Syst. Entomol. 13, 1-11. [Google Scholar]
- 41.Seifert, B. (1993) Abh. Ber. Naturkundemus. Görlitz 67(5), 9-12. [Google Scholar]
- 42.Seifert, B. (1996) Ameisen: Beobachten, Bestimmen (Naturbuch, Augsburg, Germany).
- 43.Czechowski, W., Radchenko, A. & Czechowska, W. (2002) The Ants (Hymenoptera, Formicidae) of Poland (Museum and Institute of Zoology, Polish Academy of Sciences, Warsaw).
- 44.Collingwood, C. A. (1979) The Formicidae (Hymenoptera) of Fennoscandia and Denmark (Scandinavian Science Press, Klampenborg, Denmark).
- 45.Elmes, G. W. (1983) Insectes Soc. 30, 221-234. [Google Scholar]
- 46.Francoeur, A. (1981) Can. Entomol. 113, 755-759. [Google Scholar]
- 47.Radchenko, A. G. (1995) Entomol. Rev. 74, 91-106. [Google Scholar]
- 48.Folmer, O., Black, M., Hoeh, W. Lutz, R. & Vrijenhoek, R. (1994) Mol. Mar. Biol. Biotech. 3, 294-299. [PubMed] [Google Scholar]
- 49.Simon, C., Frati, F., Beckenbach, A., Crespi, B., Liu, H. & Flook, P. (1994) Ann. Entomol. Soc. Am. 87, 651-701. [Google Scholar]
- 50.Jermiin, L. S. & Crozier, R. H. (1994) J. Mol. Evol. 38, 282-294. [DOI] [PubMed] [Google Scholar]
- 51.Tay, W. T., Cook, J. M., Rowe, D. J. & Crozier, R. H. (1997) Mol. Ecol. 6, 403-411. [Google Scholar]
- 52.Posada, D. & Crandall, K. A. (1998) Bioinformatics 14, 817-818. [DOI] [PubMed] [Google Scholar]
- 53.Swofford, D. L., Olsen, G. J., Waddell, P. J. & Hillis D. M. (1996) in Molecular Systematics, eds. Hillis, D. M., Moritz, C. & Mable, B. K. (Sinauer, Sunderland, MA), pp. 407-514.
- 54.Swofford, D. L. (1998) paup*: Phylogenetic Analysis Using Parsimony (* and Other Methods) (Sinauer, Sunderland, MA), Version 4.0.
- 55.Rambaut, A. & Grassly, N. C. (1997) Comput. Appl. Biosci. 13, 235-238. [DOI] [PubMed] [Google Scholar]
- 56.Goldman, N. (1993) J. Mol. Evol. 36, 182-198. [DOI] [PubMed] [Google Scholar]
- 57.Huelsenbeck, J. P. & Ronquist, F. (2001) Bioinformatics 17, 754-755. [DOI] [PubMed] [Google Scholar]
- 58.Huelsenbeck, J. P., Ronquist, F., Nielsen, R. & Bollback, J. P. (2001) Science 294, 2310-2314. [DOI] [PubMed] [Google Scholar]
- 59.Buckley, T. R., Arensburger, P., Simon, C. & Chambers, G. K. (2002) Syst. Biol. 51, 4-18. [DOI] [PubMed] [Google Scholar]
- 60.Shaw, K. L. (2002) Proc. Natl. Acad. Sci. USA 99, 16122-16127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Seifert, B. (1999) Insectes Soc. 46, 45-52. [Google Scholar]
- 62.Harrison, R. G. (1998) in Endless Forms: Species and Speciation, eds. Howard, D. J. & Berlocher, S. H. (Oxford Univ. Press, Oxford), pp. 19-31.
- 63.Pearson, B. (1981) in Biosystematics of Social Insects, eds. Howse, P. E. & Clement, J.-L. (Academic, London), pp. 75-84.
- 64.Norton, D. A & Carpenter, M. A. (1998) Trends Ecol. Evol. 13, 101-105. [DOI] [PubMed] [Google Scholar]
- 65.Goff, L. J., Ashen, J. & Moon, D. (1997) Evolution (Lawrence, Kans.) 51, 1068-1078. [DOI] [PubMed] [Google Scholar]
- 66.Ronquist, F. (1994) Evolution (Lawrence, Kans.) 48, 241-266. [DOI] [PubMed] [Google Scholar]
- 67.Hölldobler, B. & Wilson, E. O. (1990) The Ants (Harvard Univ. Press, Cambridge, MA).
- 68.Brian, M. V. & Brian, A. D. (1949) Trans. R. Entomol. Soc. London 100, 393-409. [Google Scholar]
- 69.Elmes, G. W. (1991) Ecol. Entomol. 16, 411-423. [Google Scholar]
- 70.West-Eberhard, M. J. (1986) Proc. Natl. Acad. Sci. USA 83, 1388-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Alford, D. V. (1975) Bumblebees (Davis-Poynter, London).
- 72.Yamane, S. (1996) in Natural History and Evolution of Paper-Wasps, eds. Turillazzi, S. & West-Eberhard, M. J. (Oxford Univ. Press, Oxford), pp. 75-97.
- 73.Elmes, G. W. & Wardlaw, J. C. (1983) Insectes Soc. 30, 134-148. [Google Scholar]
- 74.Aron, S., Passera, L. & Keller, L. (1999) Proc. R. Soc. London B 266, 173-177. [Google Scholar]
- 75.Heinze, J., Hölldobler, B. & Yamauchi, K. (1998) Behav. Ecol. Sociobiol. 42, 239-246. [Google Scholar]
- 76.Krieger, M. J. B. & Ross, K. G. (2002) Science 295, 328-332. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.